Abstract
Background
ATP-dependent chromatin remodelling complexes are responsible for establishing and maintaining the positions of nucleosomes. Chromatin remodellers are targeted to chromatin by transcription factors and non-coding RNA to remodel the chromatin into functional states. However, the influence of chromatin remodelling on shaping the functional epigenome is not well understood. Moreover, chromatin remodellers have not been extensively explored as a collective group across two-dimensional and three-dimensional epigenomic layers.
Results
Here, we have integrated the genome-wide binding profiles of eight chromatin remodellers together with DNA methylation, nucleosome positioning, histone modification and Hi-C chromosomal contacts to reveal that chromatin remodellers can be stratified into two functional groups. Group 1 (BRG1, SNF2H, CHD3 and CHD4) has a clear preference for binding at ‘actively marked’ chromatin and Group 2 (BRM, INO80, SNF2L and CHD1) for ‘repressively marked’ chromatin. We find that histone modifications and chromatin architectural features, but not DNA methylation, stratify the remodellers into these functional groups.
Conclusions
Our findings suggest that chromatin remodelling events are synchronous and that chromatin remodellers themselves should be considered simultaneously and not as individual entities in isolation or necessarily by structural similarity, as they are traditionally classified. Their coordinated function should be considered by preference for chromatin features in order to gain a more accurate and comprehensive picture of chromatin regulation.
Electronic supplementary material
The online version of this article (10.1186/s13072-019-0258-9) contains supplementary material, which is available to authorized users.
Keywords: Chromatin, Nucleosome, Chromatin remodelling, Enhancer, Promoter, Gene regulation, Epigenetics, CHD, SWI/SNF, INO80, ISWI
Background
Chromatin is a dynamic and multi-layered structure of which the core building block is the nucleosome. Nucleosomes are comprised of an octamer of histone proteins and 147 base pairs (bp) of DNA in approximately two helical turns [1]. The unique chromatin conformation of any given cell is typically maintained throughout divisions and serves as a physical barrier to transcription factors and other regulatory proteins in order to prevent promiscuous gene expression [2, 3]. Thus, chromatin structure must be modulated for regulatory factors to access DNA when required. This is largely achieved through the movement of nucleosomes by ATP-dependent chromatin remodelling complexes, which utilise ATP hydrolysis to organise nucleosomes into an active (relaxed) or repressive (compact) conformation. The process of chromatin remodelling provides means for regulating DNA structure with precision and accuracy to facilitate diverse cellular processes including transcriptional regulation, DNA repair, DNA replication and cell cycle progression [2, 4]. However, despite growing research into the molecular and biochemical mechanisms of chromatin remodelling, it is still not completely understood how remodelling complexes work together to position nucleosomes for the required chromatin function.
Beyond the physical nature of its structure, chromatin carries diverse gene regulatory information including post-translational modifications of histone proteins and DNA methylation [2, 4, 5]. These features, together with non-coding RNA (ncRNA) species, form the epigenome. Chromatin remodellers are drawn to their target regions by sequence-specific regulatory proteins or ncRNAs [6–8] and use their protein structural domains to identify epigenetic patterning and the ‘linker’ DNA between nucleosomes to identify their preferred nucleosome substrate [3, 9–11]. Yet, it is important to consider that many regulatory regions of the genome are a composite of multiple epigenetic marks; therefore, there is a multifaceted relationship between chromatin remodellers and the epigenome. For example, bivalent promoters are characterised by the trimethylation of histone 3 at both lysine 4 (H3K4me3) and lysine 27 (H3K27me3) [12–14] and could therefore potentially be ‘read’ by remodellers recognising either of these marks. Additionally, these relationships are not linear as more than one remodeller may recognise a single histone modification [15]. Uncovering the extent of overlapping and unique activity of chromatin remodeller proteins is of great interest and is essential for understanding the influence of the epigenetic signature on chromatin remodelling events at any given locus.
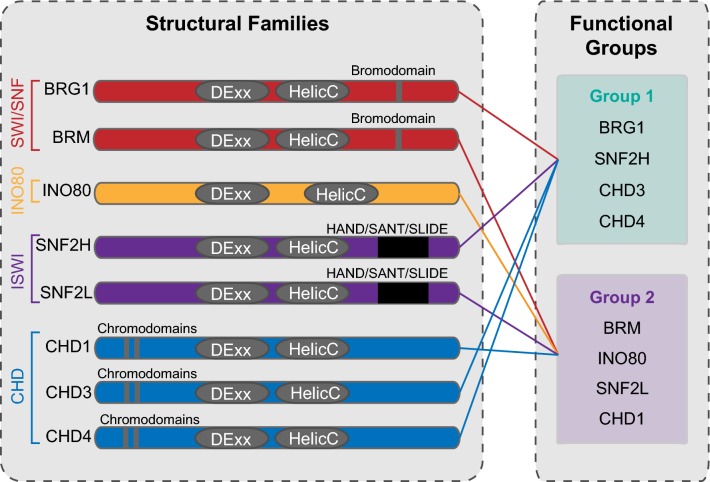
There are four structural families of chromatin remodellers: switch/sucrose non-fermenting (SWI/SNF), imitation switch (ISWI), inositol requiring 80-like (INO80-like) and chromodomain helicase DNA binding (CHD) [3, 16–18]. Chromatin remodelling complexes share common features such as an essential catalytic ATPase and a high affinity for nucleosomes [2, 3, 16–18], yet the ATPases within these complexes have evolved unique features that pertain to their specific function. SWI/SNF ATPases contain bromodomains for recognising acetylated histones; ISWI ATPases contain a HAND/SANT/SLIDE domain for recognising internucleosomal DNA, the INO80-like ATPases have a longer peptide chain between their two helicase domains that has been proposed to fit Holliday junctions and replication forks, and the CHD ATPases contain chromodomains [16, 18–22] (Fig. 1a). It is known that remodellers can have both cooperative and opposing roles at the same genomic location in yeast and mice [23–27]; however, the extent of this has not been studied extensively in human cells. Importantly, there has been no study to date examining more than three remodellers concurrently in human cells, nor with simultaneous consideration of the epigenome, transcriptome and chromatin structural states. Moreover, the influence of the epigenome on remodeller function is not well understood.
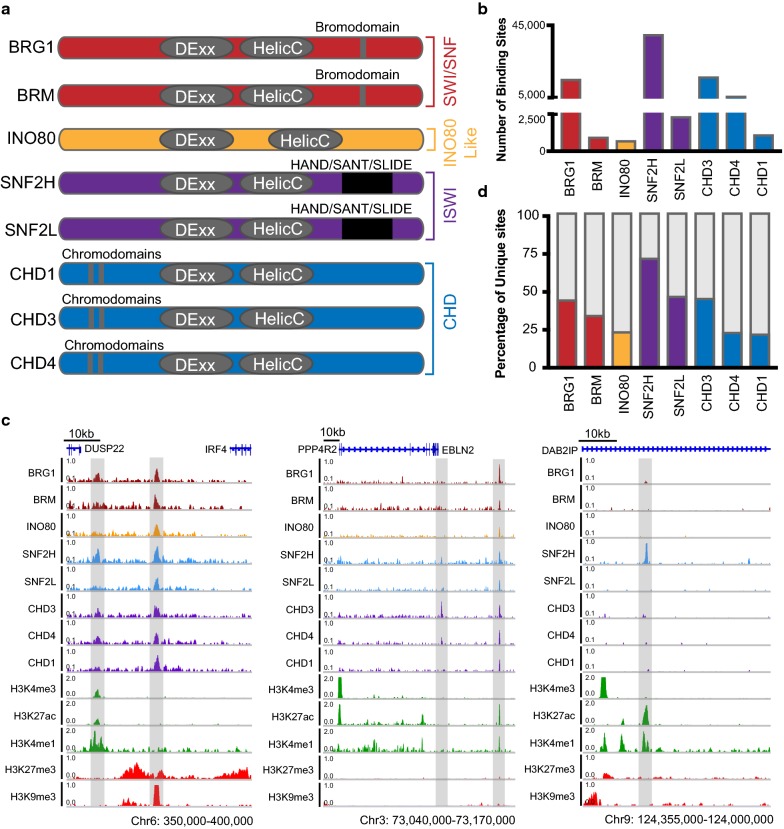
Fig. 1.
Chromatin remodeller occupancy and expression in LNCaP cells. a The domain organisation of each ATPase catalytic subunits of chromatin remodelling complexes used in this study. Each contains a SNF2-like helicase comprising of DEXDc and HELICc domains, with each subfamily containing additional domains. Key domains defining each subfamily are shown. b Number of binding sites occupied by each remodeller from MACS2 peak calling. c Genome browser view of chromatin remodeller occupancy and key histone modification tracks in LNCaP cells with overlap of chromatin remodeller binding and where SNF2H is the only bound remodeller. d Percentage of remodeller protein-binding sites unique to each remodeller (coloured) and percentage that overlaps with one or more of the other remodeller proteins (grey)
Here, we have examined the binding profiles of eight different chromatin remodeller proteins and integrated these with extensive epigenomic data, including histone modifications, DNA methylation and chromosome architectural information. Our study reveals that chromatin remodellers can be stratified into two groups based on their binding enrichment at either ‘actively marked’ or ‘repressively marked’ regions and their interactions with chromatin features of the epigenome.
