Abstract
Background
The present study aimed to perform a deep phenotypic and genotypic analysis of 15 clinical carbapenem-resistant Acinetobacter baumannii (CRAb) strains isolated in Madagascar between 2008 and 2016 from diverse sources.
Methods
CRAb isolates collected from the Clinical Biology Centre of the Institut Pasteur of Madagascar, from the neonatal unit of Antananarivo military hospital, and from intensive care units of Mahajanga Androva and Antananarivo Joseph Ravoahangy Andrianavalona (HJRA) hospitals were subjected to susceptibility testing. Whole-genome sequencing allowed us to assess the presence of antibiotic-resistance determinants, insertion sequences, integrons, genomic islands and potential virulence factors in all strains. The structure of the carO porin gene and deduced protein (CarO) were also assessed in CRAb isolates.
Results
All isolates were found to be multidrug-resistant strains. Antibiotic-resistance genes against six classes of antimicrobial agents were described. The four carbapenem-resistance genes: blaOXA-51 like, blaOXA-23, blaOXA-24, and blaOXA-58 genes were detected in 100, 53.3, 13.3, and 6.6% of the isolates, respectively. Additionally, an ISAba1 located upstream of blaOXA-23 and blaADC-like genes was observed in 53.3 and 66.7% of isolates, respectively. Further, Tn2006 and Tn2008 were found associated to the ISAba1-blaOXA-23 structure. An 8051-bp mobilizable plasmid harbouring the blaOXA-24 gene was isolated in two strains. In addition, 46.7% of isolates were positive for class 1 integrons. Overall, five sequences types (STs), with predominantly ST2, were detected. Several virulence genes were found in the CRAb isolates, among which two genes, epsA and ptk, responsible for the capsule-positive phenotype, were involved in A. baumannii pathogenesis.
Conclusions
This study revealed the presence of high-level carbapenem resistance in A. baumannii with the first description of OXA-24 and OXA-58 carbapenemases in Madagascar. This highlights the importance of better monitoring and controlling CRAb in Madagascan hospitals to avoid their spread.
Electronic supplementary material
The online version of this article (10.1186/s13756-019-0491-9) contains supplementary material, which is available to authorized users.
Keywords: Acinetobacter baumannii, Carbapenem-resistance, Antimicrobial resistance
Background
Acinetobacter baumannii is a bacterial species that can survive on moist and dry surfaces. A. baumannii is widespread in clinical environments, colonizing a variety surfaces (including medical instrumentation), surviving for instance also as a commensal on the skin or hair of hospital staff and patients. The ability of this organism to gain multiple virulence factors and survive in hospital environments for prolonged periods has led to it emerging as a successful worldwide opportunistic nosocomial pathogen. It is responsible for a wide range of infections, such as bacteraemia, sepsis, meningitis and urinary tract infections, and is therefore challenging in terms of infection control by health personnel [1, 2]. This species also has a remarkable propensity for the rapid acquisition of resistance to an extensive range of antimicrobial agents. They can exhibit a major resistance profile, including carbapenems and other β-lactam antibiotics, leaving clinicians with limited therapeutic options. Carbapenem-resistant A. baumannii (CRAb) have arisen in recent decades. The most common mechanisms for resistance involve enzymatic degradation of the drugs, modification or protection of the target and decreased permeability to or active efflux of antibiotics, often working synergistically. The resistance to carbapenems in A. baumannii has been globally related to numerous enzymes, including OXA-type carbapenemases (OXA-51, OXA-23, OXA-24 and OXA-58) and metallo-β-lactamases (IMP and NDM) [3, 4]. Major expression of OXA genes might be facilitated by insertion sequences (ISs), such as ISAba1, ISAba4 and ISAba125, which provide an additionally strong promoter [5, 6]. Furthermore, overexpression of the adeB gene, tightly regulated by the adeRS genes (two-component regulatory system of AdeABC efflux pump family), has also been described implicated in carbapenem resistance in the Acinetobacter species [7, 8]. It has also been shown that variation in porins can provide a mechanism to escape from antibacterial pressure. A. baumannii intrinsically has a smaller number and size of porins compared with other Gram-negative bacteria (GNB). The loss of membrane permeability, owing to changes in primary structure, or loss of CarO (carbapenem-associated outer-membrane protein) porin currently are the best characterised causes of intrinsic A. baumannii carbapenem resistance [9, 10]. In most of cases, these changes are the result of carO porin gene disruption by various ISs. Based on the variable domains of this porin, this channel is classified into two groups, CarOa and CarOb, where CarOb has been shown to be twice as specific for imipenem than CarOa [11].
Furthermore, A. baumannii exhibits resistance to biocides commonly used in hospitals and laboratories. In addition to the resistance issue, virulence factors have been discovered in A. baumannii and a list of virulence factors has been reported by Abbott et al. (2013) [12].
Although a huge number of studies in industrialized countries have reported on the molecular epidemiology and antimicrobial resistance profiles of A. baumannii, clinical isolates and, to a lesser extent, virulence factors of individual strains, data on resistance and virulence factors from strains isolated in sub-Saharan countries are scarce. This is especially true for Madagascar where only one study described 53 multidrug-resistant (MDR) OXA-23-producing A. baumannii strains collected from various hospitals between 2006 to 2009 [13]. The present study aimed to perform a deep phenotypic and genotypic analysis of 15 clinical CRAb strains isolated in Madagascar between 2008 and 2016. The genetic environment surrounding all carbapenem-resistance determinants, the structure of the gene encoding the CarO porin and the presence of virulence genes have been also investigated.
