Abstract
Concept detection is an integral step in natural language processing (NLP) applications in the clinical domain. Clinical concepts are detailed (e.g., “pain in left/right upper/lower arm/leg”) and expressed in diverse phrase types (e.g., noun, verb, adjective, or prepositional phrase). There are rich terminological resources in the clinical domain that include many concept synonyms. Even with these resources, concept detection remains challenging due to discontinuous and/or permuted phrase occurrences. To overcome this challenge, we investigated an approach to exploiting syntactic information. Syntactic patterns of concept phrases were mined from continuous, non-permuted forms of synonyms, and these patterns were used to detect discontinuous and/or permuted concept phrases. Experiments on 790 de-identified clinical notes showed that the proposed approach can potentially boost a recall of concept detection. Meanwhile, challenges and limitations were noticed. In this paper, we report and discuss our preliminary analysis and finding.
Introduction
Concept detection is one of the key components in clinical natural language processing (NLP) applications. The task is particularly challenging in the clinical domain because a large number of detailed concepts are involved (e.g., pain in left/right upper/lower arm/leg) and concepts are expressed in different phrase types, e.g., “pain in left upper arm” (noun phrase), “throw up”, (verb phrase), “reactive to light” (adjective phrase), and “within normal limits” (prepositional phrase). Rich terminology resources available in the domain, such as the Unified Medical Language System (UMLS)1,2, associate each clinical concept with a list of synonyms. Given such a resource as a dictionary, detection of concept may be implemented as exact string matching, also called dictionary look-up: input text is scanned, and sequences of words are looked up against synonyms in the dictionary. Dictionary look-up has several advantages in practice: Detected phrases are directly associated with detailed concepts in the dictionary; an efficient implementation facilitates fast look-up; the idea of dictionary look-up is easily understood by application users as well as system developers. Dictionary look-up, however, is susceptible to token occurrence variations, namely, discontinuous phrase occurrences and token permutation. For example, a text fragment “sensation absent in the toes, intact at the ankles” contains two clinical concepts “absent sensation” and “intact sensation”, where they are both appear in a discontinuous and/or permuted manner. This challenge is severe in clinical notes, where there can be many subtle variations of expressions.
In this study, we examined an approach to analyzing and overcoming token occurrence variations in concept detection. Our approach was based on syntactic information obtained by a parser. We used a syntactic parser provided in Stanford CoreNLP library3, and conducted a concept detection experiment on the 2014 i2b2/UTHealth corpus4. We manually reviewed concept phrases extracted by this approach and examined its advantages and limitations. The experimental results suggest the approach using syntactic information could potentially boost a recall of concept detection.
Background
Concept Detection
Detection of concept phrases has been actively studied in the clinical domain. There are two variations of this task: (i) map phrases in text to many detailed concepts, e.g., individual concepts in the UMLS Metathesaurus5,6; (ii) map phrases in text to a small number of concept groups, e.g., person names and hospital names. They pose different kinds of challenges. In this study, we focus on the former type of tasks.
In the former type of tasks, there are a large number of target concepts involved, and each concept is specific. Common synonyms of concepts are maintained in rich terminology resources. NLP systems in the clinical domain usually implement some form of dictionary look-up that facilitates mapping of phrases to detailed concepts. There are several software systems dedicated to this task in the clinical domain. MetaMap1,2 implements part-of-speech (POS) tagging and shallow parsing and applies linguistic rules to facilitate comprehensive mapping of phrases in text to UMLS Metathesaurus concepts5,6. MetaMap Lite7 is a fast implementation of basic MetaMap functions. For concept look-up, it considers every token n-grams (sequence of n tokens) starting at each token position in a sentence and looks them up in term search indices. MGREP8 is another software tool implementing fast concept mapping. It pre-computes term variations and uses a trie data structure for efficient term look-up. NOBLE9 is yet another tool for concept mapping. It uses a hash table of words to retrieve concept phrase candidates at each word position in a sentence. These and other tools have been compared for their run time speed and accuracy10-14. These studies concern different target concepts and use different evaluation corpora, and the reported results are not comparable. But they show methods based on dictionary look-up can run fast and yield accuracy comparable to elaborated concept detection systems.