Results
The degree of chromatin remodeller binding correlates with the remodeller gene expression levels in prostate cancer cells
To improve our understanding of the relationship between different chromatin remodellers, we sought to examine the genomic binding profiles of multiple chromatin remodeller proteins simultaneously from publicly available data obtained from LNCaP prostate cancer cells [28]. All eight remodellers examined are catalytic ATPases that form mutually exclusive complexes and together represent all four the structural families, unlike previous studies in mouse embryonic stem cells. The remodellers studied were BRG1 and BRM from the SWI/SNF family; SNF2H and SNF2L from the ISWI family; INO80 from the INO80-like family; and CHD1, CHD3 and CHD4 from the CHD family (Fig. 1a). We first assessed the gene expression level of each remodeller using published RNA-seq data from LNCaP cells [29] and compared this to prostate tumour samples from The Cancer Genome Atlas (TCGA) database [30] to determine the validity of using LNCaP cells as a prostate cancer model to explore chromatin remodellers. We found a high concordance in the gene expression pattern for each remodeller between these data sets, where those genes displaying higher expression in clinical samples from TCGA (n = 486) also had higher expression in the LNCaP cell line (Additional file 1: Figure S1A–B). A similar comparison was made between a normal prostate epithelial cell line (PrEC) and the normal samples from TCGA (n = 52); the expression patterns of the remodellers in PrEC cells mirrored that of the normal clinical samples (Additional file 1: Figure S1C–D). We calculated the Pearson correlation coefficient and found a strong positive linear relationship between the LNCaP cells and the TCGA tumours (Pearson R =0.8375552) and between the PrEC cells and the TCGA normal data sets (Pearson R =0.670013; Additional file 1: Figure S1E). The exception to this was SNF2L, which was significantly lower in LNCaP cells compared to the TCGA tumours.
Using the ChIP-seq binding profiles of the eight chromatin remodellers from Ye et al. [28], we next examined the number of individual binding sites for each remodeller and established they ranged from 712 for INO80 to 39,887 for SNF2H (Fig. 1b), and in total, there were 60,043 genomic regions bound by at least one chromatin remodeller. Upon visualising the remodeller binding sites, we observed several regions where multiple remodellers were bound, such as the intergenic region between DUSP22 and IRF4 on chromosome 6 and upstream of the EBLN2 gene on chromosome 3 (Fig. 1c). Additionally, there were several sites where a single remodeller was bound, such as CHD3 at the EBLN2 gene promoter and SNF2H within the DAB2IP gene (Fig. 1c). We observed that the majority of the binding sites occurred over regions marked by gene regulatory histone modifications. Over 75% of the remodeller binding sites were under 750 bp in size, corresponding to a span of ~ 1–5 nucleosomes (Additional file 1: Figure S2A–H), covering 0.93% of the genome. The degree of overlap between the remodellers varied extensively with the number of unique binding sites for each remodeller ranging from ~ 25 to ~ 50%, with the exception of SNF2H which had ~ 75% unique sites, likely paralleling the overall large number of binding sites for this remodeller (Fig. 1d).
Together, these data show that there is large variation in the number of genomic sites occupied by chromatin remodellers, with both independent and a high degree of overlapping activity between the remodeller proteins. Importantly, the high overlap in the number of binding sites containing at least two remodellers suggests there is widespread coordinated activity across the genome.
The epigenome stratifies chromatin remodellers into two groups: Group 1 is associated with ‘actively marked’ chromatin and Group 2 with ‘repressively marked’ chromatin
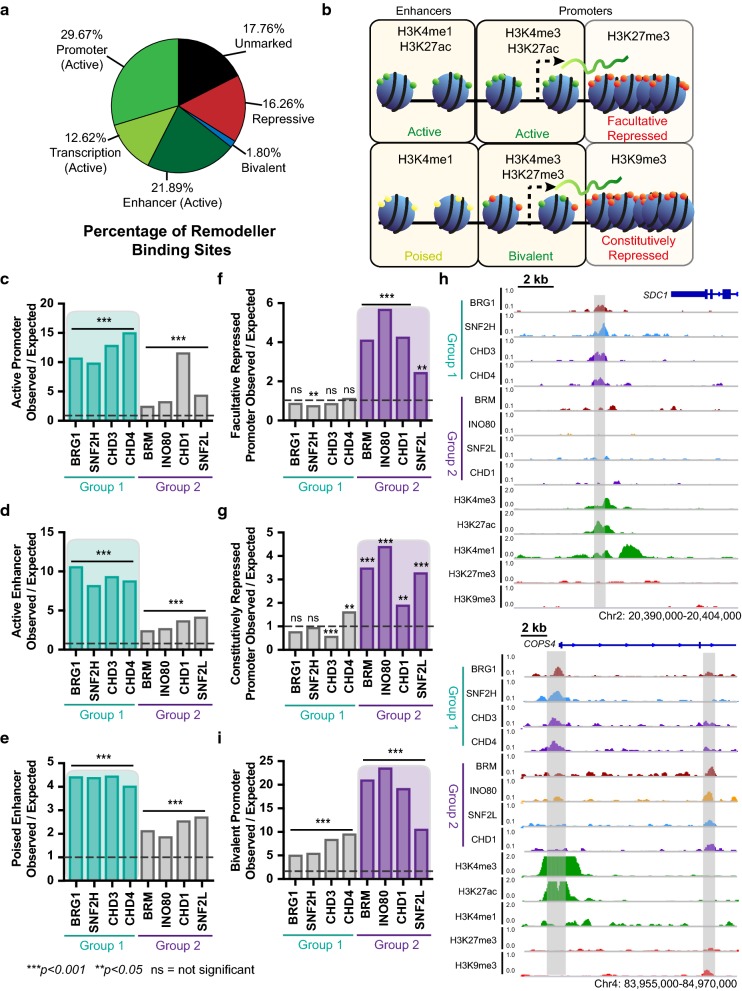
Chromatin remodellers do not exhibit sequence specificity in their binding and therefore have the potential to bind anywhere across the genome. However, as the majority of remodeller proteins contain domains targeting nucleosomes with post-translational histone tail modifications [2, 31–33], it is expected that the bulk of binding will occur within histone ‘marked’ chromatin. We compiled histone modification ChIP-seq data sets from LNCaP cells [29, 34] and performed a chromatin multivariate hidden Markov model (chromHMM) analysis to determine chromatin states based on the Epigenome Roadmap core model for chromatin state discovery [35] (Additional file 1: Figure S3A-B; see “Methods”). This analysis found that of the histone modifications examined, the ‘marked’ chromatin (active, bivalent and repressive states) encompassed 50.1% of the genome, while 49.9% is not marked with any of the histone modifications included in the analysis (see “Methods”). Of the 60,043 remodeller binding sites, 64.18% fell within ‘actively marked’ chromatin (promoters, enhancers and transcriptionally active), 16.26% fell within ‘repressively marked’ chromatin and 1.80% within bivalent chromatin. Together, 82.24% of all remodeller binding occurred within ‘marked’ chromatin, confirming that remodellers are largely localised to regulatory regions of the genome and suggesting that the primary role of remodellers is to establish and maintain gene expression (Fig. 2a).
Fig. 2.
Chromatin remodellers bind to distinct regions of chromatin in two defined groups. a Percentage of all 60,043 remodeller binding sites in each chromHMM chromatin state compiled into active promoters (promoters and flanking promoters), transcription (strong and weak and at 5′/3′ ends of genes), active enhancers (genic and other enhancers), bivalent (enhancers, promoters, and flanking promoters), repressive (polycomb, weak polycomb, zinc finger repeats and heterochromatin), and unmarked (no core marks from the Roadmap Epigenome chromatin state model). b Schematic diagram of histone modifications present at ‘active’ and ‘repressive’ promoters and enhancers. ‘Active’ histone modifications are shown in green with increased spacing between nucleosomes, while ‘repressive’ histone modifications are shown in red with compacted nucleosomes. c–g, i Enrichment of chromatin remodeller binding sites at key gene regulatory features defined by ‘active’ and ‘repressive’ histone modifications. Significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p value, ***p < 0.001 or **p < 0.05. h Genome browser view of chromatin remodeller occupancy and key histone modification tracks in LNCaP cells showing upstream of the SDC1 promoter and at the COPS4 promoter, which is occupied by all Group 1 remodellers and adjacent to exon 2 in COPS4 that is occupied by all Group 2 remodellers
As our data show that the majority of chromatin remodeller binding occurs at histone ‘marked’ chromatin, we next examined the enrichment of remodeller binding across gene regulatory features. We specifically targeted ‘actively marked’ and ‘repressively marked’ regulatory elements that are ‘marked’ with a composite of histone modifications. To define these regulatory chromatin features, we used annotated transcription start sites (TSS) from GENCODE and classified the promoter as a 2 kb region surrounding the TSS. We then separated these promoters according to the histone modifications present in LNCaP cells to classify them as active (H3K4me3 and H3K27ac), facultative repressed (H3K27me3), constitutively repressed (H3K9me3) and bivalent (H3K4me3 and H3K27me3) (Fig. 2b). Additionally, we defined putative enhancers as being at least 2 kb away from an annotated TSS and classified them as either active (H3K4me1, H3K27ac, p300 and DNaseI accessible; Additional file 1: Figure S3C) or poised (H3K4me1) (Fig. 2b), and analysed the chromatin remodellers at these regions.
We assessed whether each chromatin remodeller was bound at the gene regulatory features defined above, more (enriched) or less (depleted) than expected by chance using the Genome Association Tester (GAT) [36]. All chromatin remodeller proteins were significantly enriched at ‘actively marked’ regulatory elements, (Fig. 2c–e; Additional file 1: Figure S4A). However, it was notable that they could be stratified into two distinct groups based on the level of enrichment at these regions. BRG1, SNF2H, CHD3 and CHD4, herein called Group 1, were significantly more enriched compared to BRM, INO80, SNF2L and CHD1, herein called Group 2 (Student’s unpaired T test, active promoters p = 0.04; active enhancers p = 0.0001; poised enhancers p = 0.0004). At gene regulatory regions marked by ‘repressively marked’ epigenetic features, the facultative and constitutively repressed promoters, the majority of Group 1 remodellers were not significantly enriched, in contrast to Group 2 (Fig. 2f, g). Concomitantly, there was a significant difference between the mean enrichment scores of Group 1 and Group 2 remodellers at repressed regions (Student’s unpaired T-test, facultative repressed promoters p = 0.016; constitutively repressed promoters p = 0.02). However, statistical enrichment at a particular genomic annotated feature does not determine the extent of direct overlap of remodeller binding sites at these features. By examining the direct overlap within each group of remodellers, we found that there are common and unique binding sites (Additional file 1: Figure S4B-C); for Group 1 binding sites, only 22.6% contained two or more Group 1 remodellers (Additional file 1: Figure S4D), and for Group 2 binding sites, only 17.3% contained more than one Group 2 remodellers (Additional file 1: Figure S4E). Example regions bound by all Group 1 remodellers are illustrated upstream of the SDC1 promoter and at the COPS4 promoter, and adjacent to exon 2 of the COPS4 gene for Group 2 (Fig. 2h). Therefore, while each group of remodellers was enriched at the same genomic features (i.e. binding occurs higher than expected by chance), there are only a small percentage of regions where all the remodellers bind together, highlighting varied roles for these remodellers.