Methods
Bacterial isolates and identification
All CRAb (n = 15) isolates collected from clinical samples originating from different studies in Madagascar during the period 2008–2016 (Table 1) and conserved in the biobank of the Institut Pasteur of Madagascar were included in this study. The first identifications were carried out by API 20NE gallery tests and confirmation was conducted by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS, Bruker Daltomics, Bremen, Germany).
Table 1.
Isolate reference number, source, year of isolation and resistance phenotype of carbapenem-resistant Acinetobacter baumannii strains isolated in Madagascar between 2008 and 2016
| Isolate reference number | Source | Year of isolation | Resistance phenotype Disk diffusion method |
E-test (MICs, mg/L) | |
|---|---|---|---|---|---|
| IPM | MEM | ||||
| AB 346a | W | 2013 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, SXT, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 285a | PF | 2013 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, SXT, CIP, MEM | > 32 | > 32 |
| AB 79a | W | 2016 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, AMK, SXT, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 116a | W | 2016 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, AMK, SXT, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 184a | U | 2016 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, CIP, MEM, NAL | > 32 | > 32 |
| AB 406a | BAL | 2016 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, AMK, SXT, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 334a | BAL | 2014 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, AMK, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB4046a | UP | 2015 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, CIP, MEM, NAL | > 32 | > 32 |
| AB 141a | W | 2016 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, SXT, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 006b | RS | 2015 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 153b | RS | 2015 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 142b | F | 2015 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, SXT, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 187b | RS | 2015 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, TOB, CIP, MEM, NAL, LVX | > 32 | > 32 |
| AB 1784c | BSI | 2013 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, AMK, SXT, CIP, MEM, NAL, LVX | 12 | > 32 |
| AB 176d | SP | 2008 | TIC, TIM, PIP, TZP, CAZ, CTX, FEP, GEN, AMK, SXT, CIP, MEM, NAL, LVX | > 32 | > 32 |
MICs minimum inhibitory concentrations, TIC ticarcillin, TIM ticarcillin-clavulanic acid, PIP piperacillin, TZP piperacillin-tazobactam, CAZ ceftazidime, CTX cefotaxime, FEP cefepime, GEN gentamicin, TOB tobramycin, AMK amikacin, CIP ciprofloxacin, MEM meropenem, IPM imipenem, NAL nalidixic acid, LVX levofloxacin, W wound, PF pleural fluid, U urine, BAL bronchoalvealar lavage fluid, UP urethral pus, SP superficial pus, RS rectal swab, F faeces, RS rectal swab, BSI blood stream infection
aStrains isolated from biological samples collected at Institut Pasteur of Madagascar
bStrains collected from the neonatal unit at Antananarivo military hospital
cStrains isolated from the intensive care unit at Mahajanga Androva Hospital
dStrains isolated from the intensive care unit at Antananarivo Joseph Ravoahangy Andrianavalona (HJRA) hospital
MICs > 8 mg/L for imipenem and meropenem were considered resistant
Antimicrobial susceptibility testing
Antimicrobial susceptibilities of the isolates were determined by the disk diffusion method on Mueller-Hinton agar (Bio-Rad, France), as recommended by the Antibiogram Committee of the French Microbiology Society (ACFMS 2015). The following antimicrobial agents were tested: ticarcillin; ticarcillin/clavulanic acid; piperacillin; piperacillin/tazobactam; ceftazidime; cefotaxime; cefepime; meropenem; nalidixic acid; levofloxacin; ciprofloxacin; gentamicin; tobramycin; amikacin; and trimethoprim/sulfamethoxazole. Resistance (Minimum Inhibitory Concentrations, (MICs)) to imipenem and meropenem was determined by Etests (BioMerieux, Marcy l’Etoile, France) according to the manufacturer’s instructions and ACFMS breakpoints (strains displaying MICs > 8 mg/L for imipenem and meropenem were considered resistant).
DNA extraction and whole-genome sequencing (WGS)
DNA was extracted with the cador Pathogen 96 QIAcube HT Extraction Kit (Qiagen, Paris, France) on a Qiacube HT from 5 mL of liquid cultures grown overnight at 37 °C in Luria-Bertani infusion medium following the manufacturer’s protocol. Purity and DNA quantity were assessed using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA). Illumina sequencing libraries were prepared by with the Nextera XT DNA Sample Kit (Illumina, San Diego, USA) featuring indexed-encoded adapters from Illumina based on the manufacturer’s instructions. The libraries were pooled for sequencing on a NextSeq 500 platform (Illumina) using 2 × 150-bp runs. FqCleaner (version 3.0) was employed to eliminate adaptor sequences (1), reduce redundant or overrepresented reads (2), correct sequencing errors (3), merge overlapping paired reads (4) and discard reads with Phred scores (measure of the quality of identification of nucleobases generated by automated DNA sequencing) < 20. Sequences with < 40 times average coverage after trimming were resequenced to avoid artefacts with respect to allele calling. The Illumina sequence data were assembled utilising Spades software (version 3.9.0).
