Abstract
Conversations especially between a clinician and a patient are important sources of data to support clinical care. To date, clinicians act as the sensor to capture these data and record them in the medical record. Automatic speech recognition (ASR) engines have advanced to support continuous speech, to work independently of speaker and deliver continuously improving performance. Near human levels of performance have been reported for several ASR engines. We undertook a systematic comparison of selected ASRs for clinical conversational speech. Using audio recorded from unscripted clinical scenarios using two microphones, we evaluated eight ASR engines using word error rate (WER) and the precision, recall and F1 scores for concept extraction. We found a wide range of word errors across the ASR engines, with values ranging from 65% to 34%, all falling short of the rates achieved for other conversational speech. Recall for health concepts also ranged from 22% to 74%. Concept recall rates match or exceed expectations given measured word error rates suggesting that vocabulary is not the dominant issue.
Introduction
While our diagnostic armamentarium continues to grow, obtaining the patient’s history is still considered a critical step in delivering high quality, safe and cost-effective care. Some older studies demonstrated the gap that exists between the content of those conversations and the data recorded in the medical record. Zuckerman et al, for example, compared tape-recorded conversations with patient records and found significant omissions in crucial categories such as reason of visit and degree of disability; Romm et al compared contemporaneous notes recorded by an independent observer to the medical record and a found a 71%-73% accuracy for diagnosis, tests and information related to the current illness, and even lower accuracy for medical history.1,2 More recently, Johnson et al. attempted to support clinicians by using automatic speech recognition (ASR).3 Even more recently, Zafar et al. evaluated the performance of speaker trained ASRs and found better levels of accuracy.4 Gür, in his dissertation, described his attempts to measure the performances of two state-of-the-art automatic speech recognition engines for the task of transcribing clinical conversations.5 He found WERs over 100% for both untrained ASR engines evaluated and efforts to refine language models yielded little if any improvement. Interesting, the study found that the mean WER was lower (64.5% and 79.2%) for doctors than for patients (91.0% and 91.0%). Selected ASR engines have been evaluated on a clinical question answering task and it has been shown that domain adaptation with a language model improves the accuracy in interpreting spoken clinical questions significantly.6
ASR has improved significantly over the last 40 years.7 Particularly with the advent of deep learning approaches, automatic speech recognition (ASR) has advanced rapidly with each ASR engine reporting dramatic improvements and approaching if not exceeding human speech recognition capabilities. In March of 2017, for example, IBM announced that they had improved on their previous benchmark of 6.95% to achieve a word error rate of 5.5% on the TELEPHONE dataset though performance was poorer (10.3%) on the CallHome dataset.8 Human recognition was pegged at 5.1% and 6.8% for the TELEPHONE and CallHome datasets respectively. This is the just the most recent of a string of announcements and publications reporting performance in the range of 3.7% for short phrases to 6.8% for the TELEPHONE dataset.9,10
Recent work in the clinical domain shows promise as well. Edwards et al. achieved a WER of 16% using a neural network based ASR algorithm built for dictational speech and Chung-Cheng reported a WER of 18.3% for recognition of clinical conversation speech.11,12
Given this remarkable progress and the potential value of capturing clinical observations directly from conversations, we wanted to understand the performance of selected contemporary, readily available ASR engines when applied to conversational clinical speech and specifically the potential to extract clinical meaning from these conversations, we conducted a systematic comparison of eight ASR engines.
Methods
ASR engines were included if they employed contemporary speech recognition approaches, including examples that have reported “better than human” performance and are widely accessible (potentially for a fee).
We created clinical scenarios for the consultation part of a clinical encounter for 11 encounters likely to be seen in an ambulatory primary care practice. We constructed the scenarios based on the family practice clinical scenarios created by CMS for their “Road to 10” campaign.13 The scenarios provide a detailed description of the patient presentation but did not provide any suggested dialog or suggested descriptions. We recorded unscripted simulated clinical interviews based on these clinical scenarios with two different physicians in the provider role and seven different non-physician adults playing the role of the patient. All participants were native English speakers. The simulated encounters were recorded in a quiet office with the participants seated directly in front of two different microphones: a Razer Seiren Pro® digital microphone in omnidirectional mode (Razer, San Francisco, CA https://www.razerzone.com/gaming-audio/razer-seiren) and a Microsoft Kinect 2.0® (Microsoft, Redman, Washington). The Razer includes three 14 mm custom tuned condenser capsules in an array software configurable into four different recording patterns with 24-bit analog-to-digital converter. The Kinect includes a four-microphone array with 24-bit analog-to-digital converter and local signal processing including acoustic echo cancellation and noise suppression
The audio was digitally recorded in Windows Media Audio format (Microsoft) and encoding, in 32 bit stereo at 44,100 Hz using Window Recorder for the Razer and in Free Lossless Audio Codec or FLAC format (the xiph open source community) and encoding with 16 bit depth mono at 44,100 Hz using Microsoft Kinect Studio for the Kinect. Razer files were converted to the same format as the Kinect files. These FLAC files were submitted to the ASR engines. In preparation for the study, we assessed the potential effect of speaker segmentation on ASR engine performance and found no improvement thus, we did not incorporate speaker segmentation into the analysis approach. The recordings were professionally transcribed and annotated with metadata including speaker and time index.