We next examined bivalent chromatin, which exhibits both active and repressive epigenetic features. Given the above results, we hypothesised that both Group 1 and Group 2 remodellers would be enriched equally at bivalent chromatin. We indeed found that all remodellers were significantly enriched at bivalent promoters, and notably, we observed that Group 2 remodellers had significantly higher enrichment compared to Group 1 (Fig. 2i; Student’s unpaired T-test, p =0.0004). This is in line with previous research showing low enrichment of BRG1 and CHD4 at bivalent chromatin [27].
We next performed the equivalent enrichment analysis using the chromHMM 15 state model (see “Methods”). Again, Group 1 remodellers had a mean enrichment score significantly higher than Group 2 at ‘actively marked’ regions and a lower score for ‘repressively marked’ regions (Additional file 1: Figure S5A-I), with the exception of intragenic enhancers. All remodellers were enriched across all three bivalent states in the model (bivalent promoter, flanking bivalent promoter and bivalent enhancer), but there was no significant difference between the Group 1 and Group 2 (Additional file 1: Figure S5 J-L; Student’s unpaired T-test, p =0.118). The chromHMM model also defines states of active transcription, and we found that there is no significant enrichment of remodellers in the regions flanking active transcription and all remodellers were significantly depleted from regions of active transcription (Additional file 1: Figure S5M–O). Moreover, as there is also no significant enrichment at intragenic enhancers (Additional file 1: Figure S5D), we speculated that sites of active transcription (including the intragenic enhancers) may be due to the highly dynamic and transient nature of transcription, preventing a stable signal of the chromatin remodellers from being detected within these regions.
An interesting exception to the stratification of the remodellers described above is the presence of CHD1 at ‘actively marked’ promoters. At both annotated ‘actively marked’ promoters (Fig. 2c) as well as promoters defined by the chromHMM segmentation (Additional file 1: Figure S5A), CHD1 is found to be significantly enriched at a level comparable to the Group 1 remodellers. As CHD1 does not display the same level of significant enrichment at any other ‘actively marked’ regulatory regions, we speculate that non-epigenetic factors may be driving this high level of promoter binding.
We then took a second approach where we tested the average distribution of the ChIP-seq signal of key gene regulatory histone modifications: H3K4me3, H3K4me1, H3K27me3 and H3K9me3, across the remodeller binding sites (Additional file 1: Figure S6A–D). Our results demonstrated that the active histone marks, H3K4me3 and H3K4me1, displayed a higher average signal across the binding sites of Group 1 remodellers, and repressive histone marks exhibited a higher average signal across Group 2 remodellers. This further confirms the association of Group 1 with ‘actively marked’ regions and Group 2 with ‘repressively marked’ regions. Additionally, at ‘actively marked’ promoters and bivalent promoters defined by these histone modifications, the Pearson correlation coefficient between the remodellers in Group 1 was higher compared to Group 2 (Additional file 1: Figure S6E–F), indicating that there is more similarity in the binding pattern between Group 1 compared to Group 2 at active regions. Furthermore, we found the converse was true for repressed promoters (Additional file 1: Figure S6G–H). Taken together, these data suggest that while all remodellers are associated with ‘actively marked’ chromatin states, Group 1 remodellers have a more pronounced role at these ‘actively marked’ regions, while Group 2 remodellers play a greater role at ‘repressively marked’ regions.
Chromatin remodellers bind to AT-rich DNA and are found at unmethylated regions
We were interested to know whether DNA methylation or the DNA sequence within the remodeller binding site could also stratify the chromatin remodellers into Group 1 and 2. Although chromatin remodellers are responsible for positioning nucleosomes, the genome-wide patterning of nucleosomes is also determined in part by the DNA sequence, where there is a preference for DNA rich in ApT and TpA dinucleotides that are able to bend more sharply around the histone octamer [1, 37, 38]. We calculated the density of all four nucleotides within each chromatin remodeller binding site and found a higher density of A and T nucleotides within the remodeller binding sites for all of the remodellers, and no difference between Group 1 and Group 2 (Additional file 1: Figure S7A-B). Additionally, the ApT and TpA dinucleotides within Group 1 and Group 2 remodeller binding sites occur as frequently as all other dinucleotides in the genome, except for CpG dinucleotides (Additional file 1: Figure S7C–D). Together, this suggests that while intrinsic nucleosome positioning is determined in part by sequence composition, chromatin remodeller nucleosome targeting is not dependent on overall DNA sequence composition, nor does the sequence stratify remodellers into Group 1 and Group 2.
DNA methylation of cytosine residues occurs primarily in a CpG context and has a complex role. At promoters, it is associated with chromatin compaction, but in gene bodies it is associated with active expression [39–44]. We next examined whether DNA methylation was present at the remodeller binding sites. Overall, we detected very low levels of DNA methylation across all remodeller binding sites (Additional file 1: Figure S7E). However, we found that remodellers bound to regions with a higher CpG density had less DNA methylation within their binding sites and conversely regions with lower overall CpG density displayed higher the levels of DNA methylation (Additional file 1: Figure S7E–F).
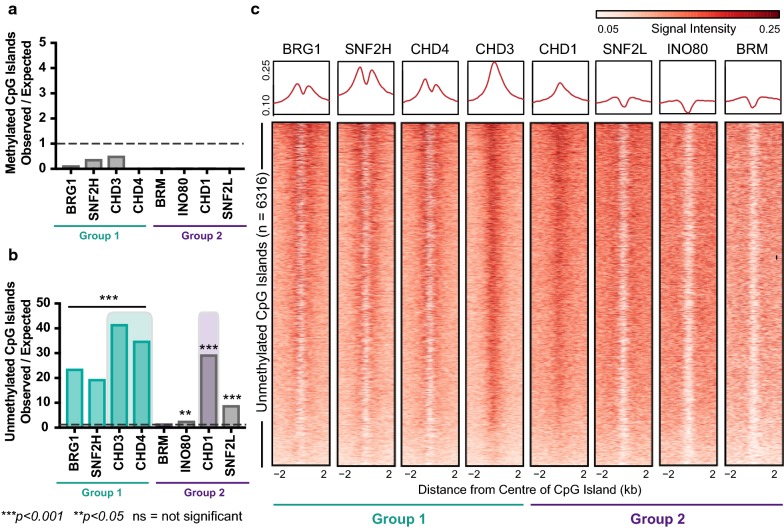
CpG islands are defined as regions of DNA with high CpG density stretching beyond 500 bp [45]. A list of annotated CpG islands was obtained from UCSC, and promoter CpG islands defined as the intersection between CpG islands and Ensembl transcription start sites (see “Methods”). These were then divided into methylated (average methylation across the CpG island above 50%) or unmethylated using whole-genome bisulphite sequencing (WGBS) data generated from LNCaP cells [46]. There was a small, but significant difference in the CpG density of the methylated and unmethylated CpG islands (one-way ANOVA, p <0.001; Additional file 1: Figure S7G). By performing GAT statistical enrichment analysis, we found that methylated promoter CpG islands are depleted of all chromatin remodellers (Fig. 3a). This was due to only 12 total remodeller peaks (the sum of: one peak for BRG1, six peaks for CHD3 and five peaks for SNF2H) overlapping methylated CpG islands. All eight remodellers displayed varying levels of positive enrichment at unmethylated promoter CpG islands (Fig. 3b), with the CHD family (both CHD3 and CHD4 from Group 1 and CHD1 from Group 2) showing the highest level of enrichment. We next examined the ChIP-seq signal intensity of the remodellers across unmethylated promoter CpG islands, extending to ± 2 kb from the centre of the island. Interestingly, we noted that there was variation in the binding pattern of remodellers. CHD1 and CHD3 bound closer to the centre of the island, BRG1, CHD4 and SNF2H bound at the borders and BRM, INO80 and SNF2L were depleted in the centre (Fig. 3c). This suggested that chromatin remodellers are associated with maintaining chromatin organisation at different parts of promoter CpG islands, and we propose that they work in coordination to maintain the accessibility status of the island; however, the remodellers do not stratify into Group 1 and Group 2 based on whether the island is methylated or unmethylated.
Fig. 3.
