Abstract
Background
The ability to digest dietary lactose is associated with lactase persistence (LP) in the intestinal lumen in human. The genetic basis of LP has been investigated in many populations in the world. Iran has a long history of pastoralism and the daily consumption of dairy products; thus, we aim to assess how LP has evolved in the Iranian population. We recruited 400 adult individuals from seven Iranian ethnic groups, from whom we investigated their lactose tolerance and screened the genetic variants in their lactase gene locus.
Results
The LP frequency distribution ranged from 0 to 29.9% in the seven Iranian ethnic groups with an average value of 9.8%. The variants, − 13910*T and − 22018*A, were significantly associated with LP phenotype in Iranians. We found no evidence of hard selective sweep for − 13910*T and − 22018*A in Persians, the largest ethnic group of Iran. The extremely low frequency of − 13915*G in the Iranian population challenged the view that LP distribution in Iran resulted from the demic diffusion, especially mediated by the spread of Islam, from the Arabian Peninsula.
Conclusions
Our results indicate the distribution of LP in seven ethnic groups across the Iranian plateau. Soft selective sweep rather than hard selective sweep played a substantial role in the evolution of LP in Iranian populations.
Electronic supplementary material
The online version of this article (10.1186/s40246-019-0195-5) contains supplementary material, which is available to authorized users.
Keywords: Lactase persistence, Iran, − 13910*T, − 22018*A, − 13915*G, Selection
Background
Lactase persistence (LP; OMIM #223100) is defined as the continued lactase enzyme activity that helps to digest lactose in dairy products in human adulthood [1]. It follows a Mendelian autosomal heritance [2] regulated by cis-acting elements of the lactase gene (LCT; OMIM *603202) [3]. Series of studies revealed five regulatory variants that are located in the 14 kb upstream of LCT in various populations: − 13910*T (rs4988235) in Europeans [4], Central Asians [5], and South Asians [6]; − 13915*G (rs41380347) in West Asians [7]; and − 13907*G (rs41525747), − 14009*G (rs869051967), and − 14010*C (rs145946881) in East Africans [8–10]. In addition, − 22018*A (rs182549) was investigated as a LP-associated variant [4, 11] in several populations [12–14], but it directed minimal enhancement of LCT promoter activity in vitro [15, 16]. The existence of independent variants underlying LP in different populations presents a paradigm of convergent evolution potentially driven by the daily consumption of large amounts of milk products after the domestication of dairy animals [17]. The co-evolution of genes for LP and milk consumption also becomes one of the most well-known gene-culture models for human evolutionary change [18, 19]. It also attracts wide interests from Neolithic archeology [20], in that the LP variant − 13910*T serves as a genetic marker in ancient DNA analyses to trace prehistoric migrations in Europe [21–23].
The genetic and archeological evidence supports the role of Iran as a domestication center of dairy animals such as goat [24, 25], cattle [26], and camel [27, 28]. The domesticated water buffaloes are also kept for milk production in Iran [29]. The long history of pastoralism and milk consumption raise the interest to explore the LP distribution as well as its genetic evolution in Iran. An early investigation of lactose intolerance revealed that the percentage of LP was 14% in Iranian adults [30]. The recent genetic screening for Iranians showed the occurrence of − 13910*T at 10% [7, 31]. The genotype-phenotype correlation was high [32], suggesting that − 13910*T might explain LP in Iranian population [33]. Despite these results, only limited samples from 42 individuals were analyzed so far. Herein, we explored a total of 400 adult individuals from seven ethnic groups living in Iran. The lactose tolerance test (LTT) was conducted to discern the LP distribution. We sequenced the relevant genomic region to identify potential variants that are associated with LP.
Materials and methods
Population samples
We recruited 400 healthy unrelated volunteers from seven Iranian ethnic groups, including Kurd (n = 138), Mazani (n = 110), Persian (n = 78), Arab (n = 26), Lur (n = 24), Azeri (n = 15), and Gilak (n = 9) (Additional file 1: Figure S1). Five-milliliter whole peripheral venous blood samples of the volunteers were collected. The study protocol was along with ethical approval and informed consent in Iran (Babol University of Medical Sciences, MUBABOL.REC.1394.354) and was also approved by the Internal Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (SMKX2017003).
Lactose tolerance test
We conducted the lactose tolerance test (LTT) in the 400 volunteers as described before [8, 34]. The volunteers were instructed to fast overnight and avoid smoking. The fasting fingertip capillary blood-glucose level at baseline was recorded with the Accu-Chek Advantage glucometer and Accu-Chek Comfort Curve Blood Glucose Test Strips (Roche, Mannheim, Germany) in the next morning. A 50-g lactose powder (Kerry Bio-Science) diluted in 250 mL of water at room temperature was given to each volunteer. The volunteers were requested to stay for the entire test duration (i.e., ~ 1 h). We measured fingertip capillary blood glucose levels in duplicates at 20-min intervals over a 1-h period. The lactase status was classified into three categories on the basis of the maximum rise in glucose level: an individual with a blood glucose level > 1.7 mmol/L was classified as LP; an individual with a blood glucose level < 1.1 mmol/L was classified as lactase non-persistence (LNP); and “lactase intermediate persistent” (LIP) was classified as an individual with a blood glucose level between 1.1 and 1.7 mmol/L.