Syntactic Parsers
Over the years, syntactic parsers have become more accurate, much faster, and more accessible to users owing to user-friendly software libraries, such as Stanford CoreNLP3. These parsers have been developed in the general NLP domain, but they have been used in the clinical domain as well15,16. Applications of syntactic parsers reported in the clinical domain includes negation detection17-19 and semantic role labeling20. There are two major kinds of syntactic parsers: constituency parsers and dependency parsers. Given a sentence, a constituency parser produces a constituent tree, in which words are grouped into constituents. A dependency parser produces a dependency graph, which represents modification relations between words. There have been tools to convert a constituent tree into a dependency graph, and a constituency parser with a convertor can also be used to produce a dependency graph.
Data and Methods
In our experiment, we used the 2014 i2b2/UTHealth corpus4, which is a collection of 790 de-identified clinical notes, consisting of longitudinal records for 301 patients. We used a subset of the UMLS Metathesarus5,6 as a concept dictionary. We focused on twelve UMLS semantic typesa that are relevant to clinical findings of our interest. We used a string matching algorithm for dictionary look-up and applied the Stanford CoreNLP library3 for syntactic analysis. This process consists of three steps: (1) pre-processing, (2) dependency pattern collection, and (3) dependency pattern matching.
Pre-processing
To compile a dictionary of concept phrases from the UMLS Metathesaurus, we selected English terms belonging to the level-0b category using the UMLS MetamorphoSys toolc. We then retrieved phrase strings and associated Concept Unique Identifiers (CUIs) from the Concept Names and Sources table (MRCONSO.RRF).
To look-up phrase strings in text, we used an existing Java implementation of Aho-Corasick algorithm21. The program facilitates efficient character string matching, and finds all occurrences of dictionary entries in given text, including overlapping occurrences. To use this program for phrase detection, dictionary entries and input text were normalized in the same manner. Specifically, letters were lower-cased and non-alphanumeric character sequences were replaced with a single white space. After the program was applied, detected character strings that did not align with word boundaries were filtered.
To obtain syntactic information, we applied a CoreNLP pipeline to input text, including tokenizer, sentence annotator, part-of-speech tagger, lemmatizer, and parser. Among different parsers provided in the library, we selected the Shift-Reduce Constituency parser because it is reportedly fast and accurated, and it would be suitable for practical applications. Derived constituent trees were converted into labeled dependency relations using a converter in the library. For the rest of the annotators, the default settings were used.
Dependency Pattern Collection
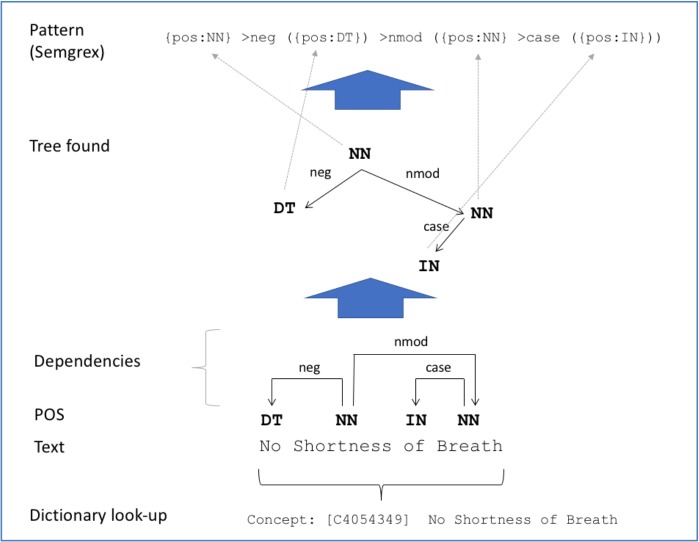
This step is described in Figure 1. In this step, two types of information from the pre-processing step were used: (i) dependency relations among tokens and (ii) concept phrases detected through dictionary look-up. In each concept phrase detected, we identified a token, from which all tokens in that concept phrase can be reached through dependency relations (e.g., “Shortness” in Figure 1). If such a token was not present, the phrase was discarded.