Chromatin remodellers do not stratify into Group 1 and Group 2 at CpG islands. a, b Enrichment of chromatin remodellers at methylated and unmethylated CpG islands. Significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p value, ***p < 0.001 or **p < 0.05. c Heatmap of remodeller ChIP-seq signal at unmethylated promoter CpG islands, ± 2 kb from the centre of the island, with average profile plots shown above the heatmap
Chromatin preferentially occupied by Group 1 remodellers is nucleosome depleted
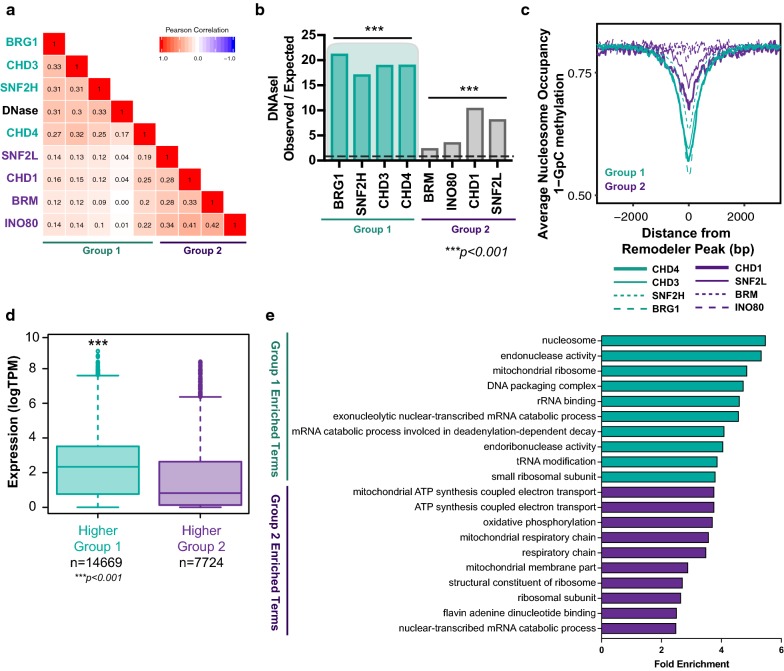
Nucleosome occupancy at regulatory features naturally represses transcription by creating an inhibitory structure, and therefore, regions of ‘actively marked’ chromatin are typically depleted of nucleosomes and have increased rates of nucleosome turnover and greater spacing between nucleosomes. Since Group 1 remodellers are enriched at ‘actively marked’ chromatin regulatory regions, we next asked whether the genomic regions corresponding to Group 1 localisation are depleted of nucleosomes. First, we examined sites of nucleosome depletion from DNaseI hypersensitivity data from LNCaP cells [47]. Analysis of the correlation between chromatin remodeller presence and DNaseI sites revealed that Group 1 remodeller occupancy is highly correlated with DNaseI hypersensitive sites (Fig. 4a), in addition to DNaseI sites being highly enriched for Group 1 remodellers (Fig. 4b). Given that DNaseI digestion does not detect all sites of ‘open’ chromatin [48], we assessed the link between chromatin remodeller binding and accessible chromatin using an alternative method—nucleosome occupancy and methylation sequencing (NOMe-seq). NOMe-seq [48–50] uses exogenous methylation of GpC dinucleotides to identify accessible chromatin regions devoid of nucleosomes or tight binding transcription factors. Importantly, this method also avoids cleavage bias found in DNaseI data [51]. We found that GpC methylation was higher at Group 1 remodeller binding sites compared to Group 2, suggesting that genomic regions occupied by Group 1 remodellers contain fewer nucleosomes than those regions occupied by Group 2 remodellers (Fig. 4c). These data are consistent with the preference of Group 1 remodellers for ‘actively marked’ chromatin states and for Group 2 remodellers to localise to regions of more ‘repressively marked’ chromatin.
Fig. 4.
Group 1 chromatin remodellers highly overlap accessible DNA regions. a The ChIP-seq signal of the chromatin remodellers and the signal from the ENCODE DNAseI peaks were determined at DNAseI called sites, and then Pearson’s correlation scores were calculated between each data set and compiled into a matrix. The matrix was ordered by hierarchical clustering using the distance between correlation scores. This resulted in the Group 1 remodellers clustering with the DNAseI data and Group 2 remodellers clustering separately. Samples are presented in the same order on both the x- and y-axis. b Enrichment of chromatin remodeller binding sites at DNAse1 sites. Significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p-value, ***p < 0.001 or **p < 0.05. c Nucleosome occupancy defined from NOMe-seq ± 2 kb from the centre of chromatin remodeller binding sites. d Boxplot of the relationship between gene expression (TPM) and remodeller binding at promoters of genes that are expressed in LNCaP cells. Expressed genes were divided into genes that have a higher enrichment of Group 1 remodellers (green) at their promoter and those that have a higher enrichment of Group 2 remodellers (purple) at their promoter. The expression of Group 1 is significantly higher (p < 0.001, Wilcox rank sum, one-tailed) than Group 2. e Enrichment scores of the top 10 GO terms for promoters with a higher signal of Group 1 remodellers (Green) or a higher signal of Group 2 remodellers (purple)
Accessible chromatin is associated with active regulatory elements and gene activity. Therefore, to determine whether chromatin remodeller binding was correlated with gene expression in LNCaP cells, the ChIP-seq read counts of all Group 1 and Group 2 remodellers were pooled. Then, the difference between the read counts (signal) for Group 1 remodellers compared to Group 2 remodellers at the promoters of expressed genes was calculated. Genes were separated into those with a higher level of Group 1 chromatin remodeller binding and those with a higher level of Group 2 remodeller binding at their promoters and the level of expression for each group plotted as transcripts per million reads (logTPM). Of the 22,393 genes that were actively expressed in LNCaP cells, 14,669 had a higher signal for Group 1 chromatin remodellers and 7724 had a higher signal for Group 2 chromatin remodellers at their promoters. Genes with a higher level of Group 1 remodellers had higher expression compared to those that had a higher expression of Group 2 remodellers (Fig. 4d), supporting the conclusion that Group 1 remodellers are associated with increased gene activity. We performed a gene ontology (GO) analysis of active genes that more highly bound by either Group 1 or Group 2 remodellers using GREAT [52]. For gene promoters with higher Group 1 binding, the most significant GO terms enriched were related to the nucleosome, transcriptional processes such as rRNA binding and tRNA modification, and mRNA processing (Fig. 4e). Enriched GO terms for active genes with a higher Group 2 remodeller signal include those associated with mitochondrial processes such as ATP synthesis and respiratory chain activity (Fig. 4e). Together, this demonstrates that the Group 1 and Group 2 remodellers maintain and associate with the regulation of different cellular pathways.
Group 1 and Group 2 remodellers are defined by chromatin architectural features
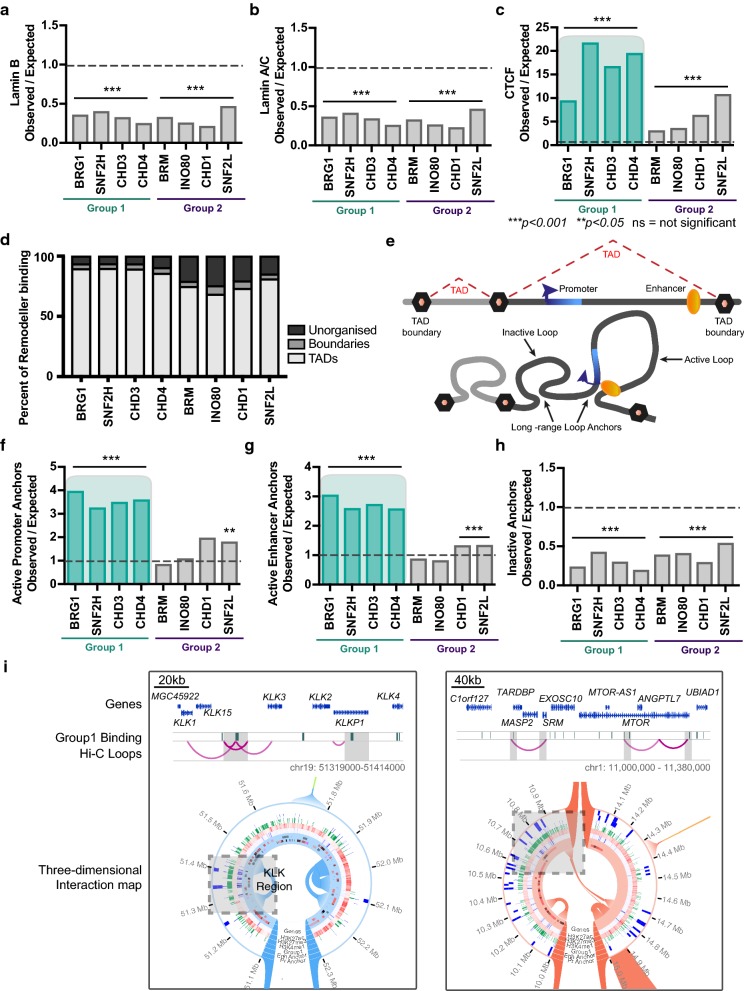
The above results determined the positioning of remodellers at regulatory features in the context of histone post-translational modifications and DNA methylation in linear genomic space. However, chromatin is organised into a higher order by the formation of topologically associated domains (TADs) [53] that function to bring distal regulatory regions together for coordinated gene expression. Architectural proteins Lamin A/C, Lamin B and CCCTC-binding factor (CTCF) contribute to the formation of this three-dimensional chromatin structure. Lamin B and Lamin A/C configure the chromatin into Lamin-associated domains (LADs), with low accessibility [54]. LADs occur at the nuclear periphery, are heavily marked with H3K9me3 and are typically associated with gene repression. Therefore, we hypothesised that Group 2 remodeller binding will co-localise with LADs. Instead, using ChIP-seq data of Lamin B and Lamin A/C binding sites [55], we found all of the chromatin remodellers were significantly depleted (p < 0.001), suggesting that the tightly compacted chromatin at the nuclear periphery does not require remodeller binding to maintain its repressive state (Fig. 5a, b). Similarly, CTCF has diverse roles in regulating chromatin and the epigenome, and it is well established that nucleosomes directly flanking CTCF-binding sites are highly ordered, displaying a strong phasing pattern [48, 49, 56]. Previous research has shown that the ISWI remodeller protein SNF2H, but not the alternative ISWI ATPase SNF2L, contributes to this highly ordered array of nucleosomes around sites of CTCF occupancy when bound to chromatin [57]. Using CTCF ChIP-seq data, we found that similar to the ‘actively marked’ DNA regulatory elements, all remodelers were enriched at CTCF-binding sites, but Group 1 remodellers were more highly enriched compared to Group 2 (Student’s unpaired T-test, p =0.018), with SNF2H displaying the highest level of enrichment (Fig. 5c).
Fig. 5.