PCR and Sanger sequencing
The genomic DNA was extracted from whole blood by a modified salting-out method [35] at Shahid Bahonar University of Kerman, Iran. We amplified and sequenced the 706 bp regulatory region for LCT in intron 13 of MCM6 referring to the previous protocol [36]. The variant − 22018*A was checked by PCR-RFLP [14]. Additionally, we sequenced − 22018*A variant region to confirm the RFLP status in 15 samples. The 683 bp of the control region 1 and 701 bp of the control region 2 for LCT were also sequenced in 400 samples [9, 37]. All sequences were checked and aligned by Lasergene (DNAStar Inc., Madison, Wisconsin, USA), and mutations were scored relative to the reference genome sequence (GRCh37/hg19).
Data analysis
To identify SNPs associated with the LP trait in the Iranian populations, we performed Fisher’s exact test with R statistical software version 3.3.2 (https://www.r-project.org/). We tested the association between LP phenotype and common SNPs (minor allele frequency > 5%) in the 400 individuals. The nucleotide diversity [38] was calculated with DNAsp v. 6.11.01 [39]. We used PHASE v.2.1.1 [40, 41] to phase haplotypes based on 18 SNPs of the regulatory region and its flanking control regions 1 and 2. The haplotypes with fewer than three occurrences were excluded [9]. The median-joining network was constructed with Network v.5.0.0.1 [42].
Whole-genome resequencing and detection of selective signals
We carried out whole-genome sequencing for 20 Persian individuals from Kerman in Southeastern Iran. The samples were randomly selected for whole-genome resequencing. The sequencing was performed on Illumina HiSeq X Ten. We referred to the GATK Best Practices for the SNP calling [43]. We retrieved the unphased SNP data of the 1 Mb (GRCh37/hg19 chr2:136,108,835-137,108,505) containing the regulatory region for LCT from the 20 Persians as well as 107 TSI (Toscani in Italia) in the 1000 Genomes Project [44] for comparison. We phased the data using SHAPEIT2 r727 [45]. For each of the SNPs, the ancestral and derived alleles were determined according to the alignments for six primates (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/ancestral_alignments/). The SNPs with ambiguously ancestral/derived states were discarded. We calculated the extended haplotype homozygosity (EHH) [46] and the integrated haplotype score (iHS) [47] with REHH 2.0 [48] and selscan software [49] (Additional file 1: Methods S1).
Results
LP in the Iranian populations
According to the LTT results, LP, LIP, and LNP accounted for 9.5%, 24%, and 66.5% of the total Iranian studied population, respectively (Table 1). The highest frequency of LP was in the Arab population (26.92%), whereas LP was not detected in the Lur population (Table 1). Differences of LP frequency were significant when tabulated by ethnicity (Fisher’s exact test, P value = 0.0004), occupation (chi-squared test, P value = 0.018), and language (chi-squared test, P value = 0.0004).
Table 1.
Phenotype frequency obtained from lactose tolerance testing among different ethnic groups in Iran
| Group | Occupation | Language | Sample Size | LP | LNP | LIP |
|---|---|---|---|---|---|---|
| Kurd | Herder | Indo-European | 138 (34.5%) | 8 (5.79%) | 92 (66.66%) | 38 (27.53%) |
| Mazani | Farmer | Indo-European | 110 (27.5%) | 9 (8.18%) | 77 (70%) | 24 (21.81%) |
| Persian | Herder | Indo-European | 78 (19.5%) | 12 (15.38%) | 52 (66.66%) | 14 (17.94%) |
| Arab | Herder | Afro-Asiatic | 26 (6.5%) | 7 (26.92%) | 13 (50.00%) | 6 (23.07%) |
| Lur | Herder | Indo-European | 24 (6.0%) | 0 (0.00%) | 15 (62.50%) | 9 (37.50%) |
| Gilak | Farmer | Indo-European | 9 (2.2%) | 1 (11.11%) | 6 (66.66%) | 2 (22.22%) |
| Azeri | Farmer | Turkic | 15 (3.8%) | 1 (6.66%) | 11 (73.33%) | 3 (20.00%) |
| Total | – | – | 400 | 38 (9.5%) | 266 (66.5%) | 96 (24.0%) |
LP variants in the Iranian populations
We identified three LP variants: − 22,018*A, − 13915*G, and − 13910*T, in the Iranian population (Table 2; Additional file 1: Table S1). All three variants were heterozygotes in the carriers. The three variants were also detected in neighboring countries (Additional file 1: Table S2). The − 22018*A allele, the most common variant (11.25%), was detected at the highest frequency in the Arab (19.23%) and the lowest in the Lur (4.16%) populations. The prevalence of − 13910*T was found in most populations (4.16–7.69%) with the exception of the Gilaks. The variant − 13915*G only occurred in one individual from both the Persians (n = 78, 1.28%) and the Arabs (n = 26, 3.84%), respectively. The co-occurrence of − 22018*A and − 13910*T was observed in all populations except the Lurs and the Gilaks (Additional file 2: Table S5). The variants of − 22018*A (Fisher’s exact test, P value = 3.725 × 10−6) and − 13910*T (Fisher’s exact test, P value = 1.509 × 10−7) were significantly associated with LP. Notably, the significant association was also detected between LP and the co-occurrence of − 13910*T and − 22018*A (Fisher’s exact test, P value = 1.73 × 10−7) (Fig. 1). In a total of 38 LP individuals, nine individuals carried both − 13910*T and − 22018*A. The nucleotide diversity of the regulatory region and the flanking control regions was highest in the LP subpopulation (0.126) and lowest in the LNP subpopulation (0.105) while the LIP individuals showed an intermediate value of 0.112 (Additional file 1: Table S3).
