Abstract
Lineage specification is an essential process in stem cell fate, tissue homeostasis and development. Microenvironmental cues provide direct and selective extrinsic signals to regulate lineage specification of stem cells. Microenvironmental milieu consists of two essential components, one being extracellular matrix (ECM) as the substratum, while the other being cell secreted exosomes and growth factors. ECM of differentiated cells modulates phenotypic expression of stem cells, while their exosomes contain phenotype specific instructive factors (miRNA, RNA and proteins) that control stem cell differentiation. This study demonstrates that osteoblasts-derived (Os-Exo) and adipocytes-derived (Ad-Exo) exosomes contain instructive factors that regulate the lineage specification of human mesenchymal stem cells (hMSCs). Analyses of exosomes revealed the presence of transcription factors in the form of RNA and protein for osteoblasts (RUNX2 and OSX) and adipocytes (C/EBPα and PPARγ). In addition, several miRNAs reported to have osteogenic and adipogenic differentiation potentials are also identified in these exosomes. Kinetic and differentiation analyses indicate that both osteoblast and adipocyte exosomes augment ECM-mediated differentiation of hMSCs into the respective lineage. The combination of osteoblast/adipocyte ECM and exosomes turned-on the lineage specific gene expressions at earlier time points of differentiation compared to the respective ECM or exosomes administered individually. Interestingly, the hMSCs differentiated on osteoblast ECM with adipogenic exosomes showed expression of adipogenic lineage genes, while hMSCs differentiated on adipocyte ECM with osteoblast exosomes showed osteogenic lineage genes. Based on these observations, we conclude that exosomes might override the ECM mediated instructive signals during lineage specification of hMSC.
Keywords: Extracellular matrix, Exosomes, lineage determination, stem cell fate, osteogenesis, adipogenesis, microenvironment
1. Introduction
Human mesenchymal stem cells (hMSCs) have the capacity to differentiate into cells of the mesodermal lineage, such as myoblasts, chondrocytes, osteoblasts and adipocytes (1). The stem cell fate is determined by sets of transcription factors and growth factors both in vivo and in vitro (2). Besides the biochemical factors, the lineage commitment of stem cell also depends on physical factors such as cytoskeletal tension and cell shape. For instance, McBeath et al have used micro-patterning method to demonstrate that the cell shape determine the hMSC’s commitment towards either osteogenic or adipogenic lineage (3). The inherent plasticity of hMSCs and their ability to ‘sense or feel’ extracellular matrix (ECM) lead to changes in the signaling cascade, inducing cell differentiation. In this way, cell-type specific ECM provides a unique and complex microenvironment that influences stem cell differentiation. In recent studies, we have demonstrated that ECM extracted from MC3T3-E1 (a mouse pre-osteoblast cell line) induced the hMSCs towards osteogenic lineage (4). Cell-secreted ECM is known to influence stem cells via its multiple physical mechanisms such as rigidity, porosity, nanotopography and mechanotransduction (5). Several studies have shown that the biomechanical property is the key player in the cell-secreted ECM mediated regulation of stem cell fate (6). Stiffness of cell-secreted ECM has also been shown to influence cell behavior, gene expression and the stem cell fate via outside-to-inside signaling mechanism associated mechanotransduction pathways (7–10). Besides these physical attributes, the ECM (in vivo) also contributes to tissue architecture by providing highly organized macromolecules and signaling factors (11). The cells dynamically synthesize a complex network of ECM, degrade and reorganize the macromolecules in a tightly controlled manner (12). The ECM (in vivo) mediated stem cell differentiation is one of the essential processes during tissue development, repair and homeostasis (13). Structural and signaling functions of the ECM (in vivo) have been attributed to structural proteins (mainly collagen), non-collagenous proteins (fibronectin, laminin, etc.,), glycosaminoglycans and proteoglycans (11). The ECM (both in vivo and in vitro) mediated signaling include ligand-cell surface receptor interaction and physical properties associated component that deliver combined influence on stem cell fate determination.
Transplantation of stem cells to the site of injury lead to their differentiation, which is influenced by the factors secreted by specific lineage cells via paracrine signaling (14). In the same context, conditioned medium has been shown to be critical in stem cell differentiation (15). In the absence of growth factors, chondrocyte conditioned medium has been shown to differentiate bone marrow-derived hMSCs into chondrocytes as an evidence for the presence of sufficient activators in the conditioned medium (16). In addition to the soluble factors, encapsulated nano-sized (40 – 100 nm diameter) vesicles called exosomes present in the conditioned medium have been identified as a key player in stem cell differentiation (17). Exosomes are multi-vesicular endosomes (MVE) secreted by many cell types. The cargo of exosomes are shown to contain both ubiquitous and cell-type specific biological molecules such as protein, RNA, long non-coding-RNA (lncRNA), micro RNA (miRNA) and fragmented DNA (18). With the current understanding, the exosomes are recognized as a component of paracrine signaling system and cellular microenvironment (19).
Both miRNA (20) and RNAs (21) have been shown to be critical component in stem cell differentiation. For example, miR-1 and miR-449 regulate cardiomyocyte progenitor differentiation via targeting Sex Determining Region Y-Box 6 (Sox6) (22). Similarly, overexpression of miR-355 has been shown to initiate chondrogenic differentiation in mouse MSCs (23). In addition to miRNA, tissue specific transcription factors have also been shown to determine lineage specification via downstream activation of cell-type specific targets. A fine balance between miRNA and specific transcription factors has been shown as the key determinant of stem cell fate (24–26). Although both cell-secreted ECM and exosomes are shown to influence the stem cell fate in vitro, it is not known whether the ECM or the exosome has the superior role in cell differentiation. We hypothesize that exosomes might have efficient control over ECM in the determination of hMSC lineage. In this report, we studied the influence of tissue specific ECM and exosomes on the differentiation of hMSCs, and thus, their role in hMSC lineage determination.
2. Materials and Methods
2.1. Cell culture
All cells used in this study [normal human osteoblasts (NHO), human subcutaneous pre-adipocytes and human mesenchymal stem cells (hMSCs)], growth medium and differentiation medium were purchased from Lonza (Singapore). Serum containing media was used in the expansion of hMSCs, NHO and pre-adipocytes, while differentiation was performed in serum-free medium. Exosomes were purified from the collected conditioned medium at day-7 of differentiation.
2.2. Mesenchymal stem cells culture and differentiation
All the experiments with hMSCs were performed within five passages of culture. The hMSCs (3 × 104 cells/cm2) were differentiated to osteoblast and adipocyte lineages with osteogenic and adipogenic induction medium, respectively. Prescribed exosomes concentrations were added during the differentiation of hMSCs at day 3, 6, 9 and 12 along with differentiation medium changes.
2.2. Osteoblasts and adipocytes culture and differentiation
Normal human osteoblasts were cultured with osteoblast growth medium (OGM). Differentiation of NHO was performed at 90% confluence with OGM containing hydrocortisone (200 nM) and β-glycerophosphate (10 mM). Briefly, NHO seeded at a density of 2 × 104/cm2 were allowed to differentiate for 7 days. The differentiated osteoblasts were confirmed by gene expression and von Kossa staining. Primary human subcutaneous pre-adipocytes obtained from Lonza were cultured with pre-adipocyte growth medium. Differentiation of pre-adipocytes was performed with adipocyte differentiation medium. Briefly, 2.5 × 104 cells/cm2 were seeded and allowed to differentiate in adipocyte differentiation medium containing insulin, dexamethasone, indomethacin and isobutyl-methylxanthanine for 7 days. Differentiation was confirmed by gene expression and lipid droplets staining with Nile red (Sigma, USA).
2.3. Exosome isolation
Exosomes were isolated from differentiated NHO and pre-adipocytes. On day 6 of osteoblast and adipocyte differentiation, the medium was changed, and the conditioned medium was collected on day 7. The conditioned medium was centrifuged sequentially at 300 × g for 5 min and at 20,000 × g for 20 min to remove cells and smaller cell debris, respectively. The collected supernatant was passed through polyethersulfone membrane filter (0.22 µm; Corning, USA) and centrifuged at 100,000 × g for 60 min. Exosome pellets were rinsed with PBS and re-centrifuged (100,000 × g for 60 min). Exosome pellets suspended in sterile PBS were stored at 4ºC until further use.
2.4. Characterization of exosomes
The exosome size and concentration were determined using NanoSight (NS300, Malvern Instruments, UK). Exosomes resuspended in PBS were observed with blue laser (405 nm) and their movement under Brownian motion was captured for 60 sec. NanoSight was used with a standard detection threshold of 3 and camera level set at 14 for all the experiments. Exosome concentration and size distribution profiles were determined by analyzing the captured video using NanoSight particle tracking software. All measurements were repeated 3 times
2.3. Scanning electron microscopy (SEM)
Exosomes suspended in 100 µl PBS were fixed with 2.5% glutaraldehyde dissolved in 0.1 M cacodylate solution (pH 7.0) for 2 hr followed by 2% osmium tetroxide for one hr. Diluted samples were added to cleaned silicon chips and treated with acetone, ethanol and distilled water. Immobilized and dried silicon chips were imaged by JEOL-7600F SEM.