Figure 1.
Dependency pattern collectionf.
Discarded cases are discussed in the next section. Using the identified phrase, we created a tree made up of labeled dependency relations and POS tags, but without the original words. Finally, a pattern was formed by using a specific notation of CoreNLP Semgrexe that specifies the tree found. Only patterns found in at least five phrases were retained.
Dependency Pattern Matching
Dependency patterns collected in the previous step were used to identify additional concept phrases that appear in text in a discontinuous and/or permuted manner.
Dependency patterns collected in the previous step were sought in parsed sentences using CoreNLP Semgrex tool. The same corpus was used for this step as the last step. Patterns match not only with the source phrases, but also with phrases that appear in text in a discontinuous and/or permuted manner.
Newly extracted phrases were sought against the dictionary and, if found, they were mapped to associated CUIs. Since tokens in these phrases may be permuted, the ordering of tokens was ignored during this dictionary search step, i.e., set equality was tested between a set of tokens from a newly detected phrase and that from a phrase in the dictionary.
Results and Discussion
Data Processing
The resultant dictionary of the twelve UMLS semantic types contains 373k normalized phrases (tokenized and lowercased strings), which are associated with 173k concepts. The dictionary look-up step identified 128,909 concept phrase occurrences (7,094 unique phrases) in the corpus of 790 notes. Among those phrases, 15,546 phrase occurrences (3,345 unique phrases) were multi-token phrases consisting of two or more tokens. Observed multi-token phrases included “chest pain”, “blood pressure”, “coronary artery disease”, and “past medical history.” Long phrases were also detected, e.g., “pupils equal, round, and reactive to light and accommodation”, “diabetes mellitus without mention of complication”, and “no evidence of coronary artery disease.” But these long phrases were much less frequent compared to short phrases.
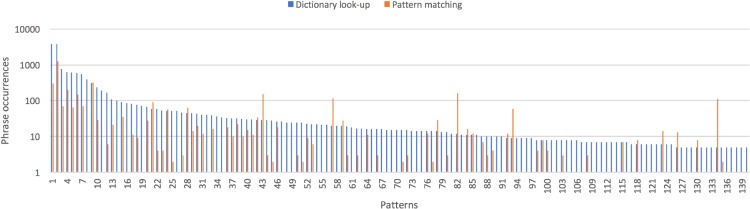
In the multi-token phrases detected, 486 dependency patterns were identified. We assumed frequently observed patterns would help extract more phrase occurrences in the subsequent pattern matching step, and also they might be more reliable, accurate patterns, compared to rarely observed patterns. Therefore, among the detected patterns, 140 patterns that were observed in five or more phrase occurrences were kept for the subsequent pattern matching step. No dependency patterns could be extracted from 4,905 concept phrase occurrences (3.8% of all the concept phrase occurrences detected, 128,909), i.e., there was no rooted tree structure in those phrases, unlike the example in Figure 1. Figure 2 shows each dependency pattern is extracted from how many source phrases (continuous and not permuted phrases) detected during dictionary look-up (labeled as “Dictionary look-up”). Figure 3 shows the same information along with the actual dependency patterns but only for the twenty patterns with the highest source phrase occurrences. The list below shows some examples of extracted dependency patterns, represented in Semgrex format, together with a few of their source phrase examples.