Table 2.
The distribution of three LP variants in Iranian ethnic groups
| Group | Size | − 22018G/A | − 13915 T/G | − 13910C/T | |||
|---|---|---|---|---|---|---|---|
| GG | GA | TT | TG | CC | CT | ||
| Kurd | 138 (34.5%) | 124 (89.85%) | 14 (10.15%) | 138 (100%) | 0 (0.00%) | 128 (92.75%) | 10 (7.24%) |
| Mazani | 110 (27.5%) | 94 (85.45%) | 16 (14.54%) | 110 (100%) | 0 (0.00%) | 106 (96.36%) | 4 (3.63%) |
| Persian | 78 (19.5%) | 72 (92.30%) | 6 (7.69%) | 77 (98.72%) | 1 (1.28%) | 72 (92.30%) | 6 (7.69%) |
| Arab | 26 (6.5%) | 21 (80.76%) | 5 (19.23%) | 25 (96.16%) | 1 (3.84%) | 24 (92.30%) | 2 (7.69%) |
| Lur | 24 (6.0%) | 23 (95.83%) | 1 (4.16%) | 24 (100%) | 0 (0.00%) | 23 (95.83%) | 1 (4.16%) |
| Gilak | 9 (2.2%) | 8 (88.88%) | 1 (11.11%) | 9 (100%) | 0 (0.00%) | 9 (100%) | 0 (0.00%) |
| Azeri | 15 (3.7%) | 13 (86.66%) | 2 (13.33%) | 15 (100%) | 0 (0.00%) | 14 (93.33%) | 1 (6.66%) |
| Total | 400 | 355 (88.75%) | 45 (11.25%) | 398 (99.50%) | 2 (0.50%) | 376 (94.02%) | 24 (5.98%) |
Fig. 1.
The genotype–phenotype correlation in the merged Iranian population
Haplotype and network analysis
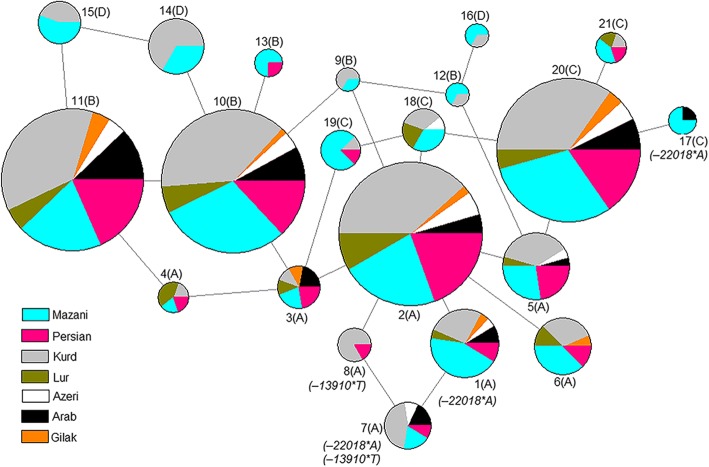
We phased the 400 sequences to get 38 haplotypes according to the 18 SNPs of the regulatory region and the flanking control regions (Additional file 1: Table S4). By excluding 17 haplotypes with fewer than three occurrences [9], we plotted 21 major haplotypes in the median-joining network (Fig. 2) according to the nomenclature proposed by Hollox et al. (2001) [37] (Additional file 1: Figure S2). All − 13910*T alleles were observed on haplotype A that is agreement with previous studies [50–52]. The − 22018*A allele was found in haplotypes A and C, suggesting a recombination event or parallel mutation occurrence [31]. Haplotype A with the − 22018*A allele was present in all groups.
Fig. 2.
Maximum parsimony neighbor joining network of 18 SNP identified in 400 samples of Iran. Mutations correspond to those in Additional file 1: Figure S2. Each circle represents a haplotype, and circle size is shown proportional to the number of individuals with a given haplotype
Detection of selective signals of − 13910*T and − 22018*A
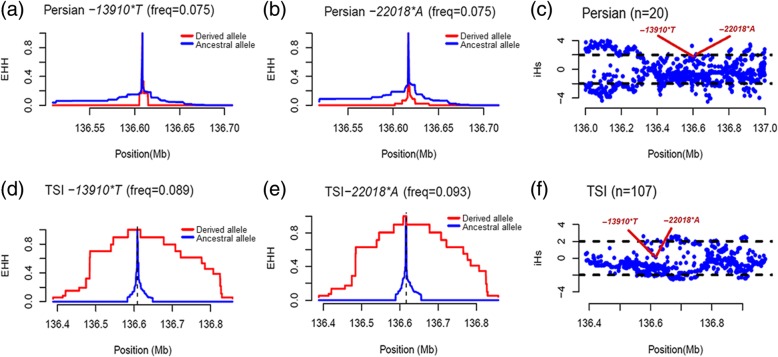
Because − 13910*T and − 22018*A are associated with LP in the Iranian populations, we tested for indications of a recent selective sweep on both alleles based on the whole genome re-sequencing data of 20 Persians. The alleles − 13910*T and − 22018*A were in complete linkage disequilibrium. The population of TSI presenting the similar characters for those two alleles was used for comparison. The allele frequency was 7.5% (3/40) in Persian (carriers classified as one LP and two LNP) and 9.11% (39/428) in TSI. We found no evidence of EHH for − 13910*T (Fig. 3a) and − 22018*A (Fig. 3b) in the Persians as compared with their ancestral alleles. The haplotypes with − 13910*T (Fig. 3d) and − 22018*A (Fig. 3e) showed EHH in TSI. No significant selective signal (i.e., |iHS| < 2) for − 13910*T and − 22018*A was detected by iHS in the Persians (Fig. 3c) and TSI (Fig. 3f).