2.4. Western blotting
Proteins extracted from exosomes and cells resolved in 10% SDS-PAGE were transferred onto nitrocellulose membrane using Novex transfer buffer (ThermoFisher, USA). The blots were blocked with 5% BSA dissolved in TBS for one hr. Specific primary antibodies were added to the blots and incubated for 16 hr at 4°C. The blots were washed with TBS containing 0.5% triton-X100 for 10 min (3X) and incubated with appropriate secondary antibody conjugated with horse radish peroxidase. The blots were developed using ECL kit (GE Healthcare, USA).
2.5. Extracellular matrix preparation
The cell-secreted ECMs were isolated as described previously (27). Briefly, the cells were grown for 7 days in the respective differentiation medium. ECM was prepared by incubating the cells with 0.02 M ammonium hydroxide for 5 min and the cell debris and genomic DNA were aspirated. The ECM was washed gently twice with sterile ice-cold PBS. The ECM containing plates were either used immediately or stored in PBS at 4ºC.
2.6. Characterization of extracellular matrix
The concentration of Type-I (ab210966) and Type-IV (ab6586) collagen, fibronectin (ab219046) and, laminin (ab119599) in the isolated ECM were quantitated using sandwich enzyme-linked immunosorbent assay (ELISA; Abcam, UK) using manufacturer’s instruction. Values are presented as mean ± SD of triplicate assays of three independent measurements. The structural and mechanical properties of the ECM were determined using atomic force microscope (AFM; Veeco Instruments, USA), as described previously (27).
2.7. Exosome labeling and uptake studies
Purified exosomes were labeled with membrane targeted green fluorescent dye PKH67 (Sigma-Aldrich, St. Louis, MO). In brief, 1 × 107 exosomes suspended in 1 ml PBS mixed with equal volume of PKH67 dye were incubated at 4ºC for 5 min. The labeling reaction was stopped by the addition of 5% BSA in PBS. The labeled exosomes purified by centrifugation (100, 000 x g for 60 min) were resuspended in PBS. For uptake studies, 1 × 107 labeled exosomes suspended in 750 µl serum free culture medium was added to pre-plated hMSCs (at least 24 h). Control group received equivalent concentration of free-dye in the medium. To block exosome uptake, hMSCs were incubated together with exosomes and chlorpromazine (Sigma, USA). Following 24 h incubation at 37ºC, exosome uptake by hMSCs was visualized with fluorescence microscope.
2.8. RNA isolation and quantitative polymerase chain reaction
Total RNA (10 µg) isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA) was reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed with gene specific TaqMan assays (Applied Biosystems, USA) (Supplementary Table-S1). Number of specific RNA copies were calculated using standard curve of known concentrations of respective genes.
2.9. Promoter constructs and luciferase activity measurement
Both an 800 bp OC and 600 bp ADPN promoters were PCR amplified from genomic DNA isolated from hMSCs. The OC promoter was cloned into KpnI/XhoI site (28), while the ADPN promoter was cloned into pGL3-basic at SacI/BglII site (29) in pGL3-basic vector (Promega, USA). The promoter clones were sequence verified prior to use. Plasmid DNA (2.5 µg) mixed with 1 µg enhancer and 1 µl of Lipofectamine 3000 (ThermoFisher, USA) was complexed by incubating at room temperature for 5 min. The DNA-Lipofectamine complex mixed with 500 µl of DMEM containing 5% serum was added dropwise to cells. Control cells received empty vector (i.e. without promoter insert). Following 24 h transfection, cellular extracts were prepared using lysis buffer and used for luciferase activity. Luciferase assay was performed as described by the manufacturer (Promega, USA). In brief, luminescence was measured in 50 µl cell lysate mixed with 100 µl luciferase substrate using GloMax luminometer (Promega, USA). The luciferase activities presented were normalized to protein concentration.
2.10. Quantitative measurement of calcium deposition and lipid formation
Osteogenic differentiated hMSCs were fixed and stained with von Kossa stain for calcium deposition (ab150687, Abcam, UK). For quantitative calcium measurement, cells were washed with PBS and the cellular calcium was extracted by decalcification using with 0.6 N HCl for 18 h at room temperature. The extracted cellular calcium was quantitated using QuantiChrom Calcium Assay Kit by following the manufacturer’s instruction (BioAssay Systems, USA). In brief, equal volume (200 µl) of reagent A and B was added to cell extract and incubated at room temperature for 5 min. Absorbance at 612 nm was measured and normalized to protein.
For quantitative measurement of lipid droplet formation during adipogenic differentiation, we used Nile Red (Sigma, USA) as a fluorescent lipid staining dye. Upon differentiation, cells were fixed with paraformaldehyde and stained with Nile Red. Fluorescent intensity of Nile Red stained cells was measured with a plate reader and normalized with total protein.
2.11. siRNA mediated knockdown
Exosomal RUNX2 (assay id # 115507) and PPARγ (assay id # 5636) specific RNA expressions were knockdown using specific siRNA kit (Applied Biosystems, USA). Eight hr prior to exosome supplementation, the cells were transfected with siRNA. One µl siRNA (5µM) mixed with 9 µl of serum-free DMEM was incubated with 10 µl DharmaFECT transfection reagent for 30 min at room temperature. The siRNA-DharmaFECT complex mixed with 80 µl of DMEM containing 10% FBS was added dropwise to the cells. Following 15 days differentiation, OC and ADPN promoter plasmids were transfected to hMSCs undergoing osteogenic and adipogenic differentiation. Luciferase assay was performed with the cell lysates as described in section 2.9.
2.12. In-vitro translation
Five µg of RNA in 25 µl of nuclease-free water wastranslated with rectic lysate (ThermoFisher, USA) at 30°C for 90 min following manufacturer instructions. The in-vitro translated proteins were resolved in a 10% SDS-PAGE and western blotting was performed with specific antibodies as described in section 2.4.
2.13. Statistical analysis
All data presented represent mean ± SD of triplicate assays. One-way analysis of variance (ANOVA) was used to determine the statistical significance (p ≤ 0.05.)
3. Results
3.1. Exosome isolation and characterization
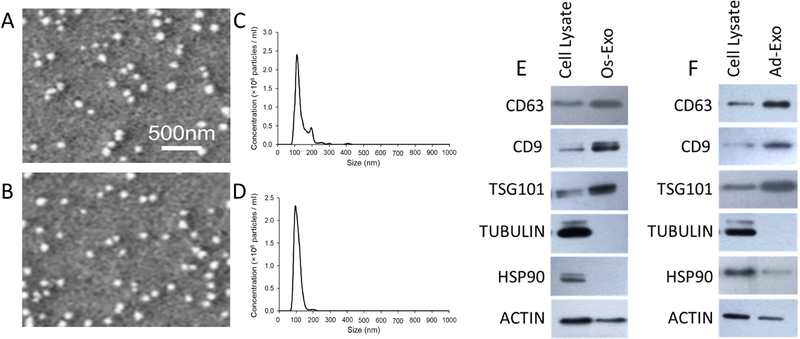
Normal human osteoblasts (NHO) and human pre-adipocytes were differentiated using conventional osteogenic and adipogenic differentiation media, respectively. The NHO differentiation was confirmed by von Kossa staining and osteocalcin (OC), Runt related transcription factor 2 (RUNX2), osterix (OSX) and osteopontin (OPN) specific gene expression (Supplementary Figure 1A and 1B), while the pre-adipocyte differentiation was confirmed by Nile Red staining and CAAT /enhancer binding protein alpha (CEBPα), lipoprotein lipase (LPL), adiponectin (ADPN) and peroxisome proliferation activator receptor gamma (PPARγ) gene expressions (Supplementary Figure 1C and 1D). Exosomes were purified from conditioned media collected from osteogenic and adipogenic differentiated cells after 7 days of differentiation. SEM analyses of osteoblastic and adipocytic exosomes revealed the presence of globular structures with a diameter in the range of 70 to 100 nm (Figures 1A and 1B). Homogeneity and size of the purified exosomes were characterized by NanoSight using Nanoparticle tracking software (NTA). Average size of osteoblast exosomes ranges from 75 to 112 nm, while adipocyte exosomes ranges from 63 to 95 nm. NanoSight images (data not shown) and the presence of single peak size distribution profile indicate that the nano-sized vesicles isolated from the conditioned media of both differentiated osteoblasts (Figures 1A and 1C) and differentiated adipocytes (Figures 1B and 1D) are highly purified (Figures 1A and 1B). Western blot analyses indicate the presence of ubiquitous exosome markers such as CD63, CD9 and TSG101 (Tumor Susceptibility Gene 101) proteins that are enriched in both osteoblast (Figure 1E) and adipocyte (Figure 1F) exosomes compared to respective cell lysates. On the other hand, tubulin and Heat Shock Protein 90 (HSP90) which belong to cytosolic fraction are depleted in the purified exosomes.