- A singular noun with a negation determiner: {pos:NN} >neg ({pos:DT})
- no change
- no nausea
- no cough
- A verb gerund with its singular object noun: {pos:VBG} >dobj ({pos:NN})
- presenting complaint
- losing weight
- taking insulin
- A singular noun modified by another singular noun with a preposition: {pos:NN} >nmod ({pos:NN} >case ({pos:IN}))
- shortness of breath
- dyspnea on exertion
- loss of consciousness
- An adjective accompanied by an infinitive verb: {pos:JJ} >xcomp ({pos:VB} >mark ({pos:TO}))
- warm to touch
- sore to touch
- unable to walk
Figure 2.
Numbers of phrase occurrences (vertical) for each dependency pattern (horizontal): Continuous and not permuted phrase occurrences observed during dictionary look-up (“Dictionary look-up”) and discontinuous and/or permuted phrase occurrences extracted during dependency pattern matching (“Pattern matching”).
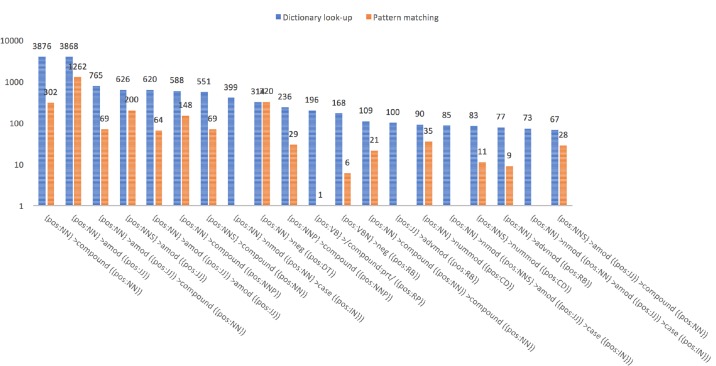
Figure 3.
Numbers of phrase occurrences (vertical) for each dependency pattern (horizontal) for the leftmost twenty patterns in Figure 2.
The collected dependency patterns were applied to the same corpus of 790 notes. In addition to the previously detected phrases, 3,862 new phrase occurrences (1,288 unique phrases) were mapped to CUIs. These phrase occurrences were previously not detected through dictionary look-up, because they involve discontinuous tokens and/or permuted tokens. The number of newly mapped phrases per pattern (“Pattern matching”) is shown in Figures 2 and 3. The list below shows examples of newly extracted phrase occurrences using the patterns shown in the above list. For instance, more occurrences of “no pain” (T033: Finding) could be identified in “no chest pain” and “no significant or definitive chest pain.”
- A singular noun with a negation determiner: {pos:NN} >neg ({pos:DT})
- no pain
- no change
- no disease
- A verb gerund with its singular object noun: {pos:VBG} >dobj ({pos:NN})
- hitting head
- losing weight
- taking insulin
- A singular noun modified by another singular noun with a preposition: {pos:NN} >nmod ({pos:NN} >case ({pos:IN}))
- none
- An adjective accompanied by an infinitive verb: {pos:JJ} >xcomp ({pos:VB} >mark ({pos:TO}))
- warm to touch
- unable to move
Manual Review of Newly Extracted Phrases
We manually reviewed detected phrases in context of their occurrences. For instance, utility of dependency patterns could be seen in an example where a dictionary entry “intact sensation” was identified in a text fragment “Vibratory sensation absent in …, intact at … ” using a collected pattern {pos:NN} >amod ({pos:JJ}) (a singular noun modified by an adjective). This phrase occurrence concerns both discontinuous and permuted tokens. The parser correctly identified a direct dependency relation between the two remote tokens, “sensation” and “intact”, and the dependency pattern for phrase extraction was applicable. Meanwhile, we found not all extraction cases were practically significant. In this section, we summarize several findings through our manual review of prevalent extraction cases.