Chromatin remodeller binding at chromatin architectural features. a–c Enrichment of chromatin remodeller binding sites at the binding sites of key chromatin architectural proteins as defined by their ChIP-seq-binding profiles. Significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p-value, ***p < 0.001 or **p <0.05. d Percentage of remodeller binding sites in TADs, TAD boundaries and unorganised chromatin. e Schematic diagram depicting TADs in linear and 3D space with an inactive loop condensing chromatin and an active loop bringing an enhancer (orange oval) and promoter (blue flag) within close proximity. f–h Enrichment of chromatin remodeller binding at anchor points of chromatin loops defined by Hi-C and histone modification data as containing an active promoter or enhancer or neither. Significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p-value, ***p <0.001 or **p < 0.05. i The KLK region on chromosome 19 and a regions on chromosome 1 containing active loops (pink arcs) with Group 1 remodeller binding sites at the long-range loop anchors shaded in grey, with complementary three-dimensional view from Rondo (Rondo.ws)
We next examined the distribution of chromatin remodellers within the context of higher-order three-dimensional chromatin structure itself using Hi-C data from LNCaP cells [29]. We divided chromatin into TADs (85.4% of the genome), TAD boundaries (2.6% of the genome) and unorganised chromatin (12.0% of the genome; Additional file 1: Figure S8A) and examined the distribution of Group 1 and Group 2 remodellers within each category. Remarkably, we found that 90% to 94% of Group 1 remodellers and 75% to 85% of Group 2 remodeller binding sites were located within TADs or at TAD boundaries, indicating that chromatin remodellers preferentially localised to sites of highly organised chromatin (Fig. 5d). We performed the GAT analysis and found a small but positive and significant enrichment of all eight of the remodellers at both TADs and TAD boundaries (Additional file 1: Figure S8B–C), which was expected as the majority of the genome is within these organised chromatin structures.
Within TADs, chromatin forms DNA loops to facilitate interactions such as those between enhancers and promoters (Fig. 5e), and these loops are in part affected by the positioning of nucleosomes [58, 59]. Our previous work has shown cancer-specific anchor points of long-range chromatin ‘loops’ are enriched for enhancers and promoters and contain a remodelled epigenetic signature, where ‘active’ marks H3K4me1, H3K4me3 and H3K27ac are increased [29]. Whether chromatin remodellers are enriched at the anchor points of these chromatin loops remains unknown. Our linear data suggest ‘active’ chromatin ‘loops’ that bring together promoters and enhancers would be more enriched for Group 1 remodellers compared to Group 2. To test this, we separated the anchor points of the long-range chromatin loops into those that contained at least one active promoter or active enhancer using the chromHMM chromatin state data (Type A anchors, ~ 20% of all anchors) and a second group that did not contain either of these regulatory elements (Type B anchors, ~ 80% of all anchors). We found that at Type A loop anchors, the remodellers continue to stratify into Group 1 and Group 2, with Group 1 chromatin remodellers significantly (p <0.001) more enriched than Group 2 (Fig. 5f, g). Interestingly, Type B loop anchors that were devoid of an active promoter or enhancer were significantly depleted (p < 0.001) of all chromatin remodellers (Fig. 5h), suggesting that these anchors do not require ongoing chromatin remodelling activity. Examples of chromatin loops with Group 1 remodeller binding and active regulatory elements are found at the KLK locus on chromosome 19, and a gene dense region on chromosome 1 (Fig. 5i). Together, this demonstrates a role for all Group 1 remodellers in chromatin three-dimensional architecture.
Discussion
Chromatin stores important epigenetic information that is inherited across cell divisions. ‘Reader’ proteins, such as ATP-dependent chromatin remodelling complexes, are required to interpret and modify chromatin when required. While several studies demonstrate that chromatin remodeller complexes have strong ties with regulatory chromatin [15, 23, 25, 27, 60, 61], little is known about how these complexes function as a group and interact with the different layers of the epigenome. We sought to address this interrelationship through the use of publically available data by examining the role of eight different chromatin remodeller proteins in LNCaP prostate cancer cells and integrating their binding profiles with multiple epigenomic data sets. This is the first extensive analysis of multiple chromatin remodellers together, with several layers of the epigenome at the same time in human cells. We found that there is substantial overlap in the binding sites of the remodellers at regulatory regions of the genome. Specifically, we found that the eight remodellers can be stratified into two distinct groups: Group 1 that was more highly enriched at ‘actively marked’ chromatin regions and Group 2 at ‘repressively marked’ chromatin regions (Fig. 6).
Fig. 6.
Schematic of chromatin remodeller families. Chromatin remodellers are divided firstly by their known structural families, which are then arranged by the groups they stratify into based on their binding enrichment from the epigenome
Our data revealed that the grouping of chromatin remodellers is remarkably consistent across all of the ‘actively marked’ and ‘repressively marked’ epigenetic features we examined but intriguingly, independent of the core modification of DNA methylation. Segregation into these two groups persists at both defined DNA regulatory elements and chromHMM chromatin states, which are a composite of histone modification marks. While Group 1 remodellers have a significantly higher enrichment than Group 2 at ‘actively marked’ regions, it is still noteworthy that all remodellers have some level of enrichment at these regions, which highlights the dynamic and complex nature of active chromatin. Previous research also demonstrates a role of Group 2 remodellers at active chromatin in embryonic mouse cells such as INO80 in maintaining open chromatin in pluripotency genes [62] and CHD1 and its role in RNA polII stalling in active gene expression [63]. We also note that we find some divergent results for the remodeler CHD1. A previous study in mouse embryonic stem cells found CHD1 to only be enriched at active promoters [27]. In our study, CHD1 had an equivalent level of enrichment at active promoters as the Group 1 remodellers, while still being enriched at ‘repressively marked’ chromatin together with the other Group 2 remodellers. These differences may be due to the different cell types, embryonic cells versus somatic cells, or may reflect differences between normal versus cancer cells. It will be interesting to interrogate these differences in future studies.
It was surprising that upon investigation of the direct overlap within Group 1 and Group 2 remodeller binding sites, we found less than 6% were in common for the entire group. This suggests that, while remodellers within the same group have high statistical enrichment at the same class of regulatory elements, they do not always bind to the exact same genomic region. For example, Group 1 remodellers demonstrate a preference for ‘actively marked’ promoters as a collective group, but they often localise to their own subset of all active promoters, showing that each remodeller potentially has a distinct and unique role. Additional data will be needed to further resolve the types of complexes each of the core remodeller proteins is capable of forming. Expansion of the existing ChIP-seq data to include various accessory subunits will help refine this analysis, in combination with fine-tuning existing definitions of DNA regulatory elements and chromHMM states. Broadly defined, the further subdivision of DNA regulatory elements and states will enable subtyping and will provide additional details to determine under which conditions, states and combinations these remodeller proteins act in a coordinated or antagonistic manner.
For the purposes of this study, we did not include all known histone modifications in our analysis, including methylation of H4K20 and H4K16ac [64–66]. Thus, the proportion of remodeller binding found at ‘unmarked’ regions may contain histone modifications not present in our analysis. However, the key marks for defining DNA regulatory elements and genes were included and therefore provide a comprehensive view of key gene regulatory chromatin features, and moreover, less than 20% of binding sites fell within the ‘unmarked’ regions. As more histone modification data become available, such as histone variants and histone modifications with a structural role, it will be interesting to determine whether these also stratify the remodellers in a similar fashion.
Interestingly, we found that remodellers consistently segregated into Group 1 and Group 2 at architectural chromatin features, such as chromatin loop anchors and CTCF sites, highlighting that chromatin remodellers are also associated with higher-order chromatin architecture. Previous to this study, BRG1 was the sole remodeller that had a demonstrated role in chromatin architecture and a well-established role in maintaining enhancer–promoter interactions [11, 60, 67]. Additionally, in MCF10A cells, BRG1 increases the stability of TAD boundaries to strengthen enhancer–promoter chromatin loops and maintain the established patterns of gene expression. Subsequent loss of BRG1 binding weakens these interactions, concomitant with a down-regulation of gene expression [68]. Our results infer that in fact, all Group 1 remodellers—BRG1, SNF2H, CHD3 and CHD4—could have individual or a combinatorial role at chromatin loop anchors that contain active promoters or enhancers. When we examined the remodellers at LADs, however, we found them to be depleted of all remodellers. We also investigated the association with the architectural protein CTCF and found that Group 1 remodellers are more enriched at CTCF-binding sites compared to Group 2. Together, our data suggest Group 1 remodellers have a prominent role at ‘active’ chromatin loops, whereas ‘inactive’ chromatin loops do not require remodeller binding to maintain their repressed state.
Our finding that the distribution of chromatin remodellers across CpG islands follows three distinct patterns (BRG1, SNF2H and CHD4 at borders; CHD3 and CHD1 in the centre and SNF2L, INO80 and BRM deplete from the centre of the island) suggests that there are different mechanisms at play for maintaining nucleosome positions at these regions. It was surprising that neither CHD3 nor CHD4 showed any level of enrichment at methylated CpG islands. These remodellers form a key part of the NuRD complex that also contains MBD2, which prefers hypermethylated promoters [42]. It is possible that once the DNA of any given genomic region becomes methylated and the chromatin is compacted, it may no longer require the remodelling complex to stay bound to the chromatin.
Previous studies that have examined two to three remodeller proteins show consistency with our findings. The overlap of BRG1 and CHD4 has previously been reported in various cell types, where they were reported to have opposing control over regulatory chromatin [23, 26, 69, 70]. For example in mice, BRG1, CHD4 and SNF2H have extensive overlap in their binding patterns and it has been implied that the sequential order of their binding is important for their correct function [23]. In our data, we found that these remodeller proteins were also enriched at the same genomic features, suggesting they may also have opposing functions in LNCaP cells. Additionally, we found overlapping enrichment patterns for CHD1 and ISWI remodeller SNF2L, which in yeast have been reported to have both competing and coordinated functions. CHD1 and SNF2L are responsible for maintaining the phasing of nucleosomes at promoters, but compete for different nucleosome spatial arrangements, impacting the kinetics of gene activation [15, 24, 25, 71]. CHD1 and SNF2L also work together at gene bodies where they are thought to maintain chromatin integrity during transcription elongation by preventing histone exchange during nucleosome turnover [72–74]. Moreover, there has been report of overlapping activity between the ATPase subunits of the NuRD remodelling complex, CHD3 and CHD4 [20], which also occurred in our data.