Figure 1: Characterization of exosomes.
Physical parameters of the purified exosomes were analyzed by SEM and NanoSight. Three separate experimental runs were performed for osteoblast and adipocyte derived exosomes. Representative SEM images are presented for osteoblast (A) and adipocyte (B) exosomes. Average size distribution profile of osteoblast (C) and adipocyte (D) exosomes were obtained from the three runs. Cell lysate and exosomes of osteoblast (E) and adipocyte (F) were scrutinized by western blot with specific antibodies.
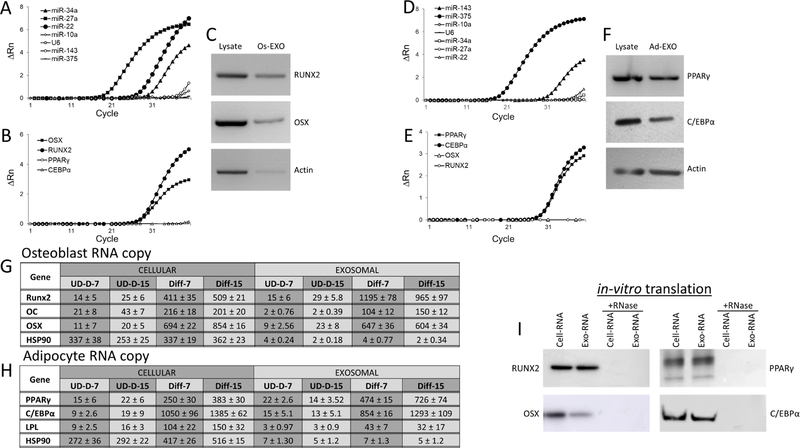
Exosomes are known to contain cell-type specific macromolecules such as miRNA, RNA and proteins. Thus, we investigated the presence of specific miRNAs and RNAs in the purified osteoblast exosomes by RT-qPCR analyses. The fluorescence amplification of targets were normalized with baseline and plotted as deltaRN (∆Rn) against the number of amplification cycles. The miRNAs (miR-34a, miR-27a and miR-22) that influence osteoblast differentiation (Figure 2A), and RUNX2 and OSX specific RNAs (Figure: 2B) were detected, whereas miR-10a, U6 snRNA, miR-143 and miR-375 (negative control) (Figure 2A) were not detected in the osteoblastic exosomes. Similarly, we were not able to detect the presence of adipocyte specific mRNA such as PPARγ and C/EBPα in the osteoblastic exosomes. In addition, RUNX2 and OSX proteins were also detected by western blot in both osteoblast cell lysates and exosomes (Figure 2C). Similarly, miRNAs influencing adipocyte differentiation (miR-143 and miR-375) (Figure 2D) and RNAs (PPARγ and C/EBPα) (Figure 2E) were detected, while miR-10a and U6 snRNA (used as negative control) (Figure 2D) were not detected in adipocyte exosomes. Western blot analysis identified PPARγ and C/EBPα protein expression in the purified adipocyte exosomes (Figure 2F). The copy number of specific mRNAs were estimated in both cell and exosomes of osteoblast and adipocyte. The osteoblasts and its exosomal RUNX2, OC, OSX and HSP90 specific transcript (i.e. mRNA) copies were determined using RT-qPCR analyses using a standard curve of known concentrations of respective genes (Figure 2G). Similarly, in adipocytes and its exosomes PPARγ, C/EBPα, LPL and HSP90 specific mRNA copies were determined (Figure 2H). Copy number analyses indicate that lineage regulating transcripts are present comparatively at higher concentrations both in cellular and exosomal contents compared to control cells during the differentiation process. At day-7 of NHO differentiation, exosomes contain 2.9 and 0.9-fold of RUNX2 and OSX transcript copies compared to cellular RNA respectively (Figure 2G). Similarly, at day-7 of adipocyte differentiation, 1.8 and 0.8-fold of PPARγ and C/EBPα transcript copies compared to cellular RNA respectively (Figure 2H). Interestingly OC and LPL RNA copies in exosomes are present at lower concentrations (0.4-fold) compared to cellular level of differentiated osteoblasts and adipocytes respectively (Figure 2G and 2H). Further, to identify whether these mRNAs have the ability to translate into respective proteins, western blot analyses of specific proteins were performed using in-vitro cell-free translation system. As shown in Figure-2I, RUNX2 and OSX proteins were detected in osteoblasts and its exosomes, while adipocytes and its exosomes exhibited the presence of PPARγ and C/EBPα proteins during in vitro translation. In contrary, Absence of these proteins in in vitro translation performed in the presence of RNase indicate these proteins are translated from the mRNA (Figure 2I).
Figure 2: Characterization of cargo of exosomes.
Osteogenic influencing miRNAs (A) and RNAs (B) abundance, and protein (RUNX2, OSX and actin) expression (C) in the purified osteoblast exosomes. Adipogenic influencing miRNAs (D) and RNA (E) abundance, and protein (PPARγ, C/EBPα and actin) expression (F) in the purified adipocyte exosomes. The miRNA and RNA expressions presented represent normalized fluorescent signals (∆Rn) plotted against the number of amplification cycles (A, B, D and E). RUNX2, OSX, PPARγ, C/EBPα and actin expressions were detected by western blot analyses using specific antibodies in respective cell lysates and purified exosomes (Os-Exo/Ad-Exo) (C and F). RUNX2, OC, OSX, PPARγ, C/EBPα, LPL and HSP90 expressions were quantitated by RT-qPCR using gene specific primers. The RNA copy numbers of osteoblasts (G) and adipocytes (H) were estimated using RT-qPCR with known concentrations of respective plasmid DNA. RNAs were isolated from undifferentiated (UD) and differentiated (Diff) at day-7 and day-15 of culture. The copy numbers are presented as mean ± S.D. The RNAs isolated from osteoblast, adipocytes and their respective exosomes were in vitro translated to proteins using cell-free translation system in the presence and absence of RNase. The presence of RUNX2, OSX, PPARγ and C/EBPα were identified by western blot analysis with specific antibodies (I).
3.2. Extracellular matrix isolation and characterization
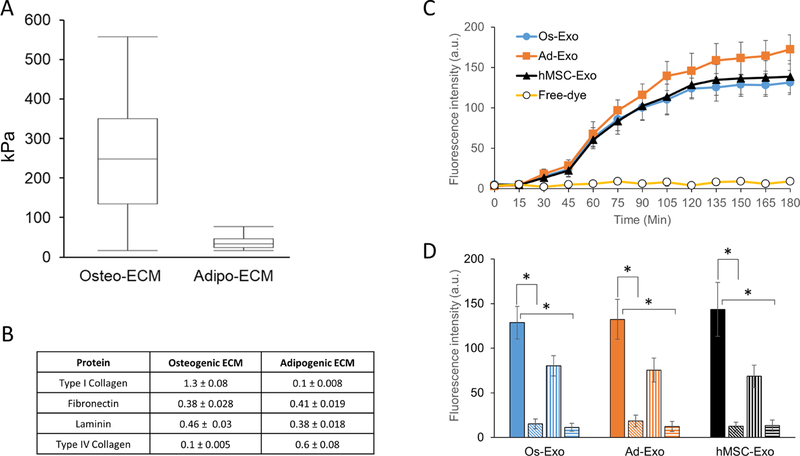
The differentiated osteoblast and adipocyte ECMs were isolated and characterized by atomic force microscope (AFM) and enzyme linked immunosorbent assay (ELISA) techniques. The AFM was used to measure the Young’s modulus of the cell-secreted ECM. Stiffness of the ECM was measured as Young’s modulus (kPa), briefly, 10 randomly selected regions of 50 × 50 µm were scanned to deduce force curve with Hertz model. The Young’s modulus estimated to be 251 ± 174 kPa and 27 ± 18 kPa for osteoblasts and adipocytes, respectively (Figure 3A). The larger error bar noticed in Young’s modulus of osteoblast might be attributed to the localized mineralization on the ECM. The cell-secreted ECM was immunostained with laminin, Type-I and Type-IV collagen specific antibodies. Presence of these proteins were established in the osteoblast and adipocyte derived ECM (Supplementary Figure 2). The soluble ECM proteins were extracted and quantitated by ELISA. Three different extractions were analyzed for Type I collagen, fibronectin, laminin and Type IV collagen content. Type I collagen and Type IV collagen content were higher in osteoblast ECM and adipocyte ECM respectively. Interestingly, the fibronectin and laminin levels were not significantly different between osteoblast and adipocyte ECMs (Figure 3B).
Figure 3: Characterization of extracellular matrix and exosome internalization.