Genome annotation and molecular analyses
All assembled genomes obtained were analysed and annotated with online tools and/or open-access databases and manual examinations. A Rapid Annotations using Subsystems Technology (RAST) server was applied for genome annotation [14]. The antimicrobial resistance gene profile was determined via KmerResistance 2.2 from the Centre for Genomic Epidemiology (CGE) [15]. Mobile elements, including ISs and class 1 integrons, were identified using the software’s ISsaga from ISFinder and Geneious 11.0.5, respectively [16, 17]. The prediction of number and visualization of genomic islands (GIs) were performed through the webserver, IslandViewer4 [18].
Distribution of virulence genes
In silico investigation of certain genes known as virulence factors in A. baumannii (AC nb: CP018861), such as cvaC (encoding for colicin V production), ptk (encoding a putative protein tyrosine kinase) and epsA (encoding a putative polysaccharide export outer-membrane protein) was performed. The genes of csu (csuA, csuB, csuC, csuD and csuE, encoding the chaperone-usher Csu fimbriae) and pga loci (pgaA, pgaB, pgaC and pgaD, encoding the polysaccharide poly-N-acetylglucosamine, PNAG), responsible for biofilm formation, were also investigated [19–21].
The complete manual annotation of all resistance and virulence determinants and their genetic environments was performed via Geneious software 11.0.5 (Biomatters, Auckland, New Zealand).
CarO amino acid sequence alignment and in silico analyses
The presence of the carO porin (AC nb: FJ652396) gene was screened for in all our CRAb isolates. The nucleotide/predicted amino acid sequences of CarO isoform proteins were determined to type the CarO (CarOa or CarOb) and evaluate gene disruption by ISs. The typing was conducted by sequence alignment of predicted CarO proteins with those already described in the NCBI database (CarOa, AC nb: ZP_05828783 and CarOb, AC nb: ADQ27797) using Geneious software. The amino acid changes observed were analysed with PROVEAN (http://provean.jcvi.org/) software, which predicts whether an amino acid substitution, insertion or deletion has any impact on the biological function of a protein.
In silico multilocus sequence typing (MLST)
The MLST scheme described by Bartual et al. [22] was performed according to the Acinetobacter baumannii MLST (Pasteur) database (https://pubmlst.org/abaumannii/). The assembled sequences of seven housekeeping genes (cnp62, fusA, gltA, pyrG, recA, rplB and rpoB) were aligned through BLAST (Basic Local Alignment Search Tool), and the aligned sequences were then extracted by comparing them to allele profiles from the A. baumannii MLST (Pasteur) database.
Core genome phylogeny analysis
To investigate genetic relatedness of the CRAb isolates evaluated in this study with those described in the literature, core genome phylogeny was performed with Harvest Suite version v1.1.2 [23]. In addition to the local CRAb panel, a set of five genomes geographically unrelated was included in the analysis. Genomic data utilised in the phylogenetic tree were downloaded from the NCBI (National Centre for Biotechnology Information) database, including the complete genome sequences of the A. baumannii isolates ATCC1778 (CP000521), ACICU (CP000863), MDR-ZJ06 (CP001937), A388 (CP024418) and AB0057 (CP001182) [24].
Nucleotide sequence accession numbers
The 6302-bp, 5551-bp and 6486-bp A. baumannii genomic sequences containing Tn2006, Tn2008 and blaOXA-24 genes and surrounding regions were deposited in the EMBL database under accession numbers LS999837.1, LS999838.1 and LS999872.1, respectively.
Results
Antimicrobial susceptibility testing
Table 1 summarizes the source, year of isolation and resistance phenotype of all CRAb isolates studied. The clinical sources from which the strains were isolated were wounds 26.7% (n = 4); rectal swabs/faeces 26.7% (n = 4); bronchoalveolar lavage fluids 13.3% (n = 2) and urine, pleural fluid, urethral pus, superficial pus and blood culture (one sample representing each at 6.7%). All isolates (100%) demonstrated resistance to at least three classes of antibiotics, hence matching with the MDR criteria [21]. The disk diffusion method indicated that all strains (100%) were resistant to ticarcillin, ticarcillin/clavulanic acid, piperacillin, piperacillin/tazobactam, cefotaxime, ceftazidime, cefepime, gentamicin and meropenem. The rate of other resistances was as follows: 93.3% (n = 14) for nalidixic acid and ciprofloxacin, 80% (n = 12) for levofloxacin, 66.6% (n = 10) for tobramycin, 40% (n = 6) for amikacin and 60% (n = 9) for trimethoprim/sulfamethoxazole. The studied isolates were resistant to imipenem and meropenem with MIC values above 32 mg/L (except AB1784, MIC to IPM = 12 mg/L).
Detection of drug-resistance genes and related ISs
The natural intrinsic blaOXA-51like (blaOXA-65, blaOXA-66, blaOXA-67 and blaOXA-69) gene, as well as three acquired oxacillinases genes, blaOXA-23, blaOXA-24 and blaOXA-58, were present in 100, 53.3% (n = 8), 13.3% (n = 2) and 6.7% (n = 1) of our CRAb isolates, respectively (Table 2). ISAba1 was detected upstream all blaOXA-23 genes, and this was later embedded in Tn2006 and Tn2008 transposons, all chromosomally located (Fig. 1).
Table 2.