We chose four performance metrics: Word Error Rate (WER) and precision, recall and F1 score for clinical concepts. We chose WER because it is the metric most often reported in the ASR literature and will allow us to compare the results in our specific use case with published results. Since our focus is on information extraction rather than creating a complete transcript, we included the precision, recall and F1 score.
Custom Python code was written to normalize the text and then use the Levenshtein algorithm as implemented in https://pypi.python.org/pypi/python-Levenshtein/0.12.0 to identify substitutions, deletions, insertions, and correct word rates using the transcript as the reference. These values were then used to compute the unweighted word error rates (WER).
Concepts were extracted from the transcripts and the output for each clinical scenario from each ASR engine microphone pair file using a commercially available NLP service and open source NLP service (CLiX NOTES, Clinithink, Bridgend CF31 1LH, UK) and biomedical annotator (https://bioportal.bioontology.org/annotator). The number of concepts were counted for each document and matched across documents to determine the precision, recall and F1 scores. The code was validated by manual feature extraction and comparison of results.
We computed the outcome measures for each conversation and then report the means of those outcomes across all conversations, scenarios and microphones.
Results
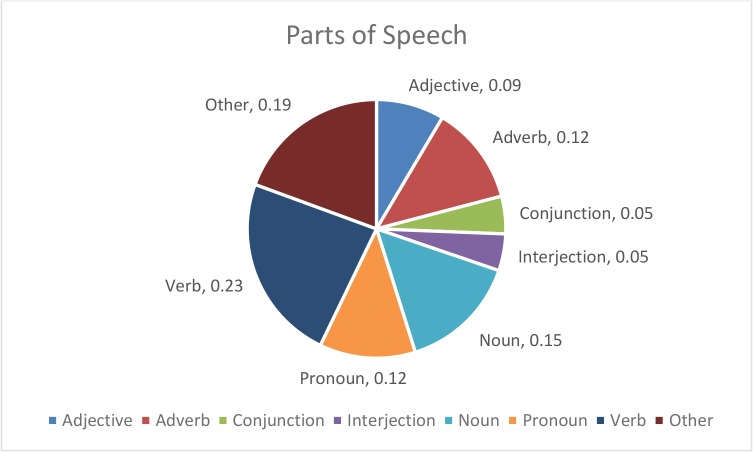
There is a total of 34 recordings of unscripted, simulated, clinical interviews averaging approximately five and one-half minutes in length and containing an average of 1,824 words each. An average of 33% of the total word count in the human transcript was unique. This was parsed into grammatical parts of speech resulting in 23.4% verbs, 19.4% other, 14.9% nouns, 12.4% adverbs, 12% pronouns, 8.5% adjectives, and 9.3% both conjunctions or interjections as illustrated in Figure 1. Eight ASR engines were tested on each of the 34 recordings, resulting in a total of 272 textual outputs. The ASR engines tested are listed in Table 1.
Figure 1—
Characterization of the grammatical parts of speech in the conversational text. Verbs representing 23.4% and nouns at 14% represent the largest parts of speech.
Table 1.