Normal human osteoblasts (NHO) and pre-adipocytes were differentiated to either osteoblasts or adipocytes on glass coverslips, as described in methods. The lysed cells were aspirated and the stiffness of the selected area (50 µm x 50 µm) deposited extracellular matrix (ECM) was examined under atomic force microscope (AFM). Young’s modulus (A) presented as mean ± S.D. for osteoblast and adipocyte ECM were calculated from 10 randomly selected regions using Hertz model. The extracted ECM was quantitated for specific proteins by ELISA. Type I collagen, fibronectin, laminin and Type IV collagen were measured for three independent extractions and presented as mean ± S.D. (B). Adhered human mesenchymal stem cells (hMSCs) were incubated with either labeled exosomes or free-dye for 24 h, as described in Materials and Methods section. Following incubation, the cells were washed with PBS, fixed with paraformaldehyde and fluorescence intensity was measured in a plate reader. Exosome uptake is presented as mean ± S.D. of fluorescence intensity of osteoblast (Os-Exo), adipocyte (Ad-Exo) and hMSC (hMSC-Exo). The hMSCs were supplemented with labeled exosomes for 2 h either at 37°C (solid bar) in the presence of 5µM (vertical lines bars) and 10µM (horizontal lines bars) of chlorpromazine. While incubation of hMSCs with exosomes at 4°C (slanted line bars) inhibited the uptake of exosomes. One-way ANOVA was used to evaluate statistical significance between groups (*p ≤ 0.005).
3.3. Exosome uptake by hMSCs
To identify whether the purified exosomes are taken up by the hMSCs, the hMSCs were incubated with fluorescence-labeled exosomes. Following 24 h incubation, the fixed cells were either visualized with fluorescent microscope or quantitatively measured using a plate reader. As shown in Supplementary Figure 4, both osteoblast and adipocyte exosomes were actively adsorbed and internalized by the hMSCs. At higher magnification, typical granular pattern of labeled exosomes were localized in the perinuclear region of hMSCs (Supplementary Figure 4). Quantitative measurement studies suggest that exosome internalization is aided at 37°C but not at 4°C; further, the fluorescence was not observed in hMSCs incubated with free dye (Figure 3C). The presence of exosome uptake by hMSCs at 37°C, but not at 4°C indicates that the exosome uptake in hMSCs are mediated by an active, but not a passive diffusion process. Since active internalization of exosomes occur through both clathrin-dependent and clathrin-independent pathways, the effect of chlorpromazine, an inhibitor of both clathrin-dependent and clathrin-independent exosome internalization (30, 31), was examined on exosome uptake in hMSCs. As shown in Figure 3D, significant inhibition (p<0.005) by chlorpromazine (5 and 10 µM) establishes that the exosome uptake is mediated by an active process in hMSCs.
3.4. Effect of exosome concentration on hMSCs differentiation on osteoblast and adipocyte ECM
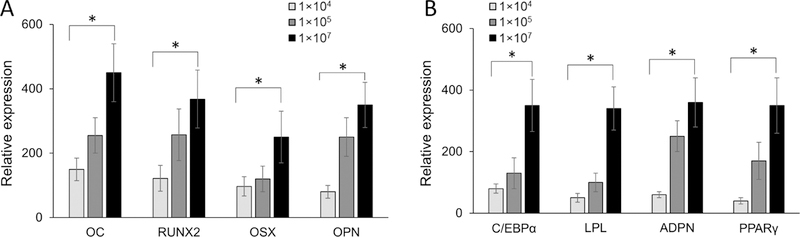
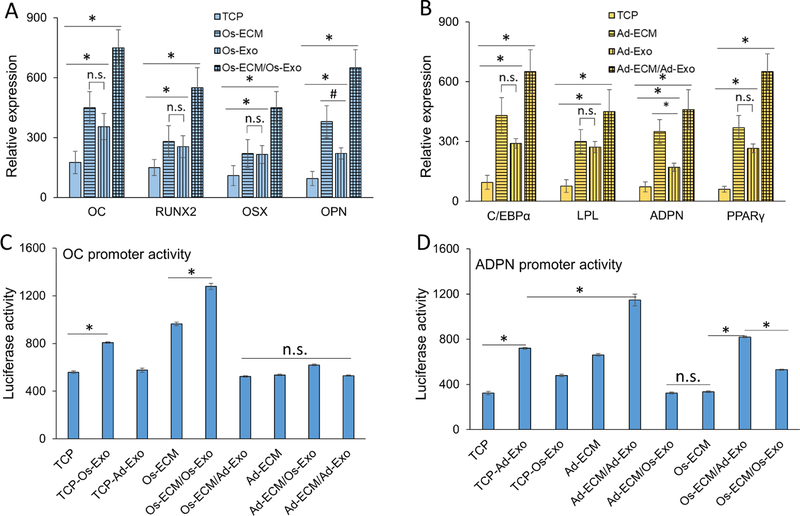
To determine the specificity of exosome effect on hMSCs differentiation, the effect of increasing exosome concentrations on hMSCs differentiated on either osteogenic or adipogenic ECM were examined. In this study, hMSCs differentiated on either osteogenic or adipogenic ECM were supplemented with different concentrations (1 × 104. 1 × 105 and 1 × 107 exosomes/ml) of the respective exosomes. For both osteogenicity and adipogenicity, we demonstrated that increasing respective exosome concentrations correspondingly increase the respective gene expression during hMSCs differentiation (Figures 4A and 4B). Increasing osteoblast exosome concentrations increased the Osteocalcin (OC) and Runt related transcription factor 2 (RUNX2) expression by 3-fold, while it increased the Osterix (OSX) and osteopontin (OPN) expression by 4.3-fold in osteogenic differentiation, (Figure 4A). Similarly, in adipogenic differentiation, increasing adipocyte exosome concentrations increased the expression of CAAT/enhancer binding protein alpha (C/EBPα) and peroxisome proliferation activator receptor gamma (PPARγ) by 4.3-fold and 8.7-fold, respectively (Figure 4B).
Figure 4: Effect of exosome concentrations on osteogenic and adipogenic gene expression in human mesenchymal stem cells.
Human mesenchymal stem cells (hMSC) were differentiated on either osteoblast (A) or adipocyte (B) extracellular matrix (ECM). The differentiation on ECM was further supplemented with different concentrations of respective exosomes. After 15 days of differentiation, the osteogenic [OC, RUNX2, OSX and OPN) and adipogenic (C/EBPα, LPL, ADPN and PPARγ) specific gene expressions were quantitated by RT-qPCR analyses using specific primers. Student t-test was used to evaluate the statistical significance (*p ≤ 0.005).
3.5. Effect of ECM and exosomes on hMSC differentiation
The ECM and exosomes were derived from normal human osteoblasts and human pre-adipocytes after 7-day differentiation in their respective differentiation medium. We investigated the role of ECM and exosomes on osteogenic and adipogenic differentiation of hMSCs. The hMSCs were differentiated either on tissue culture plate (TCP) or ECM [osteoblast ECM (Os-ECM) and adipocyte ECM (Ad-ECM)] coated plates. The differentiation was induced in the absence or in the presence of exosomes. The hMSCs were allowed to undergo osteogenic and adipogenic differentiation for 15 days in the respective differentiation medium. Exosomes were added at a concentration of 1 × 107/ml at day 3, 6, 9 and 12 during routine medium change. Gene expression analyses were performed to determine the effect of ECM and exosomes for osteogenic and adipogenic differentiation. We calculated the fold expression changes of the respective genes by considering their expression in undifferentiated hMSCs as 1 fold. The expression of OC, RUNX2, OSX and OPN were increased in osteogenic differentiation of hMSCs on TCP. The expression level of these genes was increased to a similar extent when the hMSCs were differentiated with either osteoblast ECM or osteoblast exosomes (Figure 5A). Osteoblast exosomes do not contain RNAs for OC and OPN, hence their expressions serve as an indicator of osteogenic differentiation. Interestingly, the hMSCs differentiated on osteoblast ECM supplemented with osteoblast exosomes (Os-ECM/Os-Exo) exhibited highest expression of the osteogenic genes. The key osteogenic transcription factors (RUNX2 and OSX) expressions in hMSCs differentiated on Os-ECM/Os-Exo group were increased more than 3-fold compared to that of hMSCs differentiated on TCP (Figure 5A; Supplementary Figure 5). Similarly, adipogenic differentiation of hMSCs in TCP exhibited increased expression of adipogenic genes C/EBPα, LPL, PPARγ and adiponectin (ADPN). These adipogenic genes were expressed at the highest (3-fold) level in hMSCs differentiated on adipocyte ECM supplemented with adipocyte exosomes (Ad-ECM/Ad-Exo) compared to that of hMSCs differentiated on TCP (Figure 5B; Supplementary Figure 5). Adipocyte exosomes do not contain RNAs for LPL and ADPN, hence, we used their expression levels as an indicator of adipogenic differentiation. Further, lineage specific promoter reporter constructs for osteoblasts (OC promoter) and adipocytes (ADPN promoter) were assessed during osteogenic and adipogenic differentiation at different conditions. Results indicate that both Os-ECM and Os-Exo significantly increased (1.3 to 1.5-fold, p < 0.05) OC activity compared to TCP. On the other hand, combination of Os-ECM and Os-Exo (Os-ECM/Os-Exo) activated the OC promoter to the maximum (2-fold, p < 0.05). Interestingly, addition of Ad-ECM or Ad-Exo did not significantly alter the OC promoter activity (Figure 5C). Similarly, ADPN promoter activity was investigated during adipogenic differentiation at different conditions and the results indicate that both Ad-ECM and Ad-Exo increased ADPN promoter activity (1.7 to 1.6-fold, p<0.05) compared to TCP. On the other hand, combination of Ad-ECM and Ad-Exo (Ad-ECM/Ad-Exo) activated the AADPN promoter to the maximum (3-fold, p<0.05). Interestingly, addition of osteogenic ECM or exosomes did not significantly alter the ADPN promoter activity (Figure 5D).