Genetic features of the carbapenem-resistant Acinetobacter baumannii isolates described in this study
| Isolate reference number | ST | Antimicrobial resistance genes | ISAba1 + blaOXA-23 | ISAba1 + blaADC | Number of IS | Transposon distribution |
|---|---|---|---|---|---|---|
| AB 346 | 2 | blaOXA-66; blaADC-25; aph(3′)-Ia aadA; aac(3)-Ia; aph(6)-Id; aph(3″)-Ib; sul1;sul2, tet(B) | – | + | 17 | – |
| AB 285 | 1195 | blaOXA-67/23/58; tet(39) blaADC-106; aac(3)-IId; sul2; aph(6)Id | + | – | 16 | Tn2008 |
| AB 79 | 1196 | blaOXA65/24; blaADC-52, aac(3)-IIa; aph(3″)-Ib; sul2; aph(6)Id; blaTEM1b | – | + | 13 | – |
| AB 116 | 79 | blaOXA-65/23; blaTEM-1B; aac(3)-IIa;blaADC-52; sul2; mphE; aph(3″)-Ib; aph(6)Id; msr(E) | + | + | 20 | Tn2006 |
| AB 184 | 1 | blaOXA-69; blaADC-11; ant(3″)-Ia | – | – | 7 | – |
| AB 4066 | 2 | blaOXA66/23; blaADC-25; tetB; aac(3)-Ia; aph(3′)-Ic; ant(3″)-Ia; sul2; sul1; aph(3″)-Ib; aph(6)Id | + | + | 5 | Tn2006 |
| AB 334 | 1196 | blaOXA-65/24; blaTEM-1B; aac(3)-IIa;blaADC-25; sul2; aph(3″)-Ib;; sul; aph(6)Id | – | + | 7 | – |
| AB4046 | 2 | blaOXA-66; blaADC-25; tetB; aac(3)-Ia; aph(3′)-Ia; aph(3′)-IC; aph(6)-Id; tetB; ant(3″)-Ia; sul2; sul1; aph(3″)-Ib | – | + | 5 | – |
| AB 141 | 2 | blaOXA-66/23; blaADC-25; aac(3)-Ia; ant(3″)-Ia; aph(3′)-IC; sul2; sul1; tetB; tet(32); aph(3″)-Ib, cmlA1;aph(6)Id | + | + | 5 | Tn2006 |
| AB 006 | 1 | blaOXA-69/23; blaADC-81; ant(3″)-Ia | + | – | 2 | Tn2006 |
| AB 153 | 1 | blaOXA-69/23; blaADC-10; ant(3″)-Ia | + | – | 4 | Tn2006 |
| AB 142 | 2 | blaOXA-66/23; blaADC-25; aac(3)-Ia ant(3″)-Ia; aph(3′)-Ia; aph(3″)-Ib sul1; sul2;tetB;aph(6)-Id; mphE; msr(E) | + | + | 5 | Tn2006 |
| AB 187 | 1 | blaOXA-69; blaADC-81; ant(3″)-Ia | – | – | 2 | – |
| AB 1784 | 2 | blaOXA-66; blaADC-25; tetB; aac(3)-Ia; aph(3′)-IC; ant(3″)-Ia; sul2; sul1; aph(3″)-Ib; aph(6)-Id | – | + | 4 | – |
| AB 176 | 2 | bla OXA-66/23 ; bla ADC-25 ; aac(3)-Ia; ant(3″)-Ia; aph(3”)Ia; aph(3″)-Ib; aph(6)Id; sul1; sul2; tetB | + | + | 4 | Tn2006 |
+; presence of the indicated structure; −: absence of the indicated structure; ST, sequence type; IS, insertion sequence
Fig. 1.
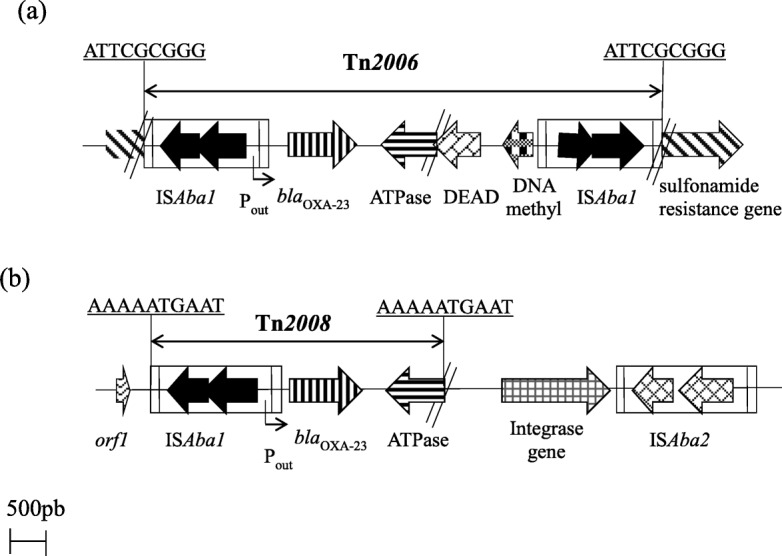
a and b Schematic representation of two transposons described in this study: (a) Tn2006; (b) Tn2008 and their genetic environment. The nine-bp target site duplication of Tn2006 and Tn2008 are represented underlined. Insertion elements are represented by ISAba1 and ISAba2. Pout: ISAba1 promoter; orf1: orf encoding for a hypothetical protein; ATPase: gene encoding the putative AAA ATPase; DEAD, gene encoding the putative DEAD helicase
The two blaOXA-24 genes flanked by the XerC/XerD recombination sites were located on a small plasmid (8Kb) designated pOXA-24_AB334. This harboured nine open-reading frames (Fig. 3), among which two replicon genes (repA and repB) and two plasmid mobilization genes (mobL and mobS) were found. Comparative analysis revealed that pOXA-24_AB334 was 99.1% identical with pABUH3a-8.2 (NZ_AYFH01000048.1) detected in the clinical A. baumannii UH 7607 strain isolated in the USA. Further, these two plasmids belong to the small Rep-3 superfamily usually described in A. baumannii [25].(Fig. 2).
