Abstract
Background
Sequence information generated from next generation sequencing is often computationally phased using haplotype-phasing algorithms. Utilizing experimentally derived allele or haplotype information improves this prediction, as routinely used in HLA typing. We recently established a large dataset of long ERMAP alleles, which code for protein variants in the Scianna blood group system. We propose the phylogeny of this set of 48 alleles and identify evolutionary steps to derive the observed alleles.
Methods
The nucleotide sequence of > 21 kb each was used for all physically confirmed 48 ERMAP alleles that we previously published. Full-length sequences were aligned and variant sites were extracted manually. The Bayesian coalescent algorithm implemented in BEAST v1.8.3 was used to estimate a coalescent phylogeny for these variants and the allelic ancestral states at the internal nodes of the phylogeny.
Results
The phylogenetic analysis allowed us to identify the evolutionary relationships among the 48 ERMAP alleles, predict 4243 potential ancestral alleles and calculate a posterior probability for each of these unobserved alleles. Some of them coincide with observed alleles that are extant in the population.
Conclusions
Our proposed strategy places known alleles in a phylogenetic framework, allowing us to describe as-yet-undiscovered alleles. In this new approach, which relies heavily on the accuracy of the alleles used for the phylogenetic analysis, an expanded set of predicted alleles can be used to infer alleles when large genotype data are analyzed, as typically generated by high-throughput sequencing. The alleles identified by studies like ours may be utilized in designing of microarray technologies, imputing of genotypes and mapping of next generation sequencing data.
Electronic supplementary material
The online version of this article (10.1186/s12967-019-1791-9) contains supplementary material, which is available to authorized users.
Keywords: Allele prediction, Next generation sequencing, Phylogeny, Scianna, ERMAP
Background
Exact matching for alleles improved survival following bone marrow transplantation [1] and reduced alloimmunization in chronically transfused patients [2–4]. Using computational algorithms, the large genotype datasets from next generation sequencing (NGS) can be phased into alleles or haplotypes [5, 6]. Using family relationships or applying experimentally confirmed allele information improves the inference accuracy, as routinely demonstrated in clinical HLA typing [7]. Blood group genes are less polymorphic than the highly variable, often shorter, HLA genes. Out of the 36 blood group systems and the genes encoding them, experimentally confirmed alleles are known for short genes only, such as ICAM4 [8] and ACKR1 [9]. For longer genes, such as ABO and ERMAP of more than 20 kb, and linked genes, such as RHD and RHCE, most haplotypes had only been computationally predicted [10–12].
The ERMAP gene, located on chromosome 1, encodes the glycoprotein carrying the antigens of the Scianna blood group system (SC; ISBT 013) in humans [13–15]. The single-pass transmembrane glycoprotein is likely involved in cell adhesion and recognized by immune cells [13, 16, 17]. The gene belongs to the butyrophilin (BTN) family which is a type 1 membrane protein of the immunoglobulin (Ig) superfamily [18]. The butyrophilin and butyrophilin-like proteins have recently been studied as potentially important immune regulators [19, 20].
We have previously assessed the nucleotide variations in the ERMAP gene and unambiguously identified 48 alleles at 21,406 nucleotides each in 50 unrelated individuals from 5 different populations [21]. We propose using the phylogeny of this set of 48 alleles and identifying evolutionary steps to derive the observed alleles [22]. We predicted unobserved alleles at every internal node and their posterior probabilities. These inferred alleles, represented by sequences identified in the nodes, are possible candidates for alleles segregating in the population. Our new approach proposes a method of utilizing not-yet-observed alleles, predicted by phylogeny, for phasing patient genotypes in clinical diagnosis and therapy.