Figure 5: Effect of extracellular matrix and exosomes on gene expression in human mesenchymal stem cell differentiation.
Human mesenchymal stem cells (hMSCs) were differentiated in either tissue culture plate (TCP, open bars) or cell type specific extracellular matrix (ECM, vertical line bars). The hMSCs differentiated in TCP (TCP/Exo, horizontal line bars) and ECM (ECM/Exo, checkered bars) were further differentiated in the presence of either osteoblast exosomes (A) or adipocyte exosomes (B). Relative gene expressions presented represent fold changes with respect to undifferentiated hMSCs. Osteogenic (OC, RUNX2, OSX and OPN) and adipogenic (C/EBPα, LPL, ADPN and PPARγ) genes were quantitated by RT-qPCR using gene specific primers. The OC (C) and ADPN (D) promoter activities were assessed during differentiation of hMSCs into osteoblast and adipocytes at different conditions, respectively. The hMSCs grown on either TCP or ECM were supplemented with exosomes as described in Materials and Methods section. The OC and ADPN promoter constructs were transfected individually into the hMSCs on day-15 of differentiation towards osteogenic and adipogenic lineage, respectively. Following 24 h transfection, luciferase activity was measured in the cell lysates. One-way ANOVA was used to evaluate statistical significance between groups (*p ≤ 0.005).
3.6. Evaluation of mineralization (osteogenic index) and lipid formation (adipogenic index) in differentiated hMSCs
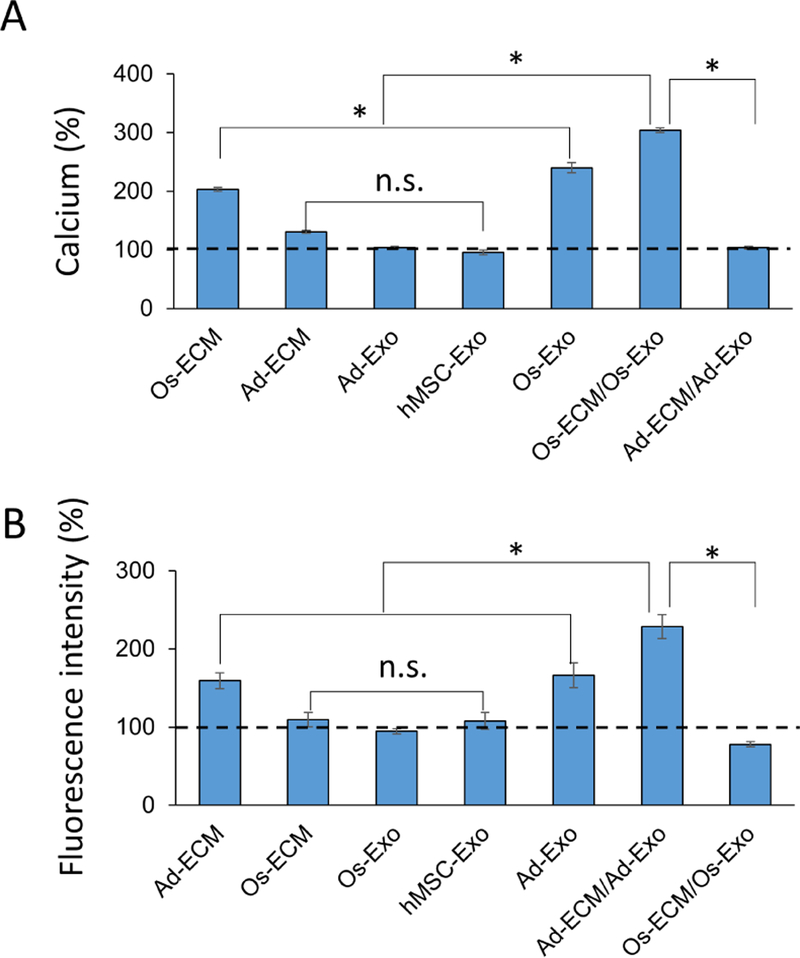
Osteoblast and adipocyte differentiation of hMSCs under different conditions (TCP, ECM, TCP/Exo and ECM/Exo) is described in section-3.4. To functionally distinguish osteoblast and adipocyte differentiation, mineralization (calcium) (osteogenic index) and lipid droplets (adipogenic index) were quantitated, respectively. Compared to TCP, the calcium levels were increased by 2.5-fold and 2.3 fold in hMSCs differentiated on osteoblast derived ECM (Os-ECM) and exosome (Os-Exo), respectively (Figure 6A). The highest calcium level (i.e., 4-fold) was measured in hMSCs differentiated on osteoblast ECM supplemented with osteoblast exosomes (Os-ECM/Os-Exo) (Figure 6A). Interestingly, addition of Ad-Exo to the hMSCs undergoing osteogenic differentiation in TCP showed a decrease in calcium deposition (25% lower, p<0.05) (Figure 6A).
Figure 6: Effect of cell specific exosomes on calcium deposition (osteogenesis) and lipid droplets (adipogenesis) on human mesenchymal stem cells.
The human mesenchymal stem cells (hMSCs) were differentiated on either tissue culture plate (TCP) or extracellular matrix (ECM). During osteogenic differentiation on osteoblast ECM (Os-ECM), the cells were also supplemented and differentiated in the presence of osteoblast exosomes (Os-ECM/Os-Exo). The deposited calcium was quantitated using calcium assay kit, normalized to protein and presented as percentage with levels on TCP as 100% (dotted line) (A). During adipogenic differentiation on adipocyte ECM (Ad-ECM), the cells were also supplemented and differentiated in the presence of adipocyte exosomes (Ad-ECM/Ad-Exo). The fluorescent intensity of the lipid droplets was measured at 505 nm, normalized to protein and presented as percentage with levels on TCP as 100% (dotted line) (B). One-way ANOVA was used to evaluate the statistical significance (*p ≤ 0.005).
Lipid droplets formed during adipogenic differentiation of hMSCs was visualized by fluorescent (Nile Red) staining. The labeled lipid droplets were quantitated by normalizing to total protein. As shown in Figure 6B, lipid droplets levels were increased by 1.3-fold and 1.2-fold in hMSCs differentiated on adipocyte ECM (Ad-ECM) and adipocyte exosomes (Ad-Exo) compared to hMSCs differentiated on TCP. The lipid droplet levels were observed to be highest (i.e., 2.1-fold) on hMSCs differentiated in adipocyte ECM supplemented with adipocyte exosomes (Ad-ECM/Ad-Exo) (Figures 6B) compared to hMSCs differentiated on TCP. Interestingly, addition of Os-Exo to the hMSCs undergoing adipogenic differentiation in TCP, showed decreased lipid staining (20% lower, p<0.05) (Figure 6B).
3.7. Exosomes speed-up differentiation
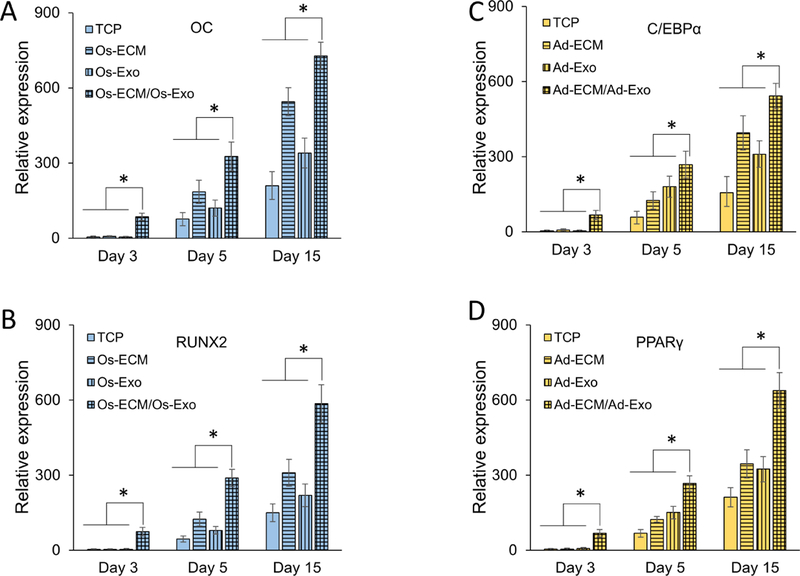
Studies were performed to investigate whether exosomes alter the kinetics of hMSC differentiation. In this study, the key lineage specific gene expressions were quantitated on the 4th, 6th, and 15th day of differentiation in the absence or presence of exosomes on TCP and ECM (i.e., TCP, TCP/Exo, ECM and ECM/Exo). As mentioned in methods section, exosomes were added on days 3, 5, 9 and 12 of differentiation. As shown in Figures 7A and 7B, within a day after osteoblast exosome supplementation, the OC and RUNX2 specific gene expressions were detected to increase by 18-fold and 10-fold in hMSCs differentiated on osteoblast ECM (i.e., Os-ECM/Os-Exo) compared to hMSCs differentiated on TCP, respectively. Although the OC and RUNX2 gene expressions were detected to increase progressively in all groups, their expressions were significantly higher in Os-ECM/Os-Exo group (Figures 7A and 7B). It is to be noted that in contrast to day-1 (10 – 18 fold), the magnitude of OC and RUNX2 gene expressions were diminished to 2 – 3 fold at 3rd and 12th day after exosome supplementation (Figures 7A and 7B; Supplementary Figure 3A).