The most frequent type of phrases extracted during the pattern matching step was one involving remote modifiers, such as “intermittent pain” found in “ intermittent right shoulder pain” or “chronic shoulder pain” found in “ chronic left shoulder pain ”. In these cases, phrases without the modifier may be in the dictionary, e.g., “right shoulder pain” and “left shoulder pain.” Yet, their modified forms, e.g., “intermittent right shoulder pain”, are not in the dictionary, and neither are discontinuous phrases found in them, e.g., “intermittent shoulder pain.” In this case, detection of discontinuous phrases would be of interest. In another case, however, detected discontinuous phrases were substrings of phrases already detected during dictionary look-up, e.g., “past history” found in “past medical history”, where “past medical history” is in the dictionary. Unlike the first case, detection of such embedded discontinuous phrases would be less interesting, since more detailed concepts can be detected using string matching.
Similar to the first case, we also observed phrases involving a remote determiner “no”, e.g., “no pain” found in “no chest pain ”, and “no clinical manifestations” found in “no current clinical ictal manifestations .” Detection of such remote “no” is significant in applications, not only in terms of concept detection, but also of negation detection17-19. Detection of this case can be especially challenging when negated concepts are coordinated, e.g., “no abdominal pain, nausea or vomiting, anorexia or weight loss.” While domain experts are able to interpret the scope of the negation, a parser has difficulty identifying the correct scope of the negation.
Phrases involving verbs were also extracted through pattern matching, e.g., “lost weight” found in “ lost so much weight ”, “hitting head” found in “ hitting his head”, etc. In some cases, extracted phrases were correct, but patterns were matched incorrectly, e.g., “vomiting diarrhea” was extracted, but falsely identified as a verb and an object during matching. Similarly, “left knee” was extracted, but falsely identified as a verb in past participle and a noun.
Failed Dependency Pattern Extraction
Dependency patterns were initially extracted from continuous concept phrases found in the dictionary. As reported above, the current approach to pattern extraction failed in 3.8% of those phrases. In these cases, tokens in an extracted phrase do not have dependency relations with one another. We commonly observed that there was actually a nearby token(s), which all the concept phrase tokens have dependency relations with. Two representative dependency patterns were found among those cases. The first type of pattern is when the concept phrase in the UMLS is “missing some part”, e.g., “history of”, which misses its adjacent word “diabetes” (“history of diabetes”). The second type, which is subtly different from the first type, is when the UMLS concept phrase is missing its head, e.g., “do not”, which misses the primary concept, like “show” (“do not show”), or “right dominant”, which misses “system” (“right dominant system”). These patterns could still be captured by Semgrex, but the extraction procedure and required patterns become more complexg.
In addition, the pre-processing procedure involves the step of converting constituent structures to labeled dependency relations. In our current configuration, dependency relations rely on the constituent structures in question. A dependency pattern cannot match with the target concept phrase if the constituent structure of that concept phrase results in a different dependency relations than what the pattern describes. For example, the constituent structure of “slow wave sleep” is supposed to be [NP [NP slow/JJ wave/NN] sleep/NN], but the constituency parser proposes a structure [NP slow/JJ wave/NN sleep/NN], which in return gives an unexpected dependency relation on the concept phrase “slow wave”. As a result, the concept phrase “slow wave” could not be matched with the extracted dependency pattern {pos: NN} > amod {pos: JJ}.
Challenges and Limitations
As stated earlier, clinical concept detection concerns a large number of detailed concepts, including those expressed in multiple tokens and in different phrase types. Moreover, a concept phrase may appear in clinical notes in a discontinuous and/or permuted manner. To overcome these challenges and improve clinical concept detection, it is of great interest to use dependency relations. While promising results were observed in our experiment, we encountered several challenges and noticed limitations of the current approach.
One limitation of the current approach is that the patterns were extracted from dictionary look-up. This may have resulted in incomplete pattern identification, e.g., “chest clear” found as “ chest was clear” or “intermittent pain” found as “pain is intermittent.” Further review of phrase occurrences in text is planned in our future study.