Methods
The sequence information for 48 ERMAP alleles was retrieved from GenBank (KX265189–KX265236) [21]. The phylogenetic tree was rooted using the chimpanzee ERMAP sequence as outgroup (GenBank number NC_006468.4; range 42,268,258 to 42,295,767). Full-length sequences were aligned using the MAFFT version 7 program [23]. All of the 72 variable sites were extracted manually from the 48 ERMAP alleles [21]. The Bayesian coalescent algorithm implemented in BEAST v1.8.3 [24] was used to estimate a coalescent phylogeny for these variants and the allelic ancestral states at the internal nodes of the phylogeny. All analysis was done using default parameters. Internal node is a theoretical representation of a common ancestor between sampled alleles and are often extant in population level studies [25]. If more than one mutational or recombinational step is required to join some nodes, predicted alleles are incorporated to complete the tree [26].
We executed 4 independent runs of the program, each using the Tamura-Nei substitution model [27], a lognormal relaxed clock model [28], and a constant-size coalescent model [29]. After 40 million generations the parameter estimates were examined and determined to have converged for each run. The allelic ancestral states at each node and their posterior probabilities were extracted manually from the maximum clade compatibility tree estimated from 9001 Markov chain Monte Carlo samples generated by the BEAST software. For the ancestral allele reconstructions, we generated a set of all possible ancestors for each node and selected the predicted allele with the highest posterior probability.
Results
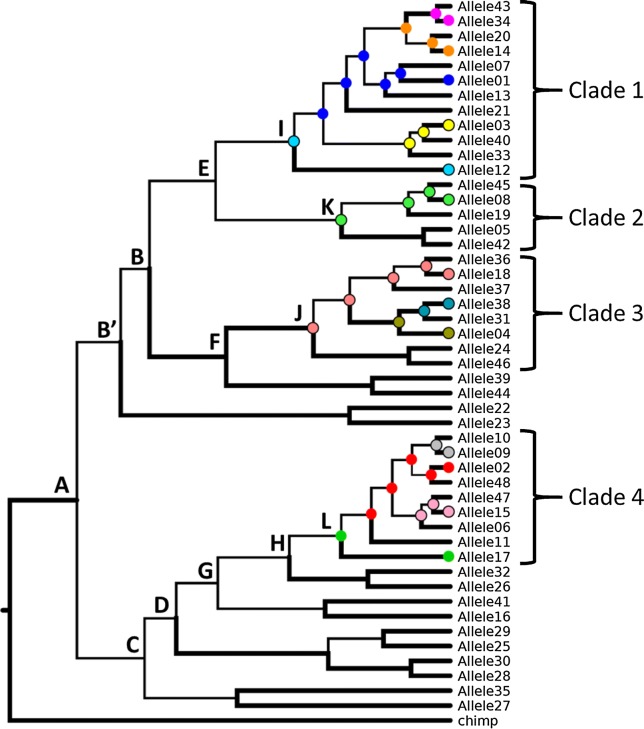
A Bayesian phylogeny of 48 previously published ERMAP alleles was calculated (Fig. 1). Based on this phylogenetic tree, we predicted alleles, many of which may be extant in the population, particularly those of greater posterior probability. Our approach applied standard methods of phylogenetic inference, ancestral character reconstruction and aimed to enrich the repertoire for a focal genomic region, of specific clinical interest.
Fig. 1.
Phylogenetic tree of 48 ERMAP alleles. The phylogeny of the 48 known ERMAP alleles was determined based on a standard Bayesian phylogenetic analysis. Branch width indicates posterior probability support (thick is ≥ 0.95 and thin is < 0.95). The colored circles represent sampled alleles that are also predicted ancestral alleles with the highest posterior probability. The 13 nodes are labelled A to L. Nodes B and B′ share the same allele with the greatest, but different, posterior probabilities (see Table 1)
Phylogeny
The Bayesian phylogenetic analysis of the 48 ERMAP alleles identified 13 nodes (Fig. 1, nodes A to L) and 4 clades (Fig. 1, clades 1 to 4). The clades comprised clusters of 5 to 12 alleles. Alleles were equally distributed between African American and Caucasian populations (Additional file 1: Fig. S1). For each clade, one observed allele was identified as the ancestral allele and had a posterior probability of more than 0.60 (nodes I to L). The remaining 9 internal nodes had 8 predicted alleles as the most probable ancestors with the highest posterior probabilities ranging from 0.235 to 0.792 (nodes A to H; Table 1). Thus, the phylogenetic tree comprised 4 confirmed alleles and 8 predicted alleles (Table 1). The most likely ancestral allele (node A; posterior probability = 0.235) for all 48 ERMAP alleles had only 4 nucleotide differences relative to our reference sequence (GenBank accession KX265235).