Figure 7: Effect of cell specific exosomes on the kinetics of human mesenchymal stem cell differentiation.
Human mesenchymal stem cell (hMSCs) were differentiated on either tissue culture plate (TCP) (solid bars) or extracellular matrix (ECM; vertical line bars). Exosomes were supplemented during differentiation (TCP/Exo; horizontal line bars) on ECM (ECM/Exo; checkered bars). Following 3, 5 and 15 days of induction, the osteogenic [OC (A) and RUNX2 (B)] and adipogenic [C/EBPα (C) and PPARγ (D)] gene expressions were quantitated by RT-qPCR analyses using specific primers. One-way ANOVA was used to evaluate the statistical significance (*p ≤ 0.005).
Similar to osteogenic differentiation, C/EBPα and PPARγ expressions were increased in hMSCs differentiated on adipocyte ECM supplemented with adipocyte exosomes (i.e., Ad-ECM/Ad-Exo) (Figures 7C and 7D). Following 1-day after adipocyte exosome supplementation, the C/EBPα and PPARγ expressions were increased by 10-fold and 20-fold in hMSCs differentiated on adipocyte ECM (Ad-ECM/Ad-Exo) compared to hMSCs differentiated on TCP, respectively (Figures 7C and 7D). Although the expression levels of both C/EBPα and PPARγ were progressively increased in all groups, the difference in their expression level are diminished at 3rd and 12th day after exosome addition (Figures 7C and 7D; Supplementary Figure 3B).
3.8. Exosomes override the ECM mediated lineage determination
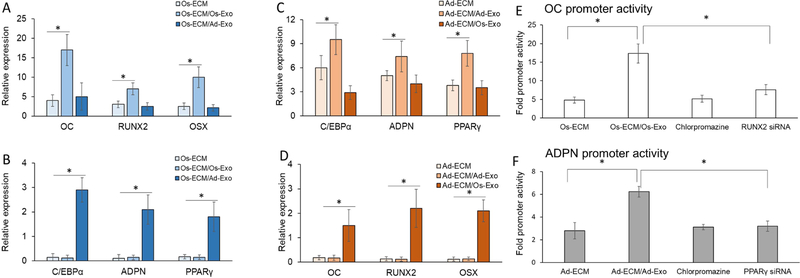
We addressed the key question of whether exosomes control the lineage determination in hMSCs. In this study, hMSCs differentiated on osteoblast ECM (Os-ECM) were supplemented with either osteoblast (Os-EMC/Os-Exo) or adipocyte exosomes (Os-ECM/Ad-Exo), while hMSCs differentiated on adipocyte ECM were supplemented with either osteoblast (Ad-EMC/Os-Exo) or adipocyte (AD-EMC/Ad-Exo) exosomes (Figure 8). Both osteogenic (OC, RUNX2 and OSX) and adipogenic (C/EBPα, ADPN and PPARγ) gene expressions were quantitated in all the conditions. All these experiments were performed using basal medium in the absence of osteogenic and adipogenic factors and other supplements. The expression levels in undifferentiated hMSCs were taken as 1-fold for calculations. In osteogenic lineage determination, osteoblast ECM (Os-ECM) increased the osteogenic gene expressions (OC, RUNX2 and OSX), which was further increased by osteoblast exosomes (Os-EXO) by 2 to 3-fold (Figure 8A; Supplementary Figure 5). In contrast, addition of adipocyte exosomes did not alter the osteogenic gene expressions in hMSCs differentiated on Os-ECM (Figure 8A). In contrast to osteogenic gene expression, the adipogenic gene expressions were very low in hMSCs differentiated on Os-ECM. However, although Os-Exo did not significantly alter, Ad-Exo increased the adipogenic gene expressions by 12 – 15 fold (Figure-8B; Supplementary Figure 5).
Figure 8: Effect of cell specific exosomes on extracellular matrix directed human mesenchymal stem cell lineage.
Human mesenchymal stem cells (hMSCs) were differentiated on either osteoblast (Os-ECM) or adipocyte (Ad-ECM) extracellular matrix (ECM). During osteogenic differentiation on Os-ECM the cells were supplemented with either osteoblast exosomes [Os-ECM/Os-Exo; (A)] or adipocyte exosomes [Os-ECM/Ad-Exo (B)], while during adipogenic differentiation on Ad-ECM the cells were supplemented with either osteoblast exosomes [Ad-ECM/Os-Exo (C)] or adipocyte exosomes [Ad-ECM/Ad-Exo (B)]. After 15 days of differentiation the osteogenic [OC, RUNX2 and OSX) and adipogenic (C/EBPα, ADPN and PPARγ) specific gene expressions were quantitated by RT-qPCR analyses using specific primers. The OC (E) and ADPN (F) promoter activities were assessed during the differentiation of hMSCs to osteoblast or adipocyte on either Os-ECM or Ad-ECM with respective exosomes. The promoter activities were also measured in the presence of 10 µM chlorpromazine supplemented with exosomes. The siRNAs targeting RUNX2 and PPARγ were transfected 8 h prior to supplementation of exosomes. One-way Anova was used to evaluate the statistical significance (*p ≤ 0.005).
Analyses of hMSCs differentiated on adipogenic ECM supplemented with Ad-Exo significantly increased, while supplementation of Os-Exo did not alter the adipogenic gene expression (Figure 8C; Supplementary Figure 5). In contrast to adipogenic genes, the osteogenic gene expressions were very low in hMSCs differentiated on Ad-Exo. However, although Ad-Exo did not significantly alter, the Os-Exo supplementation increased the osteogenic gene expressions by 6 – 10-fold in hMSCs differentiated in Ad-ECM (Figure 8D; Supplementary Figure 5).
As shown in Figure 3D, addition of chlorpromazine inhibited the internalization of exosomes in hMSCs undergoing differentiation resulting in reduced OC (60%, p<0.05) and ADPN (70%, p<0.05) promoter activity compared to Os-ECM/Os-Exo. Further, siRNA mediated knockdown of RUNX2 and PPARγ resulted in the reduced OC (50%, p<0.05) and ADPN (52%, p<0.05) promoter activity compared to Ad-ECM/Ad-Exo (Figure 8E and 8F).
4. Discussion
The cellular microenvironment, which includes the cell attachment substrate (i.e., ECM) and cellular milieu are known to dictate lineage specification of stem cells during development and differentiation (32, 33). Exosomes, a constituent of the cellular microenvironment, serve as a mode of intercellular communications during tissue formation and repair. Exosomes carry key morphogens such as Wnt and Hedgehog that are essential for tissue patterning (34–37). However, the beneficial effect of exosomes in regenerative medicine remain unexplored. Although both ECM and exosomes can regulate lineage specification, it is not clear which of these is the more critical factor for the lineage determination of hMSCs. Thus, this study was initiated to outline the role of exosomes in lineage specification of hMSCs. The results presented in the current study demonstrate that exosomes derived from osteoblasts and adipocytes influence the hMSCs differentiation. Our observations suggest that exosomes and ECM act cumulatively in the lineage specification of hMSCs. Gene knock-out studies have demonstrated the importance of the key transcription factors, RUNX2 and OSX in osteoblast differentiation (38, 39). Additionally, miRNAs such as miR-34a (40), miR-27a (41) and miR-22 (42) were reported to influence the osteoblast differentiation in different cell types. The key adipogenic transcription factors C/EBPα and PPARγ (43, 44), and miR-143 (45) and miR-375 (46) were shown to influence adipogenic differentiation. Our observations confirm the presence of miRNAs, RNAs and proteins, in the osteoblast (Os-Exo) and adipocyte (Ad-Exo) derived exosomes that modulate osteogenic and adipogenic differentiation, respectively (Figure 2). Cui et al., have recently demonstrated that exosomes derived from osteoblasts and pre-osteoblasts (MC3T3-E1) promote osteogenic differentiation of bone marrow stromal cells via miRNA perturbations in mice (47). The presence of specific miRNAs and key transcription factors in the exosomal cargoes provide signaling machinery to enhance the differentiation of hMSCs. Thus, the presence of miRNAs, and the key transcription factor specific mRNAs and proteins prompted us to investigate the role of exosomes in the determination of lineage during hMSC differentiation.