The table lists specifics of the eight ASR engines evaluated
| ASR Engine Name | Short Name | Category | Version Tested | URL |
|---|---|---|---|---|
| Bing Speech API | BING | Cloud Streaming | 1.0 | https://www.microsoft.com/cognitive-services/en-us/speech-api |
| Google Cloud Speech API | Cloud | 1.0 | https://cloud.google.com/speech/ | |
| IBM Speech To Text | IBM | Cloud Streaming | 1.0 | https://www.ibm.com/watson/developercloud/speech-to-text.html |
| Azure Media Indexer | MAVIS | Cloud | 1.0 | https://docs.microsoft.com/en-us/azure/media-services/media-services-index-content |
| Azure Media Indexer 2 Preview | MAVIS v2 | Cloud | 2.0 | https://docs.microsoft.com/en-us/azure/media-services/media-services-process-content-with-indexer2 |
| Nuance.SpeechAnywhere | Nuance | Health Focused | 3.2 | https://www.nuancehealthcaredeveloper.com/ |
| Amazon Transcribe Preview | Transcribe | Cloud | 1.0 | https://aws.amazon.com/transcribe/ |
| Mozilla DeepSpeech | DeepSpeech | Local | 0.1 | https://github.com/mozilla/DeepSpeech/wiki |
Table 2 provides a few illustrative examples of the output from two ASR engines and the human transcription process for a Kinect recording. The output from one of the ASR engines is very difficult to read with many substitutions, insertions and deletions. Medications names occurred frequently in the recordings and were almost never correctly recognized.
Table 2.
The table provides selected examples of output form human transcription, DeepSpeech and MAVISv2 ASR engines from an encounter for a pre-op clearance (scene 1).
| Human Transcription | DeepSpeech | MAVIS v2 |
| — Okay, all right, and have you ever taken any medication for it? [0:00:47.8] | om kiy rt i have y er takeng any medication for it ir have takof sombeoe redocation ortone hardi ire you taking anything today ah no not today do tor give yeusolfthing rout my taket a l right me’t tak a look here il leos lik they gave you son matopral a suxcitny am ah i tac can sometimes be a costly manicangios is se expensip that’s wha l sogood larry ave y | OK alright I Have you ever taken any medication for it I have taken some medicine medication overtime here. |
| — I have taken some medication over time, yeah. [0:00:51.2] | NOTE Confidence: 0.8099813 | |
| — All right, are you taking anything today? [0:00:53.9] | 00:00:51.780 --> 00:00:55.390 All right are you taking anything today. |
|
| — No, not today. [0:00:55.6] The doctor gave me something, but I’m not taking it. [0:00:59.5] |
NOTE Confidence: 0.8462618 | |
| — Oh, all right. [0:01:00.4] Let’s take a look here. [0:01:02.3] It looks like they gave you some metoprolol succinate. [0:01:05.3] That can sometimes be a costly medication. [0:01:11.6] |
00:00:55.760 --> 00:01:06.060 No not today doctor gave me something but I’m not taking it all right let’s take a look here looks like they gave you some mto prolo succinate. NOTE Confidence: 0.7029915 |
|
| — Yeah, it’s super expensive. [0:01:12.6] That’s why I don’t take it. [0:01:13.8] |
00:01:06.950 --> 00:01:18.650 Ah that can sometimes be a costly medication such better that’s why I’m saying it all right have |
Table 3 — Levenshtein output from one encounter. Enumerates the variation in ASR and microphone errors across one conversation.
Table 3.
Levenshtein output from one encounter. Enumerates the variation in ASR and microphone errors across one conversation.
| ASR Engine | Microphone | Correct | Deletions | Insertions | Substitutions | WER |
|---|---|---|---|---|---|---|
| transcribe | kinect | 830 | 67 | 75 | 171 | 0.29 |
| transcribe | razer | 830 | 67 | 75 | 171 | 0.29 |