In the current approach, phrases detected through dependency pattern matching were searched against synonyms in the dictionary using token set equality, i.e., token ordering was ignored when a detected phrase was compared with each synonym. Token ordering was ignored so that permuted phrases in text can still be found in the dictionary. While this approach suffices the goal in most cases, inaccurate or incorrect mapping results were still observed, e.g., “of prostate cancer” detected in “family history of prostate cancer ” was mapped to the synonym “cancer of prostate”. Similarly, dependency patterns including symbols, such as comma and colon, are subject to the same error. An ideal way of searching synonyms in the dictionary, instead of using set equality or token ordering, is to rely on dependency relations among tokens. In our experiment, dependency relations were obtained for phrases in clinical notes, but they were not available for synonym phrases in the dictionary. A syntactic parser may not be suitable for analyzing a synonym phrase by itself, without being placed in a sentence. We may look up each synonym in sentences and then apply a syntactic parser to derive dependency relations among tokens.
Figures 2 and 3 show that, for each dependency pattern, more phrases were identified during the initial dictionary look-up step, compared to the later pattern matching step, i.e., in the figure, a blue bar is taller than its peer orange bar. As expected, concepts are found more commonly among continuous and not permuted phrases than among discontinuous and/or permuted phrases. There were several exceptions seen in Figure 2, where orange bars are unexpectedly high. It turned out that they were often problematic extraction from recurrent text. For instance, the 57th pattern in Figure 2, {pos:NN} >dep ({pos:NN}), was observed twenty times during the dictionary look-up step. The dependency label “dep” used in the pattern is “unspecified dependency” when it is unable to identify an appropriate dependency label. This pattern extracted 114 phrase occurrences, including “disease history” and “smoking history”, which were repeatedly extracted from falsely split and parsed sentences (section headers). The sentence annotator and also NLP components in general need be tuned to clinical note processing.
Lastly, in our current method, we assumed a syntactic pattern consisting of POS tags and dependency relations. It may be of interest to consider lexicalized patterns including words, e.g., a POS tag along with a word. The current approach helped generalize token occurrence patterns well and reduce the number of extracted Semgrex patterns. But richer patterns involving words would help improve pattern accuracy. Analysis of such patterns may also help us understand structure and composition of clinical concept phrases better.
Conclusion
Analysis of clinical concept phrases and investigation of comprehensive mapping remains of great interest. In this study, we investigated limitations of exact string matching, commonly known as dictionary look-up. Dictionary lookup facilitates efficient mapping of phrases to a large number of granular concepts commonly required for clinical NLP applications. Despite these practical advantages, one challenge is that discontinuous and/or permuted concept phrases cannot be detected. To analyze and overcome this challenge, dependency relations of multi-token concept phrases were collected, and they were in turn used to extract additional concept phrases. A large number of concept phrase candidates were extracted in our experiment, and they were manually reviewed. The extraction result was promising in boosting a recall of dictionary look-up. Meanwhile, challenges were also noticed. In our future work, we plan to revise the method and continue investigating dependency patterns of multi-token phrases.
Acknowledgement
Deidentified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr. Ozlem Uzuner, i2b2 and SUNY. We thank Drs. Peter Li and Daniel Zisook in the KPSC Medical Informatics group for their helpful suggestion and comments on the manuscript. We also thank the current and former member of the group for helpful discussion on this topic.
Footnotes
T019: Congenital Abnormality, T020: Acquired Abnormality, T033: Finding, T037: Injury or Poisoning, T046: Pathologic Function, T047: Disease or Syndrome, T048: Mental or Behavioral Dysfunction, T049: Cell or Molecular Dysfunction, T050: Experimental Model of Disease, T184: Sign or Symptom, T190: Anatomical Abnormality, T191: Neoplastic Process.
Level 0 terms are those that do not require additional license agreements besides the UMLS license.