Mouse derived exosomes have been shown to deliver their protein cargo to human cells (48). Uptake of exosomes by the recipient cells have been demonstrated to alter transcriptome and signaling activity of the recipient cells and induce phenotypic changes (49–56). Studies by Li et al. (52) and Farahani et al. (49) have demonstrated that the exosome mediated modification of gene expression and transcriptome in the recipient cells. Transfer of mRNA from exosomes to recipient cells have been demonstrated using radio-labeled mRNA (50), exosome targeted luciferase mRNA (51) and CRE-mRNA (54). Zhang et al. have demonstrated that astrocyte derived exosomes containing PTEN targeting miRNAs reduce brain metastasis (56) confirming the delivery of miRNAs via exosomes. Although exosomes have been shown to transfer its cargoes into the cells, the mechanism of cellular delivery of exosomal content is not well understood. In general, the exosomal cargoes could be delivered through either energy-dependent or energy-independent processes. Our observations of cellular exosome uptake at 37°C and reduced uptake at 4°C indicate that exosome internalization is mediated via an energy dependent process (Figure 3D).
In earlier studies, we and others have demonstrated ECM mediated differentiation of stem cells with its physical and biological properties influencing stem cell behavior and lineage determination (4–13, 27, 53, 55). Thus, it is clear that either ECM or exosomes could individually promote the hMSCs differentiation. However, the combinatorial effect of ECM and exosomes in the hMSCs differentiation and lineage determination has never been exploited. Results presented in this study demonstrate that hMSCs differentiated on ECM supplemented with exosomes increased the rate of differentiation by enhancing key transcription factors. This conclusion is supported by the observation that the osteoblast ECM supplemented with osteoblast exosomes increased osteogenic differentiation by enhancing RUNX2 and OSX expression (Figures 5A; Supplementary Figure 5), while adipocyte ECM supplemented with adipocyte exosomes promoted adipogenic differentiation by enhancing PPARγ and CEBPα expression (Figures 5B; Supplementary Figure 5). Thus, it is likely that the cell-type specific ECM provides added influence in lineage commitment of hMSCs. In addition, the exosomal concentration dependent increase of ECM mediated differentiation establishes the combinatorial effect of ECM stimuli and exosomal cargo.
RUNX2 and PPARγ were shown to control the promoter activity and expression of OC and ADPN, respectively. Lineage specific promoter assessment studies with OC or ADPN promoters indicated that exosomes augmented the respective lineage promoters in hMSCs differentiating in the presence of growth factors supplemented medium (Figure 5C and 5D). Further, addition of chlorpromazine inhibited exosome uptake by hMSCs and resulted in the loss of exosome mediated activation of both OC and ADPN promoter activity during differentiation (Figure 8E and 8F). Our present study established that the osteogenic gene expressions are increased when the cells are differentiated on either osteoblast ECM or osteoblast ECM supplemented with osteoblast exosomes, while the adipogenic gene expression are increased in cells that are differentiated on either adipocyte ECM or adipocyte ECM supplemented with adipocyte exosomes. Adipogenic genes are expressed in cells that were differentiated on osteoblast ECM supplemented with adipocyte exosomes (Figure 8B). Similarly, osteogenic gene expressions are turned-on in cells differentiated on adipocyte ECM supplemented with osteoblast exosomes (Figure 8D). These observations indicate that despite being differentiated on a lineage specific ECM, the exosomal bio-active molecule(s) overrides and influences the phenotypic change in hMSCs. Further, the inhibition of RUNX2 and PPARγ proteins by specific siRNA during hMSC differentiation resulted in the loss of exosome mediated activation of respective lineage specific promoters. These results establish conclusion that the exosomal RNAs taken up by the hMSCs are translated by the cellular machineries into functional proteins that are regulating the hMSCs lineage. Recent reports have indicated the presence of exosomes in the ECM of different tissues withstanding the harsh conditions of ECM extraction (57–62). Isolation of ECM without exosomes would further help us to understand the role these vesicles in both regenerative medicine and pathology. We further predict that the ECM will undergo modifications based on the transcriptome of the differentiating cells. However, further studies are required to identify and establish the ECM associated modification(s) that occurs during lineage specific exosomal addition. Results and conclusions presented in this study are based on the in vitro effects of ECM prepared at specified time of differentiation. However, the cell culture and ECM preparations lack the macromolecular crowding effect usually observed in native cell/tissue due to excluded-volume effect (63–65). It may take several weeks for sufficient ECM deposition to simulate macromolecular crowding. In addition, whether the supplementation of exosomes at high concentrations as shown in the present study potentially create an excluded-volume effect, which requires further extensive studies.
5. Conclusions
Current understanding have limited knowledge on the role of specific microenviromental cues in stem cell differentiation. In this study, we have characterized the exosomes isolated from osteoblasts and adipocytes. Several key miRNAs associated with osteogenesis and adipogenesis were detected in the exosomes derived from osteoblasts and adipocytes respectively. Most importantly, key transcription factors involved in osteogenesis (RUNX2 and OSX) and adipogenesis (PPARγ and C/EBPα) were present in the exosomes. The presence of osteoblast/adipocyte specific transcription factors together with miRNAs augmented the ECM mediated differentiation of hMSCs. Results from in vitro translation and lineage specific promoter studies confirmed the translation of exosomal mRNAs into functional proteins that control the fate of hMSCs. siRNA mediated knockdown of RUNX2 and PPARγ further affirmed the importance of exosomal RNA in lineage specific promoter activation. Results presented in this study demonstrated that osteoblast derived exosomes induced osteogenic genes in spite of being presented with adipogenic ECM and vice versa. Taken together, we conclude that the exosomes might have predomination over ECM in the lineage determination of hMSCs.
Supplementary Material
ACKNOWLEDGEMENTS
Funding was provided by the Institute of Bioengineering and Nanotechnology (Biomedical Research Council, Agency for Science, Technology and Research, Singapore). The Authors (PP and BG) acknowledge the Start-Up Grant support from the Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore. VMR was supported by funding from National Institutes of Health grant (NIH/NIDDK RO1DK104791).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Caplan AI. Mesenchymal stem cells. Journal of orthopaedic research 1991;9(5):641–50. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. science 1999;284(5411):143–7. [DOI] [PubMed] [Google Scholar]
- 3.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Developmental cell 2004;6(4):483–95. [DOI] [PubMed] [Google Scholar]
- 4.Narayanan K, Leck K-J, Gao S, Wan AC. Three-dimensional reconstituted extracellular matrix scaffolds for tissue engineering. Biomaterials 2009;30(26):4309–17. [DOI] [PubMed] [Google Scholar]
- 5.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell stem cell 2009;5(1):17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reilly GC, Engler AJ. Intrinsic extracellular matrix properties regulate stem cell differentiation. Journal of biomechanics 2010;43(1):55–62. [DOI] [PubMed] [Google Scholar]
- 7.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nature reviews Molecular cell biology 2011;12(5):308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Developmental cell 2010;19(2):194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development 2010;137(9):1407–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montell DJ. Morphogenetic cell movements: diversity from modular mechanical properties. Science 2008;322(5907):1502–5. [DOI] [PubMed] [Google Scholar]
- 11.Hynes RO. The extracellular matrix: not just pretty fibrils. Science 2009;326(5957):1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nature reviews Molecular cell biology 2007;8(3):221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Midwood KS, Williams LV, Schwarzbauer JE. Tissue repair and the dynamics of the extracellular matrix. The international journal of biochemistry & cell biology 2004;36(6):1031–7. [DOI] [PubMed] [Google Scholar]
- 14.Gnecchi M, Danieli P, Malpasso G, Ciuffreda MC. Paracrine Mechanisms of Mesenchymal Stem Cells in Tissue Repair. In: Gnecchi M, editor. Mesenchymal Stem Cells: Methods and Protocols New York, NY: Springer New York; 2016. p. 123–46. [DOI] [PubMed] [Google Scholar]
- 15.Pawitan JA. Prospect of stem cell conditioned medium in regenerative medicine. BioMed research international 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alves da Silva M, Costa-Pinto A, Martins A, Correlo V, Sol P, Bhattacharya M, et al. Conditioned medium as a strategy for human stem cells chondrogenic differentiation. Journal of tissue engineering and regenerative medicine 2015;9(6):714–23. [DOI] [PubMed] [Google Scholar]
- 17.Timmers L, Lim SK, Arslan F, Armstrong JS, Hoefer IE, Doevendans PA, et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem cell research 2008;1(2):129–37. [DOI] [PubMed] [Google Scholar]