Fig. 3.
EHH and iHS for the chromosomal positions carrying the − 13910*T and − 22018*A variants. EHH plots in Persian from Kerman, Iran (a, b) and Toscani in Italy (TSI) (d, e). Chromosomes containing the derived LP-associated alleles are in red, and those with the ancestral allele are in blue. Chromosomal positions are indicated on the x-axes, and the EHH value is indicated on the y-axes. The values of iHS are plotted against the genomic position of the SNPs including 1 Mb (chr2:136000000-137000000) at MCM6 and LCT promoter in Persian (c) and TSI (f). The estimates for − 13910*T and − 22018*A are noted
Discussion
Our study depicted the distribution of LP in the Iranian populations. However, the number of recruited subjects for some ethnic groups (Gilak for example) was small. It provided an opportunity to test the culture-historical hypothesis [53] in Iran. The hypothesis suggested that LP had been under historical selection, so that it was popular in populations (e.g., nomads) where milk products served as a substantial dietary component [17]. The prevalence of LP in Iran, with an average of 9.5% in the Iranian populations, was generally lower than that in the populations from Central Asia (14%) [5, 17], Afghanistan (19%), Pakistan (38%), Turkey (30%), Saudi Arabia (81%), and Jordan (76%) [32]. Within our Iranian studied population, the herders presented the higher percentage of LP distribution than the farmers (Table 1), which is consistent with the proposed pattern of the culture-historical hypothesis [53]. In the herders, we found the LP at the highest frequency (29.9%) in the Iranian Arabs. It was compatible with previous studies revealing that the nomadic Arabs had a high frequency of LP [32, 54]. Surprisingly, the Lurs, the traditional pastoral nomads living in the Zagros Mountains [55], were characterized as LIP (37.5%) and LNP (62.5%) but not LP (Table 1). The moderate or even low level of LP frequencies in Iran could be explained by several reasons. The consumption of moderate amounts of fresh milk, averaging just 48.48 kg per person in 2013 (FAOSTAT, http://www.fao.org/faostat/en/#home), may be the main contributing factor. On a deeper level, it may reflect a complex demographic history [56–59] such as admixture among nomadic and secondary populations which was proposed in Central Asia [17].
Our analyses indicated that the − 13910*T and − 22018*A variants were significantly associated with LP in all seven Iranian ethnic groups. However, these associations may not be able to explain all LP in the Iranian populations. In the 39 LP individuals, 20 individuals do not carry − 13910*T and/or − 22018*A (Additional file 2: Table S5). The future analyses based on massive whole genome re-sequencing may have the potential to reveal certain novel LP candidate variants in Iranians. The other thing worth noting is that other non-genetic factors, such as milk allergy [60], gut microbiota [61], and dairy foods [62], should be considered in both LTT and genetic analyses. Moreover, we found no evidence of hard selective sweeps for − 13910*T and − 22018*A in the re-sequenced genomes for 20 Persian individuals (Fig. 3). This result was different from that observed in Europeans [36, 47, 63] and South Asians [6]. Indeed, when considering different demographic scenarios, selection pressure on LP in Iranians was lower than most Europeans [64]. In particular, the occurrence of − 13910*T in two distinguished LP haplotypes was observed in Iranians [31]. Both haplotypes in Iranians were dated within 3000 years [31] that was much later than the dairying practices spread from the West Asia to Europe [65], raising the possibility that the − 13910*T alleles might be introduced into Iran recently. Meanwhile, as similar in Ethiopians [9], the increasing of nucleotide diversity was observed in the Iranian LP individuals (Additional file 1: Table S3). It implied that soft sweep rather than hard sweep played substantial roles in the evolution of LP in Iranian populations.