Table 1.
Predicted alleles at internal nodes of the ERMAP phylogeny
| Node | Allelea | Sequenceb | Posterior probability | Status | GenBank number |
|---|---|---|---|---|---|
| Reference | Allele1 | ATTGGCACCAGGCCGCCGCCCTGCTTAAGCCCTGGCGTGGTACTCGTCACGGTCCGCCGGGGCCGGATTAAA | 1 | Observed | KX265235 |
| A | SPA18 | ------G--------------G--------T-------------T--------------------------- | 0.235 | Predicted | na |
| B | SPA03 | ------G--------------G--------T----------------------------------------- | 0.792c | Predicted | na |
| C | SPA06 | ------G----------A---G--------T-----------T-T--------------------------- | 0.444 | Predicted | na |
| B′ | SPA03 | ------G--------------G--------T----------------------------------------- | 0.516c | Predicted | na |
| D | SPA09 | ------G----------A---G--------T---A-A-----T-T--------------------------- | 0.608 | Predicted | na |
| E | SPA04 | ------G--------------G-------------------------------------------------- | 0.747 | Predicted | na |
| F | SPA07 | ------G-------------TG--G--G--T----------------------------------------- | 0.626 | Predicted | na |
| G | SPA10 | -C----G----------A---G--------T---A-A-----T-T--------------------------- | 0.594 | Predicted | na |
| H | SPA13 | -C----G----------A---G----T---T---A-A-----T-TC-------------------------- | 0.492 | Predicted | na |
| I | Allele12 | ---------------------G-------------------------------------------------- | 0.621 | Observed | KX265198 |
| J | Allele18 | G-----G---A--------TTG--G--G--T----------------------------------------- | 0.674 | Observed | KX265204 |
| K | Allele08 | ------G--------------G-------------T------------------------------------ | 0.888 | Observed | KX265194 |
| L | Allele17 | -C----G----------A---G----T---T---A-A---C-T-TC-------------------------- | 0.634 | Observed | KX265203 |
na not applicable
aAlleles 1, 8, 12, 17, and 18 are experimentally confirmed alleles as published previously [21]. SPA03—SPA18 are predicted alleles (see Additional file 1: Table S1)
bThe nucleotides at the 72 SNP positions with variations are shown in 5′ to 3′ orientation (Table S2 in Srivastava et al. [21])
cThe posterior probabilities differ for SPA03 depending on its position in the phylogenetic tree (see Fig. 1)
Ancestral allele prediction
From the phylogenetic tree, we extracted all possible ancestral alleles at each internal node (nodes A to L). A total of 4243 unique predicted alleles were computed and sorted according to their calculated posterior probability of being the true ancestor (Additional file 2: Excel file S1). Even though the posterior probabilities of the inferred ancestral alleles were often below the threshold for statistical significance (0.95), the posterior probabilities of the next most likely predicted alleles dropped off dramatically. The exceptions to this were at Node A (best posterior probability = 0.23, next best = 0.19), Node B′ (0.52 vs. 0.29), and Node I (0.62 vs. 0.34). In all other cases the posterior probabilities of the secondary inferred ancestral allele were less than half the greatest values.
Discussion
A phylogenetic analysis was applied to a set of 48 physically confirmed ERMAP alleles covering 5 populations worldwide [21]. We predicted 4243 unobserved alleles and their distinct posterior probabilities. The relatively small number of predicted alleles contrasted to the vastly larger number of theoretically possible alleles. The predicted alleles have a stronger support for being correct and extant in the population because they are more likely ancestral to the observed alleles. We propose the concept of detecting unobserved, likely novel, alleles based on the phylogeny of verified alleles.