| speech | kinet | 804 | 133 | 33 | 131 | 0.28 |
| speech | razer | 803 | 138 | 34 | 127 | 0.28 |
| watson | kinect | 836 | 77 | 831 | 55 | 0.29 |
| watson | razer | 836 | 77 | 831 | 55 | 0.29 |
| deepspeech | kinect | 578 | 61 | 107 | 429 | 0.56 |
| deespeech | razer | 577 | 63 | 107 | 428 | 0.56 |
| dragon | kinect | 599 | 371 | 698 | 0.44 | |
| dragon | razer | 529 | 456 | 583 | 0.51 | |
| bing | kinect | 674 | 203 | 531 | 91 | 0.42 |
| bing | razer | 674 | 203 | 531 | 91 | 0.42 |
| mavis | kinect | 746 | 126 | 261 | 96 | 0.33 |
| mavis | razer | 746 | 126 | 261 | 96 | 0.33 |
| mavis2 | kinect | 858 | 66 | 50 | 144 | 0.24 |
| mavis2 | razer | 841 | 67 | 45 | 160 | 0.25 |
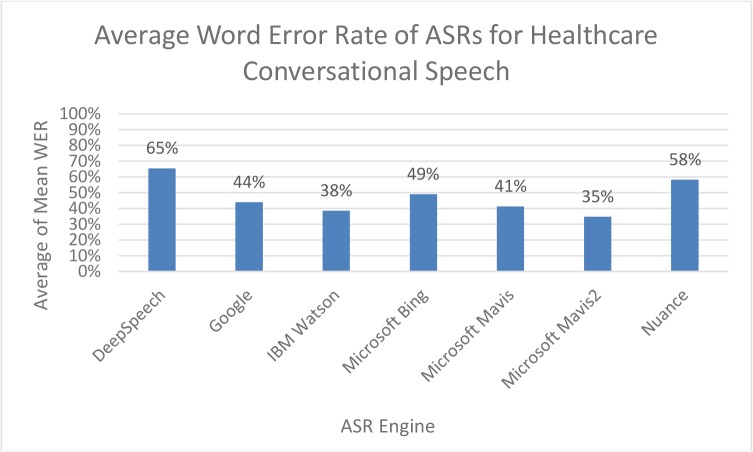
Figure 2 shows the variation in performance across the ASR engines (listed alphabetically) evaluated. The WER ranged from 35% with Microsoft’s Mavis2 engine to 65% with DeepSpeech.
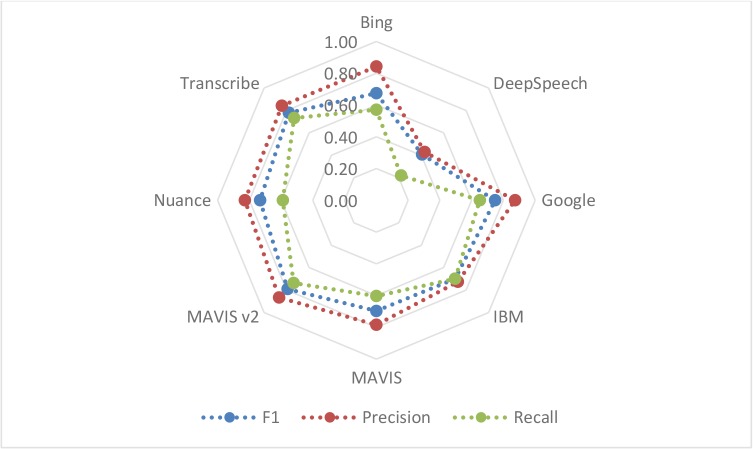
Figure 3 illustrates the precision, recall and F1 score for concept retrieval.
Figure 3--.
The radar chart visualizes the precision, recall and F1 for clinical concepts for each ASR using human transcription as the gold standard. The concepts were extracted using both a commercially available NLP engine and an open source NLP. The blue dots represent F1 scores, the green dots the recall and the red dots the precision.
Discussion
The achievable performance of contemporary ASR engines, when applied to conversational clinical speech as measured by WER and clinical concept extraction, is disappointing with WERs of approximately 50% and concept extraction rates of approximately 60%. The best performing ASR engine achieved a 35% WER and 73% recall rate. Remarkably high average word error rates were observed for some ASR engines because of large numbers of insertions and deletions which resulted in WERs of 86% for some clinical scenarios. There are a limited number of use cases where this level of performance is adequate. Insertions, for example, might lead to a symptom or finding that the patient doesn’t have being identified which creates potential for inappropriate actions that could harm the patient. Omissions may represent less risk since we anticipate that these concepts would be captured other ways including the very important role of the provider. In most use cases we anticipate, a significant number of omissions or insertions will dramatically limit the value if the user cannot have high confidence in the results. Even if clinical concepts were perfectly captured, errors in other words will limit the confidence in the result. Substitutions of “no uterus” for “normal uterus”, or the converse, could have important clinical implications. As is so often the case, for clinical care, the tolerance for errors and for wasted effort identifying and correcting those errors is limited. This intolerance is both because of the implications for the patient, and consequently the time typically required to address these errors from overburdened and expensive clinicians.
These results are reasonably consistent with Gur’s findings and the WER of 45.5% calculated for the nursing vocabulary in Souminen et al.’s recent study of nursing changeover notes that were read.14 The ASR engines continue to evolve rapidly. There was meaningful improvement from MAVIS to MAVIS v2 for example and we expect this trend to continue. The Google Brain team’s research demonstrated that a WER of 18% for clinical conversations, providing evidence that at least that level of performance and potentially better should be achievable.