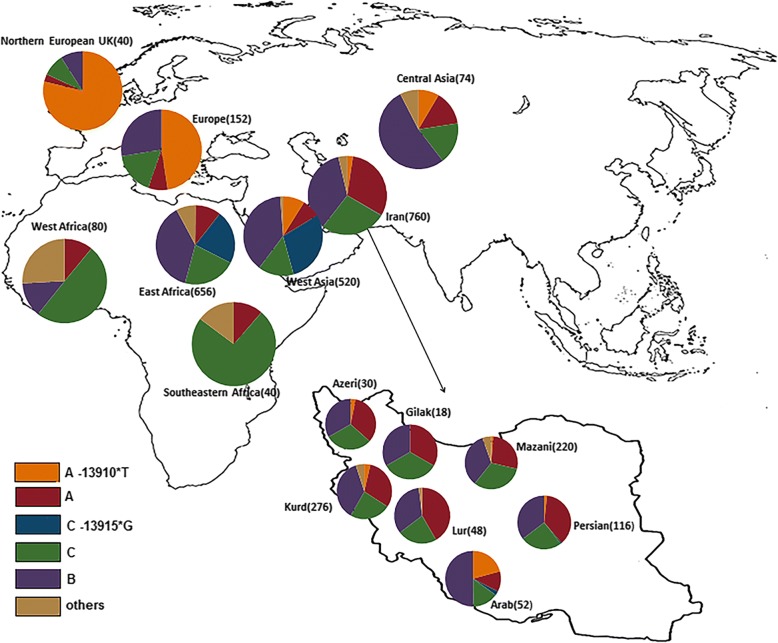
In addition, the distribution of LP variants in Iran would also provide certain clues towards the demographic history for different ethnic groups. The variant − 13915*G showed considerably high frequency in populations of Arabian Peninsula (Additional file 1: Table S2), such as Saudi (0.76) [66], Oman (0.72), and Yemeni (0.54) Arabs [67]. Southern Arabia was proposed to be the origin center of − 13915*G [67] in association with the domestication of Arabian camel (Camelus dromedarius) ~ 6000 years ago [7]. The distribution of − 13915*G was also high (10.4–17.6%) in several East African populations [7, 8]; proposing this may be the result of demic diffusion from Arabian Peninsula into East Africa, especially mediated by the spread of Islam in the last 1400 years [68, 69]. The Muslim conquest of Persia led to the fall of the Sasanian Empire (633–654 AD) [70], which also generated lots of cultural changes in Iran. Intriguingly, the Arabic-prevalent − 13915*G was almost absent in most Iranian populations except in one Arab and one Persian (Table 2; Fig. 4). In fact, a relatively high frequency of − 13910*T was detected in the Iranian Arabs (Table 2; Fig. 4). Our results suggested the demic diffusion of − 13915*G from Arabian Peninsula could be minimal in Iran, especially as compared the scenario in East Africa.
Fig. 4.
Distribution of MCM6 haplotypes in different geographic regions. The number of chromosomes is in parentheses
Conclusions
Our results indicate the distribution of LP in seven ethnic groups across the Iranian plateau. Soft selective sweep rather than hard selective sweep played substantial roles in the evolution of LP in Iranian populations. We observe the higher percentage of LP distribution in herders, thus providing evidence for the culture-historical hypothesis [53]. In the future, the integration of archeology [71, 72] and ancient DNA studies [73, 74] may uncover more details about the evolution of LP in Iran, which will also shed novel insights into “the milk revolution” [65] in the Neolithic core zone of West Asia.
Additional files
Table S1. Allele and genotype frequencies of three LP variants in Iranian ethnic groups. Table S2. Allele and genotype frequencies of the neighboring countries of Iran. Table S3. Comparison of the levels of nucleotide diversity in lactase persistent, lactase intermediate persistent and lactase nonpersistent measured across the three sequence regions from 400 Iranian Individuals. Table S4. The phased 400 sequences into 38 haplotypes based on the 18 SNPs of the regulatory region and the flanking control regions. Figure S1. Map of Iran showing approximate locations of the ethnic groups included in the present study. Figure S2. Phased Haplotypes for the LCT enhancer. Control region 1 and Control region 2, in Intron 9 and 13 of MCM6 and 1 kb Upstream of LCT from 7 ethnic groups in 400 Iranian Individuals. Method S1. Whole-Genome Resequencing and Detection of Selective Signals. (DOCX 804 kb)
Table S5. Genotype and phenotype information. (XLSX 53 kb)
Acknowledgments
H. Charati thanks the CAS-TWAS President’s Fellowship Program for Doctoral Candidates for support. A. Esmailizadeh was funded by Chinese Academy of Sciences President’s International Fellowship Initiative (No. 2016VBC051).
Funding
This work was supported by the Bureau of Science and Technology of Yunnan Province and the Animal Branch of the Germplasm Bank of Wild Species, Chinese Academy of Sciences (the Large Research Infrastructure Funding).
Availability of data and materials
All relevant data are within the paper and its additional files.
Abbreviations
- EHH
Extended haplotype homozygosity
- IHS
Integrated haplotype score
- LCT
Lactase gene
- LIP
Lactase intermediate persistent
- LNP
Lactase non-persistence
- LP
Lactase persistence
- LTT
Lactose tolerance test
- MCM6
Minichromosome maintenance complex component 6
- PCR
Polymerase chain reaction
- RFLP
Restriction fragment length polymorphism
- SNPs
Single nucleotide polymorphisms
- TSI
Toscani in Italia
Authors’ contributions
H-C, M-SP, A-E and Y-PZ conceived and designed the experiments. H-C and R-JO collected the samples and clinical information. H-C performed the experiments. H-C and XY-Y analyzed the data. H-C and M-SP wrote the paper. A-E, Y-PZ, M-AM, and W-C revised the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was along with ethical approval and informed consent in Iran (Babol University of Medical Sciences, MUBABOL.REC.1394.354) and was also approved by the Internal Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (SMKX2017003).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hadi Charati, Email: hadi@mail.kiz.ac.cn.