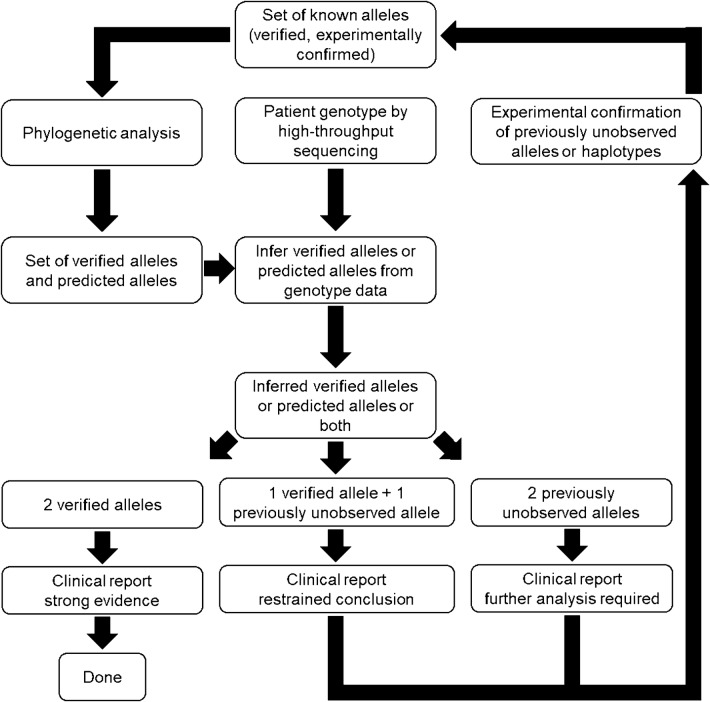
Previous computationally driven algorithms to phase NGS data such as read-backed phasing [30] and haplotype improver [31] remain very useful for phasing haplotypes and alleles in a population sample but may fail when applied to a single observation in an individual patient. Our approach utilizes predicted alleles and their posterior probabilities along with the verified alleles as templates for phasing the genotypes detected in high-throughput sequencing (Fig. 2), complementing the computationally driven algorithms. This approach increases the effective number of templates available for phasing and thus the accuracy of phased haplotypes and alleles. When a previously unobserved allele matches one of the predicted alleles, its posterior probability allows to quantify the reliability of the estimate for clinical decisions, such as in transfusion and transplantation settings, in a patient who bears a new allele. While the validation of the predicted alleles by applying our protocol was not performed in this study, the novel approach illustrates the potential use of phylogenetic data in a clinical diagnostic setting.
Fig. 2.
Algorithm to analyze genotypes and determine alleles using phylogeny data. Patient or blood donor genotype information for a particular gene is phased into alleles or haplotypes using statistical algorithms for clinical decisions. We propose a novel approach where the confidence for the inferred allele is based on verified, experimentally confirmed alleles and predicted alleles (see Fig. 1). The posterior probability of the predicted alleles is determined by a Bayesian phylogenetic analysis. Whenever a new allele is observed and experimentally confirmed, the phylogenetic analysis is in turn used to predict an updated set of alleles and their posterior probabilities. While this loop process continues, previously unobserved alleles will be encountered less frequently, as the set of confirmed allele increases
Our approach relies heavily on the accuracy of the alleles used for the phylogenetic analysis. Hence, reference sequences from online databases such as GenBank should be avoided as long as the information is not sufficiently replicated or independently verified [32]. The prevalence of the 48 alleles derived from 5 populations worldwide, but may still bias the imputation of novel alleles. Hence, addition of other alleles that are considered accurate, although computationally rather than physically derived, will strengthen the phylogenetic analysis and contribute to phasing of haplotypes and alleles, such as computed from the 1000 Genomes project [33] and similar online databases.