Fig. 3.
Phylogenetic analysis of 15 A. baumannii isolates. The phylogenetic tree was constructed with the Harverst suite for core genome alignment and visualization
Fig. 2.
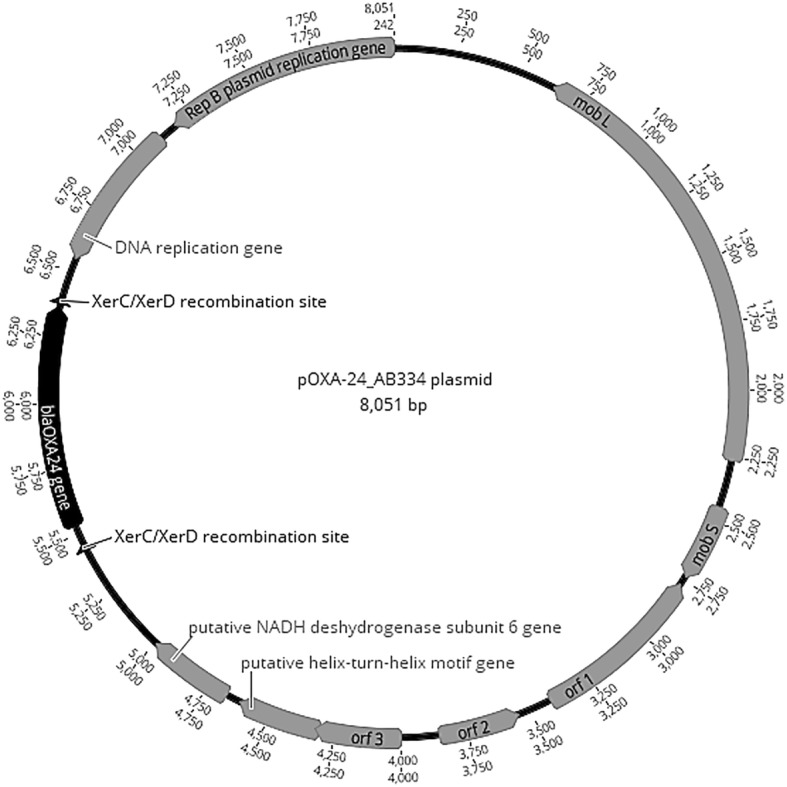
Circular map of the pOXA-24_AB334 plasmid. The two mobilization genes are represented by mobL and mobS. orf1, orf2 and orf3: genes encoding for hypothetical proteins
The KmerResistance 2.2 results revealed the presence of several others antibiotic-resistance genes (Table 2). The ampC Acinetobacter-derived cephalosporinase blaADC-like gene was established in all CRAb isolates. Sixty-six percent (n = 10) of the blaADC-positive isolates were associated with ISAba1 upstream of the blaADC genes. The variants identified were exhibiting more than 98% identity with blaADC-10, blaADC-11, blaADC-25, blaADC-52, blaADC-81 and blaADC-106 genes. All strains harboured at least one gene involved in aminoglycoside resistance (aac(3)-Ia; aac(3)-IIa; aph(3’-Ia; aph(3′)-Ic; aph(3″)-Ia; aph(3″)-Ib; aph(6)-Id, ant(3″)-Ia). Genes encoding resistance to tetracycline and/or minocycline (tet(B), tet39), sulphonamides (sul1, sul2), macrolides (msrE, mphE), β-lactams (blaTEM1b) and chloramphenicol (cmlA1) resistance were also detected.
Mobile genetic elements
Investigation of mobile genetic elements showed that 46.7% (n = 7) of isolates were positive for class 1 integrons (Table 2), which were all embedded in chromosomal genomic resistance island, AbGRI2–1. Gene cassette (GC) arrays encoded for resistance to aminoglycosides (aac(3)-Ia, ant(3″)-Ia) and for proteins with unknown functions (orfP1-orfP2-orfQ). ISs screening in all genomes analysed demonstrated a large diversity present, mainly of the IS3, IS4, IS30, IS256 and ISL3 families.
Genome annotation and GIs
The genome length for our A. baumannii isolates were approximately 4 million bp with an annotation average coverage of 50%, and the number of coding DNA sequences per genome was approximately 3500. The GIs of all genomes were detected by IslandPath-DIMOB software and their number varied from 3 to 8. The visualization of the genetic structures of these GIs revealed the presence of antibiotic resistance genes (aph(6)-Id, aph(3′)-Ib, aac(3)-Ia, sul1, blaTEM); genes encoding multidrug transporters (tetA and qacE∆1); mercuric- and arsenic-resistance determinants; transposable elements, including IS (ISAba1, IS26, ISVsa3 and IS1006); and the Tn7 transposon.