Min-Sheng Peng, Email: pengminsheng@mail.kiz.ac.cn.
Wei Chen, Email: wchenntr@163.com.
Xing-Yan Yang, Email: ynakmyxy@126.com.
Roghayeh Jabbari Ori, Email: jabbariroqayeh@yahoo.com.
Mohsen Aghajanpour-Mir, Email: mohsenaghajanpour@gmail.com.
Ali Esmailizadeh, Email: aliesmaili@uk.ac.ir.
Ya-Ping Zhang, Email: zhangyp@mail.kiz.ac.cn.
References
- 1.Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet. 2009;124:579–591. doi: 10.1007/s00439-008-0593-6. [DOI] [PubMed] [Google Scholar]
- 2.Swallow DM. Genetics of lactase persistence and lactose intolerance. Annu Rev Genet. 2003;37:197–219. doi: 10.1146/annurev.genet.37.110801.143820. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Harvey CB, Pratt WS, Sams VR, Sarner M, Rossi M, et al. The lactase persistence/non-persistence polymorphism is controlled by a cis-acting element. Hum Mol Genet. 1995;4:657–662. doi: 10.1093/hmg/4.4.657. [DOI] [PubMed] [Google Scholar]
- 4.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 5.Heyer E, Brazier L, Segurel L, Hegay T, Austerlitz F, Quintana-Murci L, et al. Lactase persistence in Central Asia: phenotype, genotype, and evolution. Hum Biol. 2011;83:379–392. doi: 10.3378/027.083.0304. [DOI] [PubMed] [Google Scholar]
- 6.Gallego Romero I, Basu Mallick C, Liebert A, Crivellaro F, Chaubey G, Itan Y, et al. Herders of Indian and European cattle share their predominant allele for lactase persistence. Mol Biol Evol. 2012;29:249–260. doi: 10.1093/molbev/msr190. [DOI] [PubMed] [Google Scholar]
- 7.Enattah NS, Jensen TG, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, et al. Independent introduction of two lactase persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranciaro A, Campbell MC, Hirbo JB, Ko WY, Froment A, Anagnostou P, et al. Genetic origins of lactase persistence and the spread of pastoralism in Africa. Am J Hum Genet. 2014;94:496–510. doi: 10.1016/j.ajhg.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones BL, Raga TO. Liebert A, Zmarz P, Bekele E, Danielsen ET, et al. Diversity of lactase persistence alleles in Ethiopia: signature of a soft selective sweep. Am J Hum Genet. 2013;93:538–544. doi: 10.1016/j.ajhg.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasinperä H, Savilahti E, Enattah NS, Kuokkanen M, Totterman N, Lindahl H, et al. A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut. 2004;53:1571–1576. doi: 10.1136/gut.2004.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halima YB, Kefi R, Sazzini M, Giuliani C, Fanti SD, Nouali C, et al. Lactase persistence in Tunisia as a result of admixture with other Mediterranean populations. Genes Nutr. 2017;12:20. doi: 10.1186/s12263-017-0573-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuchay RA, Anwar M, Thapa BR, Mahmood A, Mahmood S. Correlation of G/A –22018 single-nucleotide polymorphism with lactase activity and its usefulness in improving the diagnosis of adult type hypolactasia among North Indian children. Genes Nutr. 2013;8:145–151. doi: 10.1007/s12263-012-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Sun H, Zhang X, Wang J, Sun D, Chen F, Bai J, et al. The –22018A allele matches the lactase persistence phenotype in northern Chinese populations. Scand J Gastroenterol. 2010;45:168–174. doi: 10.3109/00365520903414176. [DOI] [PubMed] [Google Scholar]
- 15.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 16.Troelsen JT, Olsen J, Møller J, Sjostrom H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003;125:1686–1694. doi: 10.1053/j.gastro.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 17.Segurel L, Bon C. On the evolution of lactase persistence in humans. Annu Rev Genom Hum Genet. 2017;18:297–319. doi: 10.1146/annurev-genom-091416-035340. [DOI] [PubMed] [Google Scholar]
- 18.Ross CT, Richerson PJ. New frontiers in the study of human cultural and genetic evolution. Curr Opin Genet Dev. 2014;29:103–109. doi: 10.1016/j.gde.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Laland KN, Odling-Smee FJ, Myles S. How culture has shaped the human genome: bringing genetics and the human sciences together. Nat Rev Genet. 2010;11:137–148. doi: 10.1038/nrg2734. [DOI] [PubMed] [Google Scholar]
- 20.Gerbault P, Roffet-Salque M, Evershed RP, Thomas MG. How long have adult humans been consuming milk? IUBMB Life. 2013;65:983–990. doi: 10.1002/iub.1227. [DOI] [PubMed] [Google Scholar]
- 21.Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci U S A. 2007;104:3736–3741. doi: 10.1073/pnas.0607187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allentoft ME, Sikora M, Sjogren KG, Rasmussen S, Rasmussen M, Stenderup J, et al. Population genomics of bronze age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 23.Cassidy LM, Martinianoa R, Murphy EM, Teasdalea MD, Mallory J, Hartwell B, et al. Neolithic and bronze age migration to Ireland and establishment of the insular Atlantic genome. Proc Natl Acad Sci U S A. 2016;113:368–373. doi: 10.1073/pnas.1518445113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeder MA, Hesse B. The initial domestication of goats (Capra hircus) in the Zagros Mountains 10,000 years ago. Science. 2000;287:2254–2257. doi: 10.1126/science.287.5461.2254. [DOI] [PubMed] [Google Scholar]