Functional studies in human cancers have demonstrated the integrated relationships of chromatin remodellers, through the identification of synthetic–lethal relationships. Synthetic–lethal relationships occur where the cancer develops a loss of function for one protein that creates a dependency on another protein. Synthetic–lethal relationships exist for ATPases, BRG1 and BRM, in triple negative breast cancer, for accessory subunits of the SWI/SNF complex, ARID1A and ARID1B, in colorectal cancer and CHD1 with transcriptional regulator PTEN in prostate cancer [75–77]. These instances of the relationship between remodellers and their function highlight the importance of studying multiple remodellers together to provide a greater understanding of the overall mechanism of chromatin remodelling and how a chromatin state is established. It is unclear from our data how much of the remodeller binding overlap provides cooperative or antagonist function; moreover, our study does not determine the extent of competition between the remodellers for nucleosome binding. Mechanistic studies are now required, such as using gene editing approaches and in different cell model systems, to further dissect the importance and or redundancy of individual remodellers and their potential functional role in regulating chromatin, and broaden the applicability of findings to other cell types.
Conclusions
In summary, our results reveal previously unknown relationships between the remodellers, and with both the two-dimensional and three-dimensional epigenome. We propose that chromatin remodellers should be examined in the context of the different classes of remodellers we identified as Group 1 or Group 2, and not solely with consideration to existing structural families or in isolation (Fig. 6). These observations may inform decisions for future work that studies chromatin remodeller function and provides a more complete picture of chromatin remodelling action.
Methods
ChIP-seq assay and data
LNCaP chromatin remodeller ChIP-seq data are from Ye et al. [28]: GEO accession GSE72690. Histone modification LNCaP ChIP-seq data (H3K4me3, H3K4me1, H3K27ac and H3K27me3) are from Taberlay et al. [29]: GEO accession GSE73785, and Bert et al. [34]: GSE38685. CTCF ChIP-seq data is from Bert et al. [34]: GSE38685. H3K9me3, H3K36me3, Lamin B and Lamin A/C ChIP-seq data are from Du et al. [55], GSE98732. p300 ChIP-seq data is from Wang et al. [78]: GSE27824. RNA polII ChIP-seq data is from Tan et al. [79]: GSE28264. All ChIP-seq data sets were processed as previously described in Bert et al. [34], Taberlay et al. [29], Du et al. [55] and Lund et al. [80]. Histone modification and chromatin remodeller peaks were called using MACS2 [81] and Lamin domains called with the enriched domain detector (EDD) [80]. Two ChIP-seq input data sets were provided by Ye et al. [28], which were merged for calling remodeller peaks. Then, MACS2 [81] was used to call peaks on each individual input data set; any peaks overlapping with the chromatin remodellers were removed from the remodeller data sets.
Hi-C data
LNCaP Hi-C data is from Taberlay et al. [29]; GSE73785. Hi-C data were processed through the NGSANE framework v0.5.2 [82] as previously described in Taberlay et al. [29]. TADs were identified with the ‘domain-caller’ pipeline [53] as described in Taberlay et al. [29]. TADs and TAD boundaries were assessed at 40 kb resolution. The percentage of the genome covered by TADs and TAD boundaries was calculated by accumulating the number of base pairs within TADs or boundaries, divided by 3.095 × 106 and then multiplied by 100. Chromatin loops were called from contact count matrices for 10 kb resolution using a custom adaptation of Fit-Hi-C (contained in NGSANE; Buske et al. [82]) supplying iteratively corrected bias offsets calculated through HiCorrector v1.1 [83]. Chromatin loops were visualised in the WashU Epigenomics Browser [84] and Rondo (rondo.ws).
RNA-seq data
LNCaP RNA-seq data are from Taberlay et al. [29]: GSE73785, and processed clinical prostate tumour RNA-seq was downloaded from TCGA (cancergenome.nih.gov). LNCaP RNA-seq data (n = 3) were processed as described in Taberlay et al. [29]. To determine chromatin remodeller gene expression, reads mapped to hg19/GRCh37 where counted into genes using featureCounts [85] program and GENCODE v19 used as a reference transcriptome to determine the transcript per million read (TPM) value and biological triplicates were averaged. Processed RNA-seq data (n = 486 tumours) from the TCGA prostate adenocarcinoma cohort were averaged to determine chromatin remodeller expression in clinical prostate cancer samples. The log mean values for each remodeller were plotted as cancer versus normal between the cell lines (logTPM) and the normal and tumour TCGA data sets (logRKPM), along with the linear regression line of best fit. Pearson’s correlation coefficients were calculated in R.
Chromatin accessibility data
DNaseI data is from Thurman et al. [47], and processed data were downloaded from the ENCODE data portal (encodeproject.org/). DNaseI sites of accessibility from two biological replicates overlapped and the intersection from both replicates were used for downstream analysis (see Remodeller enrichment analysis). NOMe-seq data are from Valdes-Mora et al. [86]: GSE76334. NOMe-seq data were processed as previously described in Valdes-Mora et al. [86]. NOMe GpC methylation levels within remodeller binding sites were defined by first computing the methylation ratio of all GCH sites with greater than 5× coverage and then calculating the mean methylation score within each remodeller binding site. Methylation density of remodeller binding sites was plotted in R. Visualisation of nucleosome occupancy at remodeller binding sites was created using ‘methylationPlotRegions’ from the aaRon package in R, ± 3000 bp from the centre of the remodeller binding site.
DNA methylation data
WGBS sequencing data is from Pidsley et al. [46]: GSE86833. WGBS libraries were processed as previously described Pidsley et al. [46]. CpG islands and Ensembl gene coordinates were downloaded from the UCSC genome browser [87]. Promoter CpG islands were defined as the intersection between CpG islands and the 5′ ends of Ensembl genes. The promoter CpG islands were split into 40 equally sized bins and the average methylation score calculated from the WGBS data using ‘ScoreMatrixList’ from the genomation package in R. Methylated CpG islands were defined as having an average methylation score above 50%. DNA methylation levels within remodeller binding sites were defined by first computing the methylation ratio of all cytosines with greater than 5× coverage and then calculating the mean methylation score within each remodeller binding site. CpG density of methylated and unmethylated CpG islands was calculated in R. Violin and boxplots plots were created in ggplot2 in R. Remodeller binding sites were overlapped with the methylated and unmethylated islands using the GRanges package in R.
ChromHMM segmentation based on Roadmap epigenomics
The Roadmap Epigenomics chromHMM model was based on five core histone modifications (H3K4me3, H3K4me1, H3K36me3, H3K27me3, H3K9me3) and trained on 60 epigenomes [88] to categorise the genome into 15 chromatin states. There were seven chromatin states associated with ‘active’ chromatin; active promoter, flanking active promoter, transcription at 5′ and 3′ ends of a gene, strong transcription, weak transcription, active intragenic enhancers, active intergenic enhancers and poised enhancers. Three states were associated with bivalent chromatin: bivalent promoter, flanking bivalent promoter and bivalent enhancers. There were four states associated with ‘repressive’ chromatin: zinc finger genes and repeats, heterochromatin, strong polycomb and weak polycomb. The 15th state was ‘unmarked’ chromatin that did not contain any of the histone modifications in the core data set. This model was applied to histone modification ChIP-seq data from LNCaP cells using the chromHMM program (v1.10) [35], and the chromatin states were collapsed into ‘active’, ‘repressed’, ‘bivalent’ and ‘unmarked’.
Chromatin remodeller enrichment and correlation analyses
To determine whether chromatin remodeller binding sites were enriched at the site of a specific chromatin factor or chromatin regulatory element (histone modifications, CTCF sites, LADs, DNaseI sites, chromatin loop anchors, TADs, TAD boundaries, methylated and unmethylated CpG islands and chromHMM states), we used the genome association tester (GAT; v1.0) [36]. The observed-over-expected fold change and statistical significance were calculated with 10,000 iterations and determine significant if the p value was less than 0.05 or 0.001. The difference between the means of the observed/expected enrichment scores from Group 1 and Group 2 remodellers was compared using the unpaired Student’s T-test in R.
We defined the percentage of overlap between each of the remodellers, and the remodellers with TADs and TAD boundaries by intersecting the peaks identified from the ChIP-seq and Hi-C data. Histone modification average signal plots over chromatin remodeller peaks and the heatmaps of putative active enhancers and chromatin remodeller signal over CpG islands and promoters were created with SeqPlots [89] and deepTools2 [90]. Histone marks were plotted ± 2 kb from the centre of the remodeller binding sites. At CpG islands remodeller ChIP-seq signal for both average plots and heatmaps were plotted ± 2 kb from the centre of the island. Each row of the heatmap is an individual CpG island and displays the remodeller ChIP-seq signal, sorted by the average signal across all remodellers in decreasing order. Pearson’s correlation matrixes of remodellers at promoters and DNaseI sites were calculated in R.
Gene ontology enrichment
Gene promoters were defined as 2 kb surrounding the TSS of expressed genes. Read counts of the chromatin remodeller ChIP-seq data within the promoters regions of expressed genes were calculated and all Group 1 remodellers merged and separately Group 2 remodellers merged. Subtracting the total read counts of Group 2 remodellers from Group 1 was used to define which promoters had a higher signal for Group 1 and which had a higher signal for Group 2. The promoters assigned to each group were analysed for enrichment of gene ontology terms using GREAT [52], using the whole genome as background and assigned to the single nearest gene. GO terms reported are the top 10 most significant from the Molecular Function, Biological Process or Cellular Component gene set, with at least 25 observed genes in the data set.