Distribution of virulence genes
Additional file 1: Table S1 summarizes the distribution of virulence genes described in all CRAb isolates described herein. The distribution of virulence genes revealed the presence of cvaC, csuB, csuC, csuD, csuE pgaA, pgaB, pgaC and pgaD genes in all isolates assessed. The csuA was detected in 86% of strains. Interestingly, the two genes, epsA and ptk, responsible for the capsule-positive phenotype involved in the pathogenesis of A. baumannii [19] were found in all CRAb strains analysed.
Analysis of the carO porin gene and CarO isoform proteins
All strains harboured an intact structure of the carO porin gene. The predicted amino acid sequence showed that CarOa and CarOb were distributed among 20% (n = 3) and 53.3% (n = 8) of our CRAb isolates, respectively (Table 1, Additional file 1). However, a point mutation in the CarOb protein (S214 T) in AB184, AB006, AB 153 and AB 187 isolates was detected. PROVEAN analysis uncovered that this amino acid substitution had a neutral impact in terms of protein function. No disruption of the carO porin gene by an IS suggests that the CarO porin transporter was not implicated in carbapenem resistance within our isolates.
Genotype analysis and phylogenetic tree construction
In order to elucidate the different genotypes of CRAb isolates circulating in Madagascar, an MLST scheme based on the Pasteur database was employed. The results revealed that the isolates were clustered into five different genotypes or STs, mostly represented by ST2 (n = 7) followed by ST1 (n = 4). Three strains had new allelic profiles and novel ST numbers (ST1195 for AB285 and ST1196 for AB79/AB334) assigned (Table 2).
The phylogenetic tree analysis indicated that all strains belonging to the same ST (ST1, ST2 or ST1196) clustered together. The strains belonging to ST1 (n = 4) clustered with AB00057 isolated from the USA in 2004. These four strains recovered in 2015 and 2016 all originated from Antananarivo. Strains belonging to ST2 (n = 7) isolated from Antananarivo and Majunga between 2008 and 2016 clustered with MDR-ZJ06, widespread throughout China [26]. Antananarivo and Majunga are separated by 565 km and our results suggest a permanent circulation of ST2 genotype in Madagascar. The strains belonging to the new ST (ST1195 and ST1196) were phylogenetically distinct from the others belonging to ST1 and ST2 (Fig. 3).
Discussion
Carbapenem resistance in A. baumannii is based on the production of carbapenemases or synergistic effects between carbapenemases, porin modifications or loss or modification of the penicillin-binding proteins [26, 27]. In this study, 53.3, 13.3 and 6.7% of 15 CRAb isolates harboured the carbapenemases blaOXA-23, blaOXA-24 and blaOXA-58 genes, respectively (Table 2). The carbapenemases (OXA-23, OXA-24 and OXA-58, respectively) encoded by these genes have been described to confer a reduced susceptibility to carbapenems [28, 29]. Like reported elsewhere, the blaOXA-23 gene in our CRAb isolates was upregulated by an ISAba1, which provides a strong promoter [21, 30, 31]. This is the first report of OXA-24 and OXA-58 carbapenemase-producing A. baumannii in Madagascar. This latter was co-expressed with OXA-23 in the AB285 isolate.
Additionally, the OXA-51-like gene was determined in all CRAb isolates. The derived oxacillinase (OXA-51-like) confers a high level of resistance to carbapenems only when it is overexpressed [32]. Nevertheless, high-level MICs of imipenem and meropenem was exhibited by some of strains in which the OXA-51 gene was not upregulated (Table 2), and this could be explained by other mechanisms, such as the reduced expression of penicillin-binding protein (PBP2) or the loss of the particular outer-membrane proteins related to carbapenem resistance, like those highlighted by the results of Fernández-Cuenca et al. [26].
The acquisition and dissemination of the resistance genes are largely owing to the actions of mobile genetic elements (MGEs) [33]. This study described MGEs, such as transposable elements (ISAba1, Tn2006 and Tn2008), class 1 integrons, AbaR-type genomic resistance islands (AbGRI2–1) and plasmids [34]. In accordance with several others reports, these genetic structures could disseminate antibiotic resistance determinants between bacteria cell and thus compromise antibiotic treatment, including those with clinical relevance [34–36].
A striking feature of this study is the large number of antibiotic-resistance genes found (Table 2). The resistance to third-generation cephalosporins and quinolones observed was associated with the presence of an ampC gene [21] and the S83 L substitution in the gyrA gene, respectively.
Typing and phylogenetic analysis have shown that the majority of strains (11/15) assigned to ST1 and ST2 were closest to isolates belonging to globally disseminated clonal complexes I and II (CC1 and CC2), respectively. A previous study conducted in Antananarivo by Andriamanantena et al. (2010) demonstrated the circulation of two A. baumannii genotypes (A and B typed by rep-PCR) in clinical settings between 2006 and 2009 [13] but the sequence types of these two genotypes were not investigated and could therefore not be correlated with those of our isolates.
The presence of the most important virulence genes (Table 1, Additional file 1) in almost all CRAb isolates is in agreement with their clinical importance and moreover confirmed they are all potential biofilm producers.
Conclusions
The deep molecular analysis of our clinical CRAb strains allowed for the description of the newly acquired carbapenemases (OXA-24, OXA-58) not previously described in Madagascar and highlighted the potential role of mobile elements, such as transposons and plasmids, in the dissemination of carbapenem resistance. Different studies, including ours, have provided evidence for the circulation of carbapenemase-producing strains (OXA23/24/58; NDM-1/4) in Madagascar [37]. Such circulation, both in Madagascar and the Indian Ocean region, is becoming worrisome and should be better monitored and controlled in order to avoid a rise in the incidence of untreatable nosocomial infections, especially within intensive care units.