One of the primary limitations of this study is that the clinical conversations is the use of simulated clinical conversations. As described, we took several steps to limit the impact of this approach including providing specific scenarios but using an unscripted approach. Further, we believe these findings as an upper bound of the performance that is achievable today with readily accessible ASR engines because the recordings were obtained under near ideal conditions. Recordings obtained during actual clinical practice are likely to have a good deal more noise, volume fluctuation and to be of lower quality since speakers will often be more distant and not facing the microphones directly. Another limitation is that these results represent a snapshot in the evolution of these ASR engines. They continue to evolve rapidly and we expect performance to continue to evolve.
Conclusion
We believe that it is useful to understand the level of performance achievable with readily available contemporary ASR engines to guide thinking about how this technology might be used to support clinicians. The modest level of performance suggests that we need to focus on improving ASR engine performance before we can adopt these technologies for conversational speech for a broad range of clinical use cases.
Figure 1.
The bar chart shows the word error rate by ASR engine
References
- 1.Zuckerman AE, Starfield B, Hochreiter C, Kovasznay B. Validating the content of pediatric outpatient medical records by means of tape-recording doctor-patient encounters. Pediatrics. 1975 Sep 1;56(3):407–11. [PubMed] [Google Scholar]
- 2.Romm FJ, Putnam SM. The validity of the medical record. Medical care. 1981 Mar;1:310–5. doi: 10.1097/00005650-198103000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Johnson K, Poon A, Shiffman S, Lin R, Fagan LM. Q-Med: a spoken-language system to conduct medical interviews. Knowledge Systems AI Laboratory (KSL-92-09) 1992.
- 4.Zafar A, Overhage JM, McDonald CJ. Continuous speech recognition for clinicians. Journal of the American Medical Informatics Association. 1999 May 1;6(3):195–204. doi: 10.1136/jamia.1999.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gur B. Improving Speech Recognition Accuracy for Clinical Conversations (Master’s Thesis Massachusetts Institute of Technology)
- 6.Liu F, Tur G, Hakkani-Tür D, Yu H. Towards spoken clinical-question answering: evaluating and adapting automatic speech-recognition systems for spoken clinical questions. Journal of the American Medical Informatics Association. 2011 Jun 24;18(5):625–30. doi: 10.1136/amiajnl-2010-000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juang BH, Rabiner LR. Vol. 1. Atlanta Rutgers University and the University of California. Santa Barbara: Georgia Institute of Technology; 2005. Automatic speech recognition–a brief history of the technology development; p. 67. [Google Scholar]
- 8. ttps://www.ibm.com/blogs/watson/2017/03/reaching-new-records-in-speech-recognition/ Accessed March 9 2017.
- 9.Prenger RJ, Han T. 2016. Feb, Around the World in 60 Days: Getting Deep Speech to Work in Mandarin. [Google Scholar]
- 10.Eckel R. Microsoft researchers achieve speech recognition milestone. Posted September 13 2016 https://blogs.microsoft.com/next/2016/09/13/microsoft-researchers-achieve-speech-recognition-milestone/#sm.0000ato9goax0f8dziy22h7refoum Accessed: March 9 2017.
- 11.Edwards E, Salloum W, Finley GP, Fone J, Cardiff G, Miller M, Suendermann-Oeft D. Medical speech recognition: reaching parity with humans. International Conference on Speech and Computer; Springer,Cham; 2017. Sep 12, pp. 512–524. [Google Scholar]
- 12.Chiu CC, Tripathi A, Chou K, Co C, Jaitly N, Jaunzeikare D, Kannan A, Nguyen P, Sak H, Sankar A, Tansuwan J. 2017. Nov, Speech recognition for medical conversations. arXiv preprint arXiv: 1711.07274; p. 20. [Google Scholar]
- 13.Center for Medicare and Medicaid Services Department of Health and Human Services Baltimore, MD. 2017. http://www.roadto10.org/specialty-references/clinical-scenarios-family-practice/. Accessed: March 9.
- 14.Suominen H, Zhou L, Hanlen L, Ferraro G. 2. Vol. 3. JMIR medical informatics; 2015. Apr, Benchmarking clinical speech recognition and information extraction: new data, methods, and evaluations. [DOI] [PMC free article] [PubMed] [Google Scholar]