- 18.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature Reviews Immunology 2002;2(8):569–79. [DOI] [PubMed] [Google Scholar]
- 19.Bjørge IM, Kim SY, Mano J, Kalionis B, Chrzanowski W. Extracellular vesicles, exosomes and shedding vesicles in regenerative medicine–a new paradigm for tissue repair. Biomaterials science 2018. [DOI] [PubMed]
- 20.Shim J, Nam J-W. The expression and functional roles of microRNAs in stem cell differentiation. BMB reports 2016;49(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosa A, Brivanlou AH. Synthetic mRNAs: Powerful tools for reprogramming and differentiation of human cells. Cell Stem Cell 2010;7(5):549–50. [DOI] [PubMed] [Google Scholar]
- 22.Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, Doevendans PA, et al. MicroRNA-1 and-499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arteriosclerosis, thrombosis, and vascular biology 2010;30(4):859–68. [DOI] [PubMed] [Google Scholar]
- 23.Lin X, Wu L, Zhang Z, Yang R, Guan Q, Hou X, et al. MiR-335–5p Promotes Chondrogenesis in Mouse Mesenchymal Stem Cells and Is Regulated Through Two Positive Feedback Loops. Journal of Bone and Mineral Research 2014;29(7):1575–85. [DOI] [PubMed] [Google Scholar]
- 24.van de Peppel J, Strini T, Tilburg J, Westerhoff H, van Wijnen AJ, van Leeuwen JP. Identification of Three Early Phases of Cell-Fate Determination during Osteogenic and Adipogenic Differentiation by Transcription Factor Dynamics. Stem cell reports 2017;8(4):947–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stechschulte LA, Lecka-Czernik B. Reciprocal Regulation of PPARγ and RUNX2 Activities in Marrow Mesenchymal Stem Cells: Fine Balance between p38 MAPK and Protein Phosphatase 5. Current Molecular Biology Reports 2017;3(2):107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franceschi R The developmental control of osteoblast-specific gene expression: role of specific transcription factors and the extracellular matrix environment. Critical Reviews in Oral Biology & Medicine 1999;10(1):40–57. [DOI] [PubMed] [Google Scholar]
- 27.Narayanan K, Lim VY, Shen J, Tan ZW, Rajendran D, Luo S-C, et al. Extracellular matrix-mediated differentiation of human embryonic stem cells: differentiation to insulin-secreting beta cells. Tissue Engineering Part A 2013;20(1–2):424–33. [DOI] [PubMed] [Google Scholar]
- 28.Morrison N, Shine J, Fragonas J, Verkest V, McMenemy M, Eisman J. 1,25-dihydroxyvitamin D-responsive element and glucocorticoid repression in the osteocalcin gene. Science 1989;246(4934):1158–61. [DOI] [PubMed] [Google Scholar]
- 29.Kita A, Yamasaki H, Kuwahara H, Moriuchi A, Fukushima K, Kobayashi M, et al. Identification of the promoter region required for human adiponectin gene transcription: association with CCAAT/enhancer binding protein-β and tumor necrosis factor-α. Biochemical and biophysical research communications 2005;331(2):484–90. [DOI] [PubMed] [Google Scholar]
- 30.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC cancer 2011;11(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010;11(5):675–87. [DOI] [PubMed] [Google Scholar]
- 32.Adams JC, Watt FM. Regulation of development and differentiation by the extracellular matrix. Development 1993;117(4):1183–98. [DOI] [PubMed] [Google Scholar]
- 33.Peerani R, Rao BM, Bauwens C, Yin T, Wood GA, Nagy A, et al. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. The EMBO journal 2007;26(22):4744–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gradilla A-C, González E, Seijo I, Andrés G, Bischoff M, González-Mendez L, et al. Exosomes as Hedgehog carriers in cytoneme-mediated transport and secretion. Nature communications 2014;5:5649. [DOI] [PubMed] [Google Scholar]
- 35.Parchure A, Vyas N, Ferguson C, Parton RG, Mayor S. Oligomerization and endocytosis of Hedgehog is necessary for its efficient exovesicular secretion. Molecular biology of the cell 2015;26(25):4700–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nature cell biology 2012;14(10):1036. [DOI] [PubMed] [Google Scholar]
- 37.Liégeois S, Benedetto A, Garnier J-M, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. The Journal of cell biology 2006;173(6):949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komori T Regulation of bone development and maintenance by Runx2. Frontiers in bioscience: a journal and virtual library 2008;13:898–903. [DOI] [PubMed] [Google Scholar]
- 39.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002;108(1):17–29. [DOI] [PubMed] [Google Scholar]
- 40.Gámez B, Rodríguez-Carballo E, Bartrons R, Rosa JL, Ventura F. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. Journal of Biological Chemistry 2013;288(20):14264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassan MQ, Gordon JAR, Beloti MM, Croce CM, Wijnen AJv, Stein JL, et al. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24–2 cluster regulates the osteoblast differentiation program. Proceedings of the National Academy of Sciences 2010;107(46):19879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Wang S, Bian C, Yang Z, Zhou H, Zeng Y, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem cells and development 2012;21(13):2531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin F-T, Lane MD. CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proceedings of the National Academy of Sciences 1994;91(19):8757–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 1994;135(2):798–800. [DOI] [PubMed] [Google Scholar]
- 45.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, et al. MicroRNA-143 regulates adipocyte differentiation. Journal of Biological Chemistry 2004;279(50):52361–5. [DOI] [PubMed] [Google Scholar]
- 46.Ling HY, Wen GB, Feng SD, Tuo QH, Ou HS, Yao CH, et al. MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signalling. Clinical and Experimental Pharmacology and Physiology 2011;38(4):239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui Y, Luan J, Li H, Zhou X, Han J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS letters 2016;590(1):185–92. [DOI] [PubMed] [Google Scholar]
- 48.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology 2007;9(6):654–9. [DOI] [PubMed] [Google Scholar]
- 49.Farahani M, Rubbi C, Liu L, Slupsky JR, Kalakonda N. CLL exosomes modulate the transcriptome and behaviour of recipient stromal cells and are selectively enriched in miR-202–3p. PloS one 2015;10(10):e0141429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature communications 2015;6:7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire CA, Chen JW, et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS nano 2014;8(1):483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li CC, Eaton SA, Young PE, Lee M, Shuttleworth R, Humphreys DT, et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA biology 2013;10(8):1333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin CQ, Bissell MJ. Multi-faceted regulation of cell differentiation by extracellular matrix. The FASEB Journal 1993;7(9):737–43. [DOI] [PubMed] [Google Scholar]
- 54.Ridder K, Keller S, Dams M, Rupp A-K, Schlaudraff J, Del Turco D, et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS biology 2014;12(6):e1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roskelley CD, Srebrow A, Bissell MJ. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Current opinion in cell biology 1995;7(5):736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang W-C, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015;527(7576):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van der Merwe Y, Faust AE, Steketee MB. Matrix bound vesicles and miRNA cargoes are bioactive factors within extracellular matrix bioscaffolds. Neural regeneration research 2017;12(10):1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Z, McClure MJ, Zhao J, Ramey AN, Asmussen N, Hyzy SL, et al. MicroRNA Contents in Matrix Vesicles Produced by Growth Plate Chondrocytes are Cell Maturation Dependent. Scientific reports 2018;8(1):3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rilla K, Mustonen A-M, Arasu UT, Härkönen K, Matilainen J, Nieminen P. Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix Biology 2017. [DOI] [PubMed]
- 60.Than UTT, Guanzon D, Leavesley D, Parker T. Association of extracellular membrane vesicles with cutaneous wound healing. International journal of molecular sciences 2017;18(5):956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huleihel L, Bartolacci JG, Dziki JL, Vorobyov T, Arnold B, Scarritt ME, et al. Matrix-bound nanovesicles recapitulate extracellular matrix effects on macrophage phenotype. Tissue Engineering Part A 2017;23(21–22):1283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huleihel L, Hussey GS, Naranjo JD, Zhang L, Dziki JL, Turner NJ, et al. Matrix-bound nanovesicles within ECM bioscaffolds. Science advances 2016;2(6):e1600502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C, Loe F, Blocki A, Peng Y and Raghunath M Applying macromolecular crowding to enhance extracellular matrix deposition and its remodeling in vitro for tissue engineering and cell-based therapies. Advanced drug delivery reviews, 2011; 63(4–5): 277–290. [DOI] [PubMed] [Google Scholar]
- 64.Lareu RR, Arsianti I, Subramhanya HK, Yanxian P and Raghunath M, In vitro enhancement of collagen matrix formation and crosslinking for applications in tissue engineering: A preliminary study. Tissue Engineering, 2007; 13(2): 385–391. [DOI] [PubMed] [Google Scholar]
- 65.Ang XM, Lee MHC, Blocki A, Chen C, Ong LLS, Asada HH, et al. Macromolecular Crowding Amplifies Adipogenesis of Human Bone Marrow-Derived Mesenchymal Stem Cells by Enhancing the Pro-Adipogenic Microenvironment. Tissue Engineering. Part A, 2014; 20(5–6), 966–981. 10.1089/ten.tea.2013.0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.