Additional file
Additional file 1: Figure S1. A) Gene expression of each remodeller protein in LNCaP cells from RNA-seq (mean and SE; n = 3). B) Gene expression of remodeller proteins in TCGA data set of 486 prostate epithelial tumours (mean and SE; n = 486). C) Gene expression of each remodeller in PrEC cells from RNA-seq (mean and SE; n = 3), D) Gene expression of remodellers in TCGA data set of normal prostate epithelial tissue (mean and SE; n = 52). E) Scatter plot of logRPKM values for TCGA data versus logTPM values for LNCaP and PrEC RNA-seq data. The PrEC comparison for PrEC and normal TCGA is shown in green, and the LNCaP compared to TCGA tumours is shown in red. Lines are linear regression line of best fit, and Pearson’s correlation coefficient between the two normal data sets (cor = 0.670013) and the two cancer data sets (cor = 0.8375552, SNF2L is excluded as an outlier) is shown under the plot. Figure S2. A–H) Histograms of chromatin remodeller binding sites, binned at 150 bp widths. Vertical line indicates the point of 750 bp (~ 5 nucleosomes). Figure S3. A) Heat map of ChromHMM emission profile based on the learned model from the epigenome roadmap. B) Heat map of chromatin state enrichment generated from the chromHMM analysis, where the percentage of each state in the genome is represented in the first column and the remaining columns are the enrichment of each chromatin state over annotated genomic features. C) Heatmap of H3K27ac, H3K4me1, H3K4me3 and p300 ChIP-seq signal and DNaseI signal at putative active enhancers, ± 2.5 kb from the centre of each enhancer, sorted by H3K27ac and H3K4me1 signal. Figure S4. A) Heatmap H3K4me3, RNA polII, and chromatin remodeller ChIP-seq signal and DNaseI signal at refseq-annotated promoters. Signal is plotted ± 2 kb from the transcription start site (TSS) and sorted by H3K4me3 signal. B, D) Venn diagram of chromatin remodeller binding site overlaps for Group 1 and Group 2 remodellers, respectively. C) Percentage of all Group 1 remodeller binding sites that are unique to each Group 1 remodeller and contain multiple Group 1 remodellers. E) Percentage of all Group 2 remodeller binding sites that are unique to each Group 2 remodeller and contain multiple Group 2 remodellers. Figure S5. A–O) Enrichment of chromatin remodeller binding sites across the 15 state chromHMM model based on the Epigenome roadmap (see Methods). Significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p-value, ***p < 0.001 or **p < 0.05. Figure S6. A–D) Genome-wide average distribution of ChIP-seq signals for histone modifications H3K4me3, H3K4me1, H3K27me3 and H3K9me3 ± 2 kb from the centre of chromatin remodeller binding sites. E–H) Pearson’s correlation score matrix of chromatin remodeller ChIP-seq signal at active, bivalent, facultative and constitutive promoters. Each matrix was ordered by hierarchical clustering. Figure S7. A–B) Violin plots of nucleotide frequency within remodeller binding sites for Group 1 and Group 2. C–D) Dinucleotide frequency within chromatin remodeller Group 1 and Group 2 binding sites. E) DNA methylation density within chromatin remodeller binding sites for each remodeller. F) CpG density within chromatin remodellers binding sites for each remodeller protein. G) CpG density of unmethylated CpG islands, methylated CpG islands and the whole genome. A significant difference in CpG density was detected between each of the groups (one-way ANOVA, ***p < 0.001). Figure S8. A) The genome divided into TADs (85.4%), TAD boundaries (2.6%) and unorganised chromatin (12.0%) using TADs and boundaries called to a 40 kb resolution from Hi-C data. B–C) GAT enrichment of chromatin remodellers at TADs and TAD boundaries, where significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p-value, ***p < 0.001 or **p < 0.05.
Authors’ contributions
KAG, SJC and PCT were involved in conceptualisation; KAG, CMG, QD and KS were involved in formal analysis; SJC and PCT were involved in funding acquisition; QD and JS were involved in investigation; KAG was involved in writing—original draft; KAG, MPM, JAK, SJC and PCT were involved in writing—reviewing and editing; KAG and MPM were involved in visualisation; JAK, CS, SJC and PCT were involved in supervision; SJC and PCT are senior authors. All authors read and approved the manuscript.
Acknowledgements
We would like to thank the Garvan Institute of Medical Research for the use of the computing resources.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All sequencing data sets used in this study have been previously published and are available through Gene Expression Omnibus (ncbi.nlm.nih.gov/geo/). GEO accession numbers are as follows: chromatin remodeller ChIP-seq GSE72690; histone modification ChIP-seq data GSE73785, GSE38685, and GSE98732; CTCF ChIP-seq GSE38685; Lamin B and Lamin A/C ChIP-seq GSE98732; p300 ChIP-seq GSE27824; RNA polII ChIP-seq GSE28264; Hi-C GSE73785; RNA-seq GSE73785; NOMe-seq GSE76334; and whole-genome bisulphite sequencing GSE86833. TCGA data are available from the data portal at cancergenome.nih.gov, and ENCODE DNaseI data are available from the ENCODE data portal at encodeproject.org/.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
K.A.G. is supported by an Australian Postgraduate Award (APA) and Research Excellence Award from UNSW Sydney. Q.D. is supported by an APA from UNSW Sydney. S.J.C. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow #1063559. P.C.T. is a NHMRC Career Development Fellow #1109696. This work was supported by grants from Cure Cancer Australia Foundation Project Grant #1060713 to P.C.T. and NHMRC Project Grants #1011447 and #1070418 to S.J.C. and C.S. and #1051757 to S.J.C. and P.C.T.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BRG1
Brahma-related gene 1
- BRM
Brahma
- CHD
chromodomain helicase DNA binding
- chromHMM
chromatin multivariate hidden Markov model
- CTCF
CCCTC-binding factor
- H3K27ac
histone 3 lysine 27 acetylation
- H3K27me3
histone 3 lysine 27 trimethylation
- H3K4me1
histone 3 lysine 4 monomethylation
- H3K4me3
histone 3 lysine 4 trimethylation
- H3K9me3
histone 3 lysine 9 trimethylation
- Hi-C
high-throughput chromosome conformation capture
- INO80
inositol requiring 80 complex
- SNF2
sucrose non-fermenting 2
- SWI/SNF
switch/sucrose non-fermenting
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Giles KA, Taberlay PC. Mutations in Chromatin remodeling factors. Reference Module in Biomedical Sciences. 2018.
- 3.Johnson CN, Adkins NL, Georgel P. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol. 2005;83(4):405–417. doi: 10.1139/o05-115. [DOI] [PubMed] [Google Scholar]
- 4.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15(11):703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 5.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15(2):172–183. doi: 10.1016/S0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 6.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514(7520):102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cajigas I, Leib DE, Cochrane J, Luo H, Swyter KR, Chen S, et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Dev. 2015;142(15):2641–2652. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12(10):1094–1098. doi: 10.1080/15476286.2015.1063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154(3):490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Local A, Huang H, Albuquerque CP, Singh N, Lee AY, Wang W, et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat Genet. 2018;50(1):73–82. doi: 10.1038/s41588-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 14.van Galen P, Viny AD, Ram O, Ryan RJ, Cotton MJ, Donohue L, et al. A multiplexed system for quantitative comparisons of chromatin landscapes. Mol Cell. 2016;61(1):170–180. doi: 10.1016/j.molcel.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zentner GE, Tsukiyama T, Henikoff S. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet. 2013;9(2):e1003317. doi: 10.1371/journal.pgen.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18(7):407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha A, Wittmeyer J, Cairns BR. Mechanisms for nucleosome movement by ATP-dependent chromatin remodeling complexes. Results Probl Cell Differ. 2006;41:127–148. doi: 10.1007/400_005. [DOI] [PubMed] [Google Scholar]
- 18.Skulte KA, Phan L, Clark SJ, Taberlay PC. Chromatin remodeler mutations in human cancers: epigenetic implications. Epigenomics. 2014;6(4):397–414. doi: 10.2217/epi.14.37. [DOI] [PubMed] [Google Scholar]
- 19.Morrison AJ. Genome maintenance functions of the INO80 chromatin remodeller. Philos Trans R Soc Lond Ser B Biol Sci. 2017;372(1731):20160289. doi: 10.1098/rstb.2016.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmeister H, Fuchs A, Erdel F, Pinz S, Grobner-Ferreira R, Bruckmann A, et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017;45(18):10534–10554. doi: 10.1093/nar/gkx711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning BJ, Yusufzai T. The ATP-dependent chromatin remodeling enzymes CHD6, CHD7, and CHD8 exhibit distinct nucleosome binding and remodeling activities. J Biol Chem. 2017;292(28):11927–11936. doi: 10.1074/jbc.M117.779470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618(1–2):30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris SA, Baek S, Sung MH, John S, Wiench M, Johnson TA, et al. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21(1):73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281(24):16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ocampo J, Chereji RV, Eriksson PR, Clark DJ. The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res. 2016;44(10):4625–4635. doi: 10.1093/nar/gkw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis CD, Griffin CT. The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Mol Cell Biol. 2012;32(7):1312–1320. doi: 10.1128/MCB.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Dieuleveult M, Yen K, Hmitou I, Depaux A, Boussouar F, Bou Dargham D, et al. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016;530(7588):113–116. doi: 10.1038/nature16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Z, Chen Z, Sunkel B, Frietze S, Huang TH, Wang Q, et al. Genome-wide analysis reveals positional-nucleosome-oriented binding pattern of pioneer factor FOXA1. Nucleic Acids Res. 2016;44(16):7540–7554. doi: 10.1093/nar/gkw659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taberlay PC, Achinger-Kawecka J, Lun AT, Buske FA, Sabir K, Gould CM, et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 2016;26(6):719–731. doi: 10.1101/gr.201517.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research N The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, et al. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46(8):2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- 32.Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280(51):41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 34.Bert SA, Robinson MD, Strbenac D, Statham AL, Song JZ, Hulf T, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell. 2013;23(1):9–22. doi: 10.1016/j.ccr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Ernst J, Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc. 2017;12(12):2478–2492. doi: 10.1038/nprot.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heger A, Webber C, Goodson M, Ponting CP, Lunter G. GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics. 2013;29(16):2046–2048. doi: 10.1093/bioinformatics/btt343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20(3):267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelfman S, Cohen N, Yearim A, Ast G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013;23(5):789–799. doi: 10.1101/gr.143503.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stirzaker C, Song JZ, Ng W, Du Q, Armstrong NJ, Locke WJ, et al. Methyl-CpG-binding protein MBD2 plays a key role in maintenance and spread of DNA methylation at CpG islands and shores in cancer. Oncogene. 2017;36(10):1328–1338. doi: 10.1038/onc.2016.297. [DOI] [PubMed] [Google Scholar]