Additional file
Table S1. Distribution of porins, efflux pumps and virulence genes among the fifteen carbapenems-resistant Acinetobacter baumannii strains. (DOCX 17 kb)
Acknowledgements
We would like to thank Andriamanantena Tahiry Sylviane for providing the Acinetobacter baumannii AB176 strain. All authors would like to thank Astrid Knoblauch for her help in proofreading the article.
Funding
This study is based upon a postdoctoral internship supported by Postdoctoral Grant CALMETTE and YERSIN from Institut Pasteur Department of International Affairs.
Availability of data and materials
All data generated or included during this study are featured in this published article.
Abbreviations
- A. baumannii
Acinetobacter baumannii
- ABC
ATP-binding cassette
- AC nb
Accession number
- ACFMS
Antibiogram Committee of the French Microbiology Society
- ADC
Acinetobacter-derived cephalosporinase
- AMK
Amikacin
- BLAST
Basic Local Alignment Search Tool
- CarO
Carbapenem-associated outer-membrane
- CAZ
Ceftazidime
- CGE
Centre for genomic epidemiology
- CIP
Ciprofloxacin
- CRAb
Carbapenem-resistant Acinetobacter baumannii
- CRISPRs
Clustered regularly interspaced short palindromic repeats
- csu
Chaperone-usher
- CTX
Cefotaxime
- epsA
Polysaccharide export outer membrane protein
- FEP
Cefepime
- GCs
Gene cassettes
- GEN
Gentamicin
- GIs
Genomic islands
- GNB
Gram-negative bacteria
- HJRA
Joseph Ravoahangy Andrianavalona (HJRA) hospital
- IPM
Imipenem
- ISs
Insertion sequences
- LVX
Levofloxacin
- MDR
Multidrug-resistance
- MEM
Meropenem
- MFS
Major facilitator superfamily
- MGE
Mobile genetic element
- MICs
Minimum Inhibitory Concentrations
- MLST
Multilocus sequence typing
- NAL
Nalidixic acid
- NCBI
National Centre for Biotechnology Information
- PIP
Piperacillin
- PNAG
Poly-N-acetylglucosamine
- ptk
Protein tyrosine kinase
- RAST
Rapid annotations using subsystems technology
- RND
Resistance nodulation division
- SMR
Small multidrug resistance
- STs
Sequences type
- TIC
Ticarcillin
- TIM
Ticarcillin-clavulanic acid
- TOB
Tobramycin
- TZP
Piperacillin-tazobactam
- WGS
Whole genome sequencing
Authors’ contributions
PLST, MANR and CK carried out the molecular genetic analysis. PLST drafted and wrote the manuscript. VA, AR, ERH, ZZA and FR provided the strains. VE performed whole-genome sequencing. JMC designed the study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pierrette Landrie Simo Tchuinte, Phone: +261 34 42 494 83, Email: simolandrie@yahoo.fr, Email: pierrette@pasteur.mg.
Mamitiana Alain Noah Rabenandrasana, Email: rnoah@pasteur.mg.
Carole Kowalewicz, Email: kowalewicz.carole@hotmail.com.
Volasoa Herilalaina Andrianoelina, Email: volasoa@pasteur.mg.
Andriniaina Rakotondrasoa, Email: aina@pasteur.mg.
Zafitsara Zo Andrianirina, Email: zozand03@yahoo.fr.
Vincent Enouf, Email: vincent.enouf@pasteur.fr.
Elisoa Hariniaina Ratsima, Email: elisoa@pasteur.mg.
Frédérique Randrianirina, Email: frederique@pasteur.mg.
Jean-Marc Collard, Email: jmcollard@pasteur.mg.
References
- 1.Molina L, Udaondo Z, Duque E, Fernández M, Molina-Santiago C, Roca A, et al. Antibiotic resistance determinants in a Pseudomonas putida strain isolated from a hospital. PLoS One. 2014;9(1):e81604. doi: 10.1371/journal.pone.0081604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vijayakumar S, S BA, Kanthan K, Veeraraghavan B. Whole-genome shotgun sequences of seven colistin-resistant Acinetobacter baumannii isolates from bacteraemia. J Glob Antimicrob Resist. 2018;12:155–156. doi: 10.1016/j.jgar.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Hujer KM, Hujer AM, Endimiani A, Thomson JM, Adams MD, Goglin K, et al. Rapid determination of quinolone resistance in Acinetobacter spp. J Clin Microbiol. 2009;47(5):1436–1442. doi: 10.1128/JCM.02380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horii T, Muramatsu H, Iinuma Y. Mechanisms of resistance to fluoroquinolones and carbapenems in Pseudomonas putida. J Antimicrob Chemother. 2005;56(4):643–647. doi: 10.1093/jac/dki254. [DOI] [PubMed] [Google Scholar]
- 5.Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 Carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16(1):35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes BS, Amyes SGB. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J Med Microbiol. 2012;61(8):1103–1108. doi: 10.1099/jmm.0.044156-0. [DOI] [PubMed] [Google Scholar]
- 7.Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, et al. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother. 2010;65(8):1589–1593. doi: 10.1093/jac/dkq218. [DOI] [PubMed] [Google Scholar]
- 8.Yoon EJ, Chabane YN, Goussard S, Snesrud E, Courvalin P, Dé E, Grillot-Courvalin C. Contribution of Resistance-Nodulation-Cell Division Efflux Systems to Antibiotic Resistance and Biofilm Formation in Acinetobacter baumannii. MBio. 2015;6(2):1–13. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mussi MA, Limansky AS, Viale AM. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob Agents Chemother. 2005;49(4):1432–1440. doi: 10.1128/AAC.49.4.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catel-Ferreira M, Coadou G, Molle V, Mugnier P, Nordmann P, Siroy A, et al. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J Antimicrob Chemother. 2011;66(9):2053–2056. doi: 10.1093/jac/dkr267. [DOI] [PubMed] [Google Scholar]