In our previously published set of long range ERMAP alleles with 72 single nucleotide polymorphisms (SNPs), the number of theoretically possible alleles was 272 [21]. However, it is known that the majority of the haplotype diversity is constituted by only few common haplotypes, which is constant in a given population [34]. Our algorithm restricts the possible ERMAP alleles from 272 to 4243 only, some associated with greater probability of being correct, but all as potential precursors of the experimentally verified extant alleles. With only 72 variable nucleotide positions in our set of 48 ERMAP alleles [21], the vast majority of positions remained uninformative (21,334 of 21,406 nucleotides: 99.66%).
Our observation contrasts with the 2353 SNPs, including 66 out of our 72 SNPs, reported for this DNA stretch covering the ERMAP gene [35], most of them being rare and often not validated to the extent needed for clinical decision making. Increasing the sample size will result in the confirmation of many or most of the previously reported 2353 SNPs and also the identification of novel SNPs in this DNA stretch. However, many of these SNPs will be specific for a small number of individuals resulting in a small global allele frequency.
While initially disregarding recombination as a major contributor, the subsequent analysis of the ERMAP sequences using the ClonalFrameML software [36] was also unable to detect any recombination event among the 48 confirmed alleles. This observation could be explained by the small sample size, which will resolve with the accumulation of more data. Our observation may, however, be an actual feature of ERMAP alleles in the population, because it is similar to the ABO gene, for which the detected recombinant alleles are also of low frequency [37]. As ERMAP alleles caused by recombination will eventually be found, they can be incorporated in the set of alleles used to compute the phylogenetic analysis.
Summary
By applying a Bayesian phylogenetic approach to 48 alleles, more than 21 kb long and all experimentally verified, we predicted a large set of not-yet-observed alleles of the ERMAP blood group gene. We propose a strategy of using these predicted alleles and their associated probabilities of correctness in clinical diagnostics such as designing of microarray technologies, imputing of genotypes and mapping of NGS data.
Additional files
Additional file 1. Table S1. Predicted ERMAP alleles with posterior probability of greater than 0.10. Figure S2. Distribution of alleles in 5 ethnic groups. The number of alleles observed in 50 individuals, as previously reported in Srivastava et al. (Table S2) [21], are shown for the clades in the phylogenetic tree (see Fig. 1).
Additional file 2. Excel file S1. List of 4243 predicted alleles of ERMAP gene.
Authors’ contributions
WAF developed the study plan. KRW designed and performed computer modeling. Data were analyzed and discussed by all authors. KS and WAF wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Harvey Gordon Klein for critical review of the manuscript; and Elizabeth Jane Furlong for English edits. We acknowledge the use of the High Performance Computing (HPC) cluster at the Office of Cyber Infrastructure and Computational Biology (OCICB), National Institute of Allergy and Infectious Diseases (NIAID), Bethesda MD. The data and the new algorithm have been presented at the AABB Annual Meeting on October 9, 2017 [22].
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Data availability
All data analyzed in this study has been extracted from GenBank database (KX265189–KX265236). Additional file 2: Excel file S1 lists all the 4243 predicted alleles of ERMAP gene.
Ethics approval and consent to participate
Not applicable.
Funding statement
This work was supported by the Intramural Research Program (Project ID Z99 CL999999) of the NIH Clinical Center.
Statement of disclaimer
The views expressed do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the U.S. Federal Government.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kshitij Srivastava, Email: srivastavak@mail.nih.gov.