- 25.Amills M, Capote J, Tosser-Klopp G. Goat domestication and breeding: a jigsaw of historical, biological and molecular data with missing pieces. Anim Genet. 2017;48:631–644. doi: 10.1111/age.12598. [DOI] [PubMed] [Google Scholar]
- 26.Arbuckle BS, Price MD, Hongo H, Öksüz B. Documenting the initial appearance of domestic cattle in the eastern Fertile Crescent (northern Iraq and western Iran) J Archaeol Sci. 2016;72:1–9. doi: 10.1016/j.jas.2016.05.008. [DOI] [Google Scholar]
- 27.Wapnish P. Camel caravans and camel pastoralists at tell jemmeh. JANES. 1981;13(101–21):104–105. [Google Scholar]
- 28.Meyers ME. The Oxford encyclopedia of archaeology in the near east. Oxford: Oxford University Press; 1997. [Google Scholar]
- 29.Safari A, Hossein-Zadeh NG, Shadparvar AA, Abdollahi Arpanahi R. A review on breeding and genetic strategies in Iranian buffaloes (Bubalus bubalis) Trop Anim Health Prod. 2018;50:707–714. doi: 10.1007/s11250-018-1563-1. [DOI] [PubMed] [Google Scholar]
- 30.Sadre M, Karbasi K. Lactose intolerance in Iran. Am J Clin Nutr. 1979;32:1948–1954. doi: 10.1093/ajcn/32.9.1948. [DOI] [PubMed] [Google Scholar]
- 31.Enattah NS, Trudeau A, Pimenoff V, Maiuri L, Auricchio S, et al. Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Hum Genet. 2007;81:615–625. doi: 10.1086/520705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itan Y, Jones BL, Ingram CJ, Swallow DM, Thomas MG. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol. 2010;10:36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alizadeh M, Sadr-Nabavi A. Evaluation of a genetic test for diagnose of primary hypolactasia in northeast of Iran (Khorasan) Iran J Basic Med Sci. 2012;15:1127–1130. [PMC free article] [PubMed] [Google Scholar]
- 34.Arola H. Diagnosis of hypolactasia and lactose malab- sorption. Scand J Gastroenterol Suppl. 1994;202:26–35. doi: 10.3109/00365529409091742. [DOI] [PubMed] [Google Scholar]
- 35.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res. 1988;16:12–15. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebert A, López S, Jones BL, Montalva N, Gerbault P, Lau W, et al. World-wide distributions of lactase persistence alleles and the complex effects of recombination and selection. Hum Genet. 2017;136:1445–1453. doi: 10.1007/s00439-017-1847-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollox EJ, Poulter M, Zvarik M, Ferak V, Krause A, Jenkins T, et al. Lactase haplotype diversity in the Old World. Am J Hum Genet. 2001;68:160–172. doi: 10.1086/316924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 40.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 43.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 46.Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 47.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gautier M, Klassmann A, Vitalis R. rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol Ecol Resour. 2017;17:78–90. doi: 10.1111/1755-0998.12634. [DOI] [PubMed] [Google Scholar]
- 49.Szpiech ZA, Hernandez RD. Selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 2014;31:2824–2827. doi: 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coelho M, Luiselli D, Bertorelle G, Lopes AI, Seixas S, Destro-Bisol G, et al. Microsatellite variation and evolution of human lactase persistence. Hum Genet. 2005;117:329–339. doi: 10.1007/s00439-005-1322-z. [DOI] [PubMed] [Google Scholar]
- 52.Poulter M, Hollox E, Harvey CB, Mulcare C, Peuhkuri K, Kajander K, et al. The causal element for the lactase persistence/nonpersistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann Hum Genet. 2003;67:298–311. doi: 10.1046/j.1469-1809.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 53.Simoons FJ. Primary adult lactose intolerance and the milking habit: a problem in biologic and cultural interrelations. II. A culture historical hypothesis. Am J Dig Dis. 1970;15:695–710. doi: 10.1007/BF02235991. [DOI] [PubMed] [Google Scholar]
- 54.Cook GC, Al-Torki MT. High intestinal lactase concentrations in adult Arabs in Saudi Arabia. Br Med J. 1975;3:135–136. doi: 10.1136/bmj.3.5976.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rashidvash V. Iranian People: Iranian Ethnic Groups IJHSS. 2013;3:15. [Google Scholar]
- 56.Nasidze I, Quinque D, Rahmani M, Alemohamad SA, Stoneking M. Concomitant replacement of language and mtDNA in South Caspian populations of Iran. Curr Biol. 2006;16:668–673. doi: 10.1016/j.cub.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 57.Terreros MC, Rowold DJ, Mirabal S, Herrera RJ. Mitochondrial DNA and Y-chromosomal stratification in Iran: relationship between Iran and the Arabian peninsula. Hum Genet. 2011;56:235–246. doi: 10.1038/jhg.2010.174. [DOI] [PubMed] [Google Scholar]