- 43.Szyf M, Pakneshan P, Rabbani SA. DNA demethylation and cancer: therapeutic implications. Cancer Lett. 2004;211(2):133–143. doi: 10.1016/j.canlet.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 44.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21(5):655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. Critical evaluation of the illumina methylationepic BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014;24(9):1421–1432. doi: 10.1101/gr.163485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22(12):2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lay FD, Kelly TK, Jones PA. Nucleosome occupancy and methylome sequencing (NOMe-seq) Methods Mol Biol. 2018;1708:267–284. doi: 10.1007/978-1-4939-7481-8_14. [DOI] [PubMed] [Google Scholar]
- 51.He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2014;11(1):73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Steensel B, Belmont AS. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169(5):780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du Q, Bert AS, Armstrong JN, Caldon EC, Song JZ, Nair SS, et al. Replication timing and epigenome remodelling are associated with the nature of chromosomal rearrangements in cancer. Nat Commun. 2019;10(1):416. doi: 10.1038/s41467-019-08302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiechens N, Singh V, Gkikopoulos T, Schofield P, Rocha S, Owen-Hughes T. The chromatin remodelling enzymes SNF2H and SNF2L position nucleosomes adjacent to CTCF and other transcription factors. PLoS Genet. 2016;12(3):e1005940. doi: 10.1371/journal.pgen.1005940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nizovtseva EV, Clauvelin N, Todolli S, Polikanov YS, Kulaeva OI, Wengrzynek S, et al. Nucleosome-free DNA regions differentially affect distant communication in chromatin. Nucleic Acids Res. 2017;45(6):3059–3067. doi: 10.1093/nar/gkw1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brennan LD, Forties RA, Patel SS, Wang MD. DNA looping mediates nucleosome transfer. Nat Commun. 2016;7:13337. doi: 10.1038/ncomms13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:14648. doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hodges HC, Stanton BZ, Cermakova K, Chang CY, Miller EL, Kirkland JG, et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol. 2018;25(1):61–72. doi: 10.1038/s41594-017-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, et al. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14(5):575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skene PJ, Hernandez AE, Groudine M, Henikoff S. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife. 2014;3:e02042. doi: 10.7554/eLife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA. H4K20 methylation regulates quiescence and chromatin compaction. Mol Biol Cell. 2013;24(19):3025–3037. doi: 10.1091/mbc.e12-07-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jorgensen S, Schotta G, Sorensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41(5):2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 67.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27(24):2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barutcu AR, Lajoie BR, Fritz AJ, McCord RP, Nickerson JA, van Wijnen AJ, et al. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016;26(9):1188–1201. doi: 10.1101/gr.201624.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao H, Lukin K, Ramirez J, Fields S, Lopez D, Hagman J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci USA. 2009;106(27):11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimono Y, Murakami H, Kawai K, Wade PA, Shimokata K, Takahashi M. Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J Biol Chem. 2003;278(51):51638–51645. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- 71.Lieleg C, Ketterer P, Nuebler J, Ludwigsen J, Gerland U, Dietz H, et al. Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density. Mol Cell Biol. 2015;35(9):1588–1605. doi: 10.1128/MCB.01070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 2012;19(9):884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alen C, Kent NA, Jones HS, O’Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell. 2002;10(6):1441–1452. doi: 10.1016/S1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 74.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20(3):259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 75.Kelso TWR, Porter DK, Amaral ML, Shokhirev MN, Benner C, Hargreaves DC. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. eLife. 2017;6:e30506. doi: 10.7554/eLife.30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111(8):3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao D, Lu X, Wang G, Lan Z, Liao W, Li J, et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017;542(7642):484–488. doi: 10.1038/nature21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32(2):399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lund E, Oldenburg AR, Collas P. Enriched domain detector: a program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res. 2014;42(11):e92. doi: 10.1093/nar/gku324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buske FA, French HJ, Smith MA, Clark SJ, Bauer DC. NGSANE: a lightweight production informatics framework for high-throughput data analysis. Bioinformatics. 2014;30(10):1471–1472. doi: 10.1093/bioinformatics/btu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li W, Gong K, Li Q, Alber F, Zhou XJ. Hi-Corrector: a fast, scalable and memory-efficient package for normalizing large-scale Hi-C data. Bioinformatics. 2015;31(6):960–962. doi: 10.1093/bioinformatics/btu747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou X, Lowdon RF, Li D, Lawson HA, Madden PA, Costello JF, et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods. 2013;10(5):375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 86.Valdes-Mora F, Gould CM, Colino-Sanguino Y, Qu W, Song JZ, Taylor KM, et al. Acetylated histone variant H2A.Z is involved in the activation of neo-enhancers in prostate cancer. Nat Commun. 2017;8(1):1346. doi: 10.1038/s41467-017-01393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stempor P, Ahringer J. SeqPlots—interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res. 2016;1:14. doi: 10.12688/wellcomeopenres.10004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. A) Gene expression of each remodeller protein in LNCaP cells from RNA-seq (mean and SE; n = 3). B) Gene expression of remodeller proteins in TCGA data set of 486 prostate epithelial tumours (mean and SE; n = 486). C) Gene expression of each remodeller in PrEC cells from RNA-seq (mean and SE; n = 3), D) Gene expression of remodellers in TCGA data set of normal prostate epithelial tissue (mean and SE; n = 52). E) Scatter plot of logRPKM values for TCGA data versus logTPM values for LNCaP and PrEC RNA-seq data. The PrEC comparison for PrEC and normal TCGA is shown in green, and the LNCaP compared to TCGA tumours is shown in red. Lines are linear regression line of best fit, and Pearson’s correlation coefficient between the two normal data sets (cor = 0.670013) and the two cancer data sets (cor = 0.8375552, SNF2L is excluded as an outlier) is shown under the plot. Figure S2. A–H) Histograms of chromatin remodeller binding sites, binned at 150 bp widths. Vertical line indicates the point of 750 bp (~ 5 nucleosomes). Figure S3. A) Heat map of ChromHMM emission profile based on the learned model from the epigenome roadmap. B) Heat map of chromatin state enrichment generated from the chromHMM analysis, where the percentage of each state in the genome is represented in the first column and the remaining columns are the enrichment of each chromatin state over annotated genomic features. C) Heatmap of H3K27ac, H3K4me1, H3K4me3 and p300 ChIP-seq signal and DNaseI signal at putative active enhancers, ± 2.5 kb from the centre of each enhancer, sorted by H3K27ac and H3K4me1 signal. Figure S4. A) Heatmap H3K4me3, RNA polII, and chromatin remodeller ChIP-seq signal and DNaseI signal at refseq-annotated promoters. Signal is plotted ± 2 kb from the transcription start site (TSS) and sorted by H3K4me3 signal. B, D) Venn diagram of chromatin remodeller binding site overlaps for Group 1 and Group 2 remodellers, respectively. C) Percentage of all Group 1 remodeller binding sites that are unique to each Group 1 remodeller and contain multiple Group 1 remodellers. E) Percentage of all Group 2 remodeller binding sites that are unique to each Group 2 remodeller and contain multiple Group 2 remodellers. Figure S5. A–O) Enrichment of chromatin remodeller binding sites across the 15 state chromHMM model based on the Epigenome roadmap (see Methods). Significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p-value, ***p < 0.001 or **p < 0.05. Figure S6. A–D) Genome-wide average distribution of ChIP-seq signals for histone modifications H3K4me3, H3K4me1, H3K27me3 and H3K9me3 ± 2 kb from the centre of chromatin remodeller binding sites. E–H) Pearson’s correlation score matrix of chromatin remodeller ChIP-seq signal at active, bivalent, facultative and constitutive promoters. Each matrix was ordered by hierarchical clustering. Figure S7. A–B) Violin plots of nucleotide frequency within remodeller binding sites for Group 1 and Group 2. C–D) Dinucleotide frequency within chromatin remodeller Group 1 and Group 2 binding sites. E) DNA methylation density within chromatin remodeller binding sites for each remodeller. F) CpG density within chromatin remodellers binding sites for each remodeller protein. G) CpG density of unmethylated CpG islands, methylated CpG islands and the whole genome. A significant difference in CpG density was detected between each of the groups (one-way ANOVA, ***p < 0.001). Figure S8. A) The genome divided into TADs (85.4%), TAD boundaries (2.6%) and unorganised chromatin (12.0%) using TADs and boundaries called to a 40 kb resolution from Hi-C data. B–C) GAT enrichment of chromatin remodellers at TADs and TAD boundaries, where significant enrichment is defined as a score above one and significantly depleted as below one with Benjamini–Hochberg adj p-value, ***p < 0.001 or **p < 0.05.
Data Availability Statement
All sequencing data sets used in this study have been previously published and are available through Gene Expression Omnibus (ncbi.nlm.nih.gov/geo/). GEO accession numbers are as follows: chromatin remodeller ChIP-seq GSE72690; histone modification ChIP-seq data GSE73785, GSE38685, and GSE98732; CTCF ChIP-seq GSE38685; Lamin B and Lamin A/C ChIP-seq GSE98732; p300 ChIP-seq GSE27824; RNA polII ChIP-seq GSE28264; Hi-C GSE73785; RNA-seq GSE73785; NOMe-seq GSE76334; and whole-genome bisulphite sequencing GSE86833. TCGA data are available from the data portal at cancergenome.nih.gov, and ENCODE DNaseI data are available from the ENCODE data portal at encodeproject.org/.