- 11.Siroy A, Molle V, Lemaître-guillier C, Vallenet D, Pestel-caron M, Cozzone AJ, et al. Channel Formation by CarO , the Carbapenem Resistance-Associated Outer Membrane Protein of Acinetobacter baumannii Channel Formation by CarO , the Carbapenem Resistance-Associated Outer Membrane Protein of Acinetobacter baumannii 2005;49(12):4876–83. [DOI] [PMC free article] [PubMed]
- 12.Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. Carbapenem resistance in Acinetobacter baumannii: laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Anti-Infect Ther. 2013;11(4):395–409. doi: 10.1586/eri.13.21. [DOI] [PubMed] [Google Scholar]
- 13.Andriamanantena TS, Ratsima E, Rakotonirina HC, Randrianirina F, Ramparany L, Carod JF, et al. Dissemination of multidrug resistant Acinetobacter baumannii in various hospitals of Antananarivo Madagascar. Ann Clin Microbiol Antimicrob. 2010;9:2–7. doi: 10.1186/1476-0711-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clausen PTLC, Zankari E, Aarestrup FM, Lund O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J Antimicrob Chemother. 2016;71(9):2484–2488. doi: 10.1093/jac/dkw184. [DOI] [PubMed] [Google Scholar]
- 16.Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011;12(3):R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, et al. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun. 2010;78(9):3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darvishi M. Virulence factors profile and antimicrobial resistance of Acinetobacter baumannii strains isolated from various infections recovered from immunosuppressive patients. Biomed Pharmacol J. 2016;9(3):1057–1062. doi: 10.13005/bpj/1048. [DOI] [Google Scholar]
- 21.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter reed Army medical. Antimicrob Agents Chemother. 2006;50(12):4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartual SG, Seifert H, Hippler C, Rodríguez-Valera F, Domı MA. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):1–15. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, Huang Y, Yu L, Liu Y, Niu W, Zou D, et al. Isolation and whole-genome sequence analysis of the imipenem Heteroresistant Acinetobacter baumannii clinical isolate HRAB-85. Int J Infect Dis. 2017;62:94–101. doi: 10.1016/j.ijid.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Lean SS, Yeo CC. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: good, bad, who knows? Front Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Cuenca F, Martínez-Martínez L, Conejo MC, Ayala JA, Perea EJ, Pascual A. Relationship between β-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J Antimicrob Chemother. 2003;51(3):565–574. doi: 10.1093/jac/dkg097. [DOI] [PubMed] [Google Scholar]
- 27.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect [Internet] 2006;12(9):826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 28.Poirel L, Bonnin RA, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life. 2011;63(12):1061–1067. doi: 10.1002/iub.532. [DOI] [PubMed] [Google Scholar]
- 29.Héritier C, Poirel L, Nordmann P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect. 2006;12(2):123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H, Pi BR, Yang Q, Yu YS, Chen YG, Li LJ, et al. Dissemination of imipenem-resistant Acinetobacter baumannii strains carrying the ISAba1-blaOXA-23 genes in a Chinese hospital. J Med Microbiol. 2007;56(8):1076–1080. doi: 10.1099/jmm.0.47206-0. [DOI] [PubMed] [Google Scholar]
- 31.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258(1):72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 32.Héritier C, Poirel L, Fournier P, Claverie J, Raoult D, Nordmann P, et al. Characterization of the naturally occurring Oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49(10):4174–4179. doi: 10.1128/AAC.49.10.4174-4179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4):1–61. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagano M, Martins AF, Barth AL. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Brazilian J Microbiol. 2016;47(4):785–792. doi: 10.1016/j.bjm.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigro SJ, Farrugia DN, Paulsen IT, Hall RM. A novel family of genomic resistance islands, AbGRI-2, contributing to aminoglycoside resistance in Acinetobacter baumannii isolates belonging to global clone 2. J Antimicrob Chemother. 2013;68(3):554–557. doi: 10.1093/jac/dks459. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Gao J, Zhang H, Ying C. Spread of the blaOXA-23-containing Tn2008 in carbapenem-resistant Acinetobacter baumannii isolates grouped in CC92 from China. Front Microbiol. 2017;8:1–6. doi: 10.3389/fmicb.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holman AM, Allyn J, Miltgen G, Lugagne N, Traversier N, Picot S, et al. Surveillance of carbapenemase-producing Enterobacteriaceae in the Indian Ocean region between January 2010 and December 2015. Med Mal Infect. 2017;47(5):333–339. doi: 10.1016/j.medmal.2017.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Distribution of porins, efflux pumps and virulence genes among the fifteen carbapenems-resistant Acinetobacter baumannii strains. (DOCX 17 kb)
Data Availability Statement
All data generated or included during this study are featured in this published article.