- 58.Grugni V, Battaglia V, Hooshiar Kashani B, Parolo S, Al-Zahery N, Achilli A, et al. Ancient migratory events in the Middle East: new clues from the Y-chromosome variation of modern Iranians. PLoS One. 2012;7:e41252. doi: 10.1371/journal.pone.0041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derenko M, Malyarchuk B, Bahmanimehr A, Denisova G, Perkova M, Farjadian S, et al. Complete mitochondrial DNA diversity in Iranians. PLoS One. 2013;8:e80673. doi: 10.1371/journal.pone.0080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heine RG, AlRefaee F, Bachina P, De Leon JC, Geng L, Gong S, et al. Lactose intolerance and gastrointestinal cow's milk allergy in infants and children-common misconceptions revisited. World Allergy Organ J. 2017;10:41. doi: 10.1186/s40413-017-0173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lukito W, Malik SG, Surono IS, Wahlqvist ML. From lactose intolerance to lactose nutrition. Asia Pac J Clin Nutr. 2015;1:S1–S8. doi: 10.6133/apjcn.2015.24.s1.01. [DOI] [PubMed] [Google Scholar]
- 62.Szilagyi A. Adaptation to lactose in lactase non persistent people: effects on intolerance and the relationship between dairy food consumption and evolution of diseases. Nutrients. 2015;7:6751–6779. doi: 10.3390/nu7085309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerbault P, Moret C, Currat M, Sanchez-Mazas A. Impact of selection and demography on the diffusion of lactase persistence. PLoS One. 2009;4:e6369. doi: 10.1371/journal.pone.0006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Curry A. The milk revolution. Nature. 2013;500:20–22. doi: 10.1038/500020a. [DOI] [PubMed] [Google Scholar]
- 66.Imtiaz F, Savilahti E, Sarnesto A, Trabzuni D, Al-Kahtani K, Kagevi I, et al. The T/G –13915 variant upstream of the lactase gene (LCT) is the founder allele of lactase persistence in an urban Saudi population. J Med Genet. 2007;44:e89. doi: 10.1136/jmg.2007.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al-Abri AR, Al-Rawas O, Al-Yahyaee S, Al-Habori M, Al-Zubairi AS, Bayoumi R. Distribution of the lactase persistence-associated variant alleles –13910* T and –13915* G among the people of Oman and Yemen. Hum Biol. 2012;84:271–286. doi: 10.3378/027.084.0310. [DOI] [PubMed] [Google Scholar]
- 68.Priehodová E, Abdelsawy A, Heyer E, Cerný V. Lactase persistence variants in Arabia and in the African Arabs. Hum Biol. 2014;86:7–18. doi: 10.3378/027.086.0101. [DOI] [PubMed] [Google Scholar]
- 69.Ingram CJ, Elamin MF, Mulcare CA, Weale ME, Tarekegn A, Raga TO et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum Genet. 2007;120:779–788. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- 70.Akram AI, al-Mehri AB. The Muslim Conquest of Persia. 2009. Ch: 1 ISBN 978-0-19-597713-4.
- 71.Dunne J, Evershed RP, Salque M, Cramp L, Bruni S, Ryan K, et al. First dairying in green Saharan Africa in the fifth millennium BC. Nature. 2012;486:390–394. doi: 10.1038/nature11186. [DOI] [PubMed] [Google Scholar]
- 72.Evershed RP, Payne S, Sherratt AG, Copley MS, Coolidge J, Urem-Kotsu D, et al. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature. 2008;455:528–531. doi: 10.1038/nature07180. [DOI] [PubMed] [Google Scholar]
- 73.Gamba C, Jones RE, Teasdale DM, McLaughlin LR, Gonzalez-Fortes G, Mattiangeli V, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5:52–57. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sverrisdóttir OÓ, Timpson A, Toombs J, Lecoeur C, Froguel P, Carretero JM, et al. Direct estimates of natural selection in Iberia indicate calcium absorption was not the only driver of lactase persistence in Europe. Mol Biol Evol. 2014;31:975–983. doi: 10.1093/molbev/msu049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Allele and genotype frequencies of three LP variants in Iranian ethnic groups. Table S2. Allele and genotype frequencies of the neighboring countries of Iran. Table S3. Comparison of the levels of nucleotide diversity in lactase persistent, lactase intermediate persistent and lactase nonpersistent measured across the three sequence regions from 400 Iranian Individuals. Table S4. The phased 400 sequences into 38 haplotypes based on the 18 SNPs of the regulatory region and the flanking control regions. Figure S1. Map of Iran showing approximate locations of the ethnic groups included in the present study. Figure S2. Phased Haplotypes for the LCT enhancer. Control region 1 and Control region 2, in Intron 9 and 13 of MCM6 and 1 kb Upstream of LCT from 7 ethnic groups in 400 Iranian Individuals. Method S1. Whole-Genome Resequencing and Detection of Selective Signals. (DOCX 804 kb)
Table S5. Genotype and phenotype information. (XLSX 53 kb)
Data Availability Statement
All relevant data are within the paper and its additional files.