Kurt R. Wollenberg, Email: wollenbergk@niaid.nih.gov
Willy A. Flegel, Phone: (301) 594-7401, Email: waf@nih.gov
References
- 1.Tay GK, Witt CS, Christiansen FT, Charron D, Baker D, Herrmann R, Smith LK, Diepeveen D, Mallal S, McCluskey J, et al. Matching for MHC haplotypes results in improved survival following unrelated bone marrow transplantation. Bone Marrow Transplant. 1995;15(3):381–385. [PubMed] [Google Scholar]
- 2.Chou ST, Liem RI, Thompson AA. Challenges of alloimmunization in patients with haemoglobinopathies. Br J Haematol. 2012;159(4):394–404. doi: 10.1111/bjh.12061. [DOI] [PubMed] [Google Scholar]
- 3.Tournamille C, Meunier-Costes N, Costes B, Martret J, Barrault A, Gauthier P, Galacteros F, Nzouekou R, Bierling P, Noizat-Pirenne F. Partial C antigen in sickle cell disease patients: clinical relevance and prevention of alloimmunization. Transfusion. 2010;50(1):13–19. doi: 10.1111/j.1537-2995.2009.02382.x. [DOI] [PubMed] [Google Scholar]
- 4.Allen ES, Srivastava K, Hsieh MM, Fitzhugh CD, Klein HG, Tisdale JF, Flegel WA. Immunohaematological complications in patients with sickle cell disease after haemopoietic progenitor cell transplantation: a prospective, single-centre, observational study. Lancet Haematol. 2017;4(11):e553–e561. doi: 10.1016/S2352-3026(17)30196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat Rev Genet. 2011;12(10):703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd SS, Steele EJ, Dawkins RL. Analysis of Haplotype Sequences. In: Kulski JK, editor. Next Generation Sequencing-Advances, Applications and Challenges. InTechOpen; 2016. pp. 345–368.
- 7.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SGE. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava K, Almarry NS, Flegel WA. Genetic variation of the whole ICAM4 gene in Caucasians and African Americans. Transfusion. 2014;54(9):2315–2324. doi: 10.1111/trf.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmid P, Ravenell KR, Sheldon SL, Flegel WA. DARC alleles and Duffy phenotypes in African Americans. Transfusion. 2012;52(6):1260–1267. doi: 10.1111/j.1537-2995.2011.03431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calafell F, Roubinet F, Ramirez-Soriano A, Saitou N, Bertranpetit J, Blancher A. Evolutionary dynamics of the human ABO gene. Hum Genet. 2008;124(2):123–135. doi: 10.1007/s00439-008-0530-8. [DOI] [PubMed] [Google Scholar]
- 11.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, Albracht D, Kremitzki M, Rock S, Kotkiewicz H, Kremitzki C, Wollam A, Trani L, Fulton L, Fulton R, Matthews L, Whitehead S, Chow W, Torrance J, Dunn M, Harden G, Threadgold G, Wood J, Collins J, Heath P, Griffiths G, Pelan S, Grafham D, Eichler EE, Weinstock G, Mardis ER, Wilson RK, Howe K, Flicek P, Hubbard T. Modernizing reference genome assemblies. PLoS Biol. 2011;9(7):e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, Fulton RS, Kremitzki M, Magrini V, Markovic C, McGrath S, Steinberg KM, Auger K, Chow W, Collins J, Harden G, Hubbard T, Pelan S, Simpson JT, Threadgold G, Torrance J, Wood JM, Clarke L, Koren S, Boitano M, Peluso P, Li H, Chin CS, Phillippy AM, Durbin R, Wilson RK, Flicek P, Eichler EE, Church DM. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27(5):849–864. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su YY, Gordon CT, Ye TZ, Perkins AC, Chui DH. Human ERMAP: an erythroid adhesion/receptor transmembrane protein. Blood Cells Mol Dis. 2001;27(5):938–949. doi: 10.1006/bcmd.2001.0465. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Foltz L, Sha Y, Madlansacay MR, Cain C, Lindemann G, Vargas J, Nagy D, Harriman B, Mahoney W, Schueler PA. Cloning and characterization of human erythroid membrane-associated protein, human ERMAP. Genomics. 2001;76(1–3):2–4. doi: 10.1006/geno.2001.6600. [DOI] [PubMed] [Google Scholar]
- 15.Wagner FF, Poole J, Flegel WA. Scianna antigens including Rd are expressed by ERMAP. Blood. 2003;101(2):752–757. doi: 10.1182/blood-2002-07-2064. [DOI] [PubMed] [Google Scholar]