Dependency patterns corresponding to the three example phrases may be represented as, respectively, ({pos: NN} > /nmod:of/ {}) $+ ({pos: IN} < case {}) and ({pos: /VB./} < aux{})$+ ({pos: RB} < neg{}), and ({pos: JJ} < amod{})$+ ({pos: JJ} < amod{}).
Please refer to the Semgrex Javadoc for the Semgrex symbols used:
https://nlp.stanford.edu/nlp/javadoc/javanlp/edu/stanford/nlp/semgraph/semgrex/SemgrexPattern.html
References
- 1.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson AR, Lang F-M, Mork JG. Using MetaMap: A Tutorial. Presented at the: BioNLP Workshop. 2014. Jun 27, https nlm.nih.gov/Publications/Papers/14.06.27.MetaMapTutorial.pptx.
- 3.Manning CD, Surdeanu M, Bauer J, Finkel J, Bethard SJ, McClosky D. The Stanford CoreNLP Natural Language Processing Toolkit. Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations; 2014. pp. 55–60. [Google Scholar]
- 4.Stubbs A, Uzuner O. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus. JBiomedInform. 2015;58(Suppl):S20–29. doi: 10.1016/j.jbi.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32(4):281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demner-Fushman D, Rogers WJ, Aronson AR. MetaMap Lite: an evaluation of a new Java implementation of MetaMap. J Am Med Inform Assoc JAMIA. 2017;24(4):841–844. doi: 10.1093/jamia/ocw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah NH, Bhatia N, Jonquet C, Rubin D, Chiang AP, Musen MA. Comparison of concept recognizers for building the Open Biomedical Annotator. BMCBioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tseytlin E, Mitchell K, Legowski E, Corrigan J, Chavan G, Jacobson RS. NOBLE - Flexible concept recognition for large-scale biomedical natural language processing. BMC Bioinformatics. 2016;17:32. doi: 10.1186/s12859-015-0871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc JAMIA. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanenblatt MA, Coden A, Sominsky IL. 2010. The ConceptMapper Approach to Named Entity Recognition; pp. 546–551. [Google Scholar]
- 12.Leaman R, Islamaj Dogan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinforma Oxf Engl. 2013;29(22):2909–2917. doi: 10.1093/bioinformatics/btt474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Divita G, Zeng QT, Gundlapalli AV, Duvall S, Nebeker J, Samore MH. Sophia: A Expedient UMLS Concept Extraction Annotator. AMIA Annu Symp Proc AMIA Symp. 2014;2014:467–476. [PMC free article] [PubMed] [Google Scholar]
- 14.Funk C, Baumgartner W, Garcia B, et al. Large-scale biomedical concept recognition: an evaluation of current automatic annotators and their parameters. BMC Bioinformatics. 2014;15:59. doi: 10.1186/1471-2105-15-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan J, Yang EW, Jiang M, et al. Syntactic parsing of clinical text: guideline and corpus development with handling ill-formed sentences. J Am Med Inform Assoc JAMIA. 2013;20(6):1168–1177. doi: 10.1136/amiajnl-2013-001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen R, Elhadad M. Syntactic dependency parsers for biomedical-NLP. AMIA Annu Symp Proc AMIA Symp. 2012;2012:121–128. [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Lowe HJ. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform Assoc JAMIA. 2007;14(3):304–311. doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn S, Wu S, Chute CG. Dependency Parser-based Negation Detection in Clinical Narratives. AMIA Jt Summits Transl Sci Proc AMIA Jt Summits Transl Sci. 2012;2012:1–8. [PMC free article] [PubMed] [Google Scholar]
- 19.Mehrabi S, Krishnan A, Sohn S, et al. DEEPEN: A negation detection system for clinical text incorporating dependency relation into NegEx. J Biomed Inform. 2015;54:213–219. doi: 10.1016/j.jbi.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc JAMIA. 2013;20(5):922–930. doi: 10.1136/amiajnl-2012-001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.hankcs. Aho-Corasick Double Array Trie. https://github.com/hankcs/AhoCorasickDoubleArrayTrie.