- 16.Velliquette RW. Review: the Scianna blood group system. Immunohematology. 2005;21(2):70–76. [PubMed] [Google Scholar]
- 17.Ye T-Z, Gordon CT, Lai Y-H, Fujiwara Y, Peters LL, Perkins AC, Chui DHK. Ermap, a gene coding for a novel erythroid specific adhesion/receptor membrane protein. Gene. 2000;242(1–2):337–345. doi: 10.1016/s0378-1119(99)00516-8. [DOI] [PubMed] [Google Scholar]
- 18.Afrache H, Gouret P, Ainouche S, Pontarotti P, Olive D. The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics. 2012;64(11):781–794. doi: 10.1007/s00251-012-0619-z. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DA, Reith W, Trowsdale J. Regulation of Immunity by Butyrophilins. Annu Rev Immunol. 2016;34:151–172. doi: 10.1146/annurev-immunol-041015-055435. [DOI] [PubMed] [Google Scholar]
- 20.Di Marco Barros R, Roberts NA, Dart RJ, Vantourout P, Jandke A, Nussbaumer O, Deban L, Cipolat S, Hart R, Iannitto ML, Laing A, Spencer-Dene B, East P, Gibbons D, Irving PM, Pereira P, Steinhoff U, Hayday A. Epithelia use butyrophilin-like molecules to shape organ-specific gammadelta T cell compartments. Cell. 2016;167(1):203–218. doi: 10.1016/j.cell.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava K, Lee E, Owens E, Rujirojindakul P, Flegel WA. Full-length nucleotide sequence of ERMAP alleles encoding Scianna (SC) antigens. Transfusion. 2016;56(12):3047–3054. doi: 10.1111/trf.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava K, Wollenberg KR, Flegel WA. Use of 48 ERMAP alleles, at 21,406 nucleotides each, to predict haplotypes for genotype prediction from next generation sequencing data (abstract) Transfusion. 2017;57(Supplement S3):44A. [Google Scholar]
- 23.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21(2):255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 26.Lam JC, Roeder K, Devlin B. Haplotype fine mapping by evolutionary trees. Am J Hum Genet. 2000;66(2):659–673. doi: 10.1086/302756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 28.Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst Biol. 2007;56(3):453–466. doi: 10.1080/10635150701420643. [DOI] [PubMed] [Google Scholar]
- 29.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161(3):1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long Q, MacArthur D, Ning Z, Tyler-Smith C. HI: haplotype improver using paired-end short reads. Bioinformatics. 2009;25(18):2436–2437. doi: 10.1093/bioinformatics/btp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Koyuturk M, Maxwell S, Xiang M, Veigl M, Cooper RS, Tayo BO, Li L, LaFramboise T, Wang Z, Zhu X, Chance MR. Discovery of common sequences absent in the human reference genome using pooled samples from next generation sequencing. BMC Genomics. 2014;15:685. doi: 10.1186/1471-2164-15-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Genomes Project C A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 35.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11(2):e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsson ML, Chester MA. Polymorphism and recombination events at the ABO locus: a major challenge for genomic ABO blood grouping strategies. Transfus Med. 2001;11(4):295–313. doi: 10.1046/j.1365-3148.2001.00320.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Predicted ERMAP alleles with posterior probability of greater than 0.10. Figure S2. Distribution of alleles in 5 ethnic groups. The number of alleles observed in 50 individuals, as previously reported in Srivastava et al. (Table S2) [21], are shown for the clades in the phylogenetic tree (see Fig. 1).
Additional file 2. Excel file S1. List of 4243 predicted alleles of ERMAP gene.
Data Availability Statement
All data analyzed in this study has been extracted from GenBank database (KX265189–KX265236). Additional file 2: Excel file S1 lists all the 4243 predicted alleles of ERMAP gene.