Abstract
Background
Epstein–Barr virus (EBV) is a universal herpes virus which can cause a life-long and largely asymptomatic infection in the human population. However, the exact pathogenesis of the EBV infection is not well known.
Objective
A comprehensive bioinformatics prediction was carried out for investigating the molecular properties of the BGLF2 and to afford a foundation for future research of the role and instrument of BGLF2 in the course of EBV infection.
Materials and Methods
A 1011-base-pair sequence of BGLF2 gene from the Epstein-Barr virus (EBV) Akata strain genome was amplified using polymerase chain reaction and was further characterized by cloning, sequencing, and subcellular localization in the COS-7 cells.
Results
The bioinformatics analysis demonstrated that EBV BGLF2 gene encodes a putative BGLF2 polypeptide which contains a conservative Herpes_UL16 domain. It was established that the polypeptide shows a close relationship with the Herpes UL16 tegument protein family and is extremely conserved among its homologues proteins encoded by UL16 genes. Multiple sequence alignments of the nucleic acid and amino acid sequence showed that the gene product of EBV BGLF2 contains a comparatively higher homology with the BGLF2-like proteins of the subfamily Gammaherpesvirinae than that of other subfamilies of the herpes virus. Moreover, the phylogenetic analyses suggested that EBV BGLF2 has a close genetic relationship with the member of Gammaherpesvirinae; in particular with the members of Cercopithecine herpesvirus 15 and Callitrichine herpesvirus 3. An antigen epitope analysis indicated that BGLF2 contains several potential B-cell epitopes. In addition, the secondary structure, as well as the three dimensional structure prediction suggests that BGLF2 consists of the both α-helix and β-strand. Besides, the subcellular localization prediction revealed that BGLF2 localizes in both nucleus and cytoplasm.
Conclusions
Illustrating the relevance of the molecular properties and genetic evolution of EBV, BGLF2 will offer the perspectives for further study on the role and mechanism of the BGLF2 in course of EBV infection. These works will also conduct our understanding of the EBV at the molecular level as well as enriching the herpesvirus database.
Keywords: BGLF2, Cloning, Bioinformatics analysis, Epstein-Barr virus, Molecular property
1. Background
Epstein–Barr virus (EBV), also named as human herpesvirus 4 (HHV-4), is a ubiquitous herpesvirus with the property of being universally spread in the human population, inducing a perpetual, and to a large extent asymptomatic infection. It was shown that EBV is associated with the infectious mononucleosis and various malignancies, including Burkitt Lymphoma (BL), Hodgkin Lymphoma (HL), post-transplant lympho-proliferative disease (PTLD), as well as an epithelial tumour, T/NK cell lymphomas, and nasopharyngeal carcinoma (NPC) (1-3). However, the exact pathogenic mechanism of the EBV infection is still less understood.
As a potential tegument protein, EBV BGLF2 is important for re-envelopment in the cytoplasm or plasma membrane, which can be detected in the mature EBV virions. BGLF2 is also related to EBV reactivation via stimulation of p38 mitogen-activated protein kinase (1-3). However, the accurate functions of BGLF2 gene and its product BGLF2 are not well known.
2. Objectives
For sake of deciphering the role of BGLF2, the BGLF2 gene was amplified from the DNA template of the EBV Akata strain by employing polymerase chain reaction (PCR) followed by cloning, sequencing, and expressing in the COS-7 cells. Subsequently, a comprehensive bioinformatics prediction was carried out to investigate the molecular properties of the BGLF2 and to offer a foundation for future investigation of the role and the mechanism of BGLF2 in the course of EBV infection. The prediction included various bioinformatics tools such as clone manager professional suite 8.0, Conserved Domains, SignalP-4.0, DNAstar 7.0, NetPhos 2.0, Bioedit 7.0, PSIpred, and CPHmodels 3.2.
3. Materials and Methods
3.1. Cloning and Sequencing of EBV BGLF2
The primers for PCR amplification of EBV Akata strain BGLF2 (accession No. KC207813) were designed using clone manager professional suite 8.0 and afforded by Sangon biotech (Shanghai, China). The forward primer BGLF2-F: 5′-TTGAATTCCATGGCATCCGCCGCGAACAG-3′ is complementary to the initial 20 nucleotides of BGLF2 and introduced with an EcoRI restriction site (underlined) for cloning. The reverse primer BGLF2-R: 5′-AAGGATCCTCAATAAGAATGTAAGACCTGAC G-3′ anneals to the 23 terminal nucleotides of BGLF2 and introduced with a BamHI restriction site (underlined).
The BGLF2 gene was amplified with KOD-plus-Neo DNA polymerase (TOYOBO) via PCR using the EBV-BAC (bacterial artificial chromosome) of Akata strain (AK-BAC) (a generous gift from Dr. Teru Kanda) as the template DNA (4). PCR profiles implicated initial pre-denaturation for 10 min at 94 °C followed by 30 cycles of denaturation at 95 °C for 50 s, annealing at 54 °C for 45 s, and extension at 72 °C for 70 s. The final extension step was implemented at 72 °C for 10 min. Then, the PCR product was purified and digested with EcoRI and BamHI and inserted into the corresponding digested eucaryotic yellow fluorescent protein expression vector pEYFP-C1 (Clontech, USA) to generate pEYFP-BGLF2. The existence of the proper fragment in the attained plasmid was then verified by the restriction analysis, PCR, and sequencing (5).
3.2. Western Blot Analysis
To further verify the validity of pEYFP-BGLF2, COS-7 cells were seeded at 2.5×105 cells per well in a 12-well plate and at 80% confluence after overnight incubation cells were transiently transfected with the expressing plasmid pEYFP-BGLF2 or empty vector pEYFP-C1 by the standard calcium phosphate precipitation process. The transfection efficiencies of the different plasmids in COS-7 cells were evaluated by enhanced yellow fluorescent protein (EYFP). Twenty-four hours later the transfected cells were harvested and fractionated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and subsequent Western blot analysis was carried out as described previously using monoclonal antibody (mAb) against YFP (Santa Cruz Biotechnology, Santa Cruz, CA) (6-12).
3.3. Plasmid Transfection and Fluorescence Microscopy
Transfection and fluorescence microscopy procedures were carried out as reported in our previous researches (6-8, 13-15). For duplicate the assay, each transfection was manipulated at least three times. Results presented are from one representative assay. Results were analyzed by a Zeiss Axiovert 200 M microscope (Germany). All the photomicrographs were captured at an enlargement of ×400. Each picture shows an enormous majority of the cells with analogous subcellular localizations. The light-translucent images indicate cellular morphology. Fluorescence pictures of the EYFP tagged fusion proteins are indicated in pseudocolor green. Figures were refined with Adobe Photoshop software.
3.4. Nucleotide Sequence Bioinformatics Analysis of EBV BGLF2
To determine the nucleotide sequence similarity, the National Center for Biotechnology Information (NCBI) nucleotide Basic Local Alignment Search Tool (16) was applied. Subsequently, Clustal V in the MegAlign program of DNAStar (version 7.0, DNAStar, Inc.) was employed to analyze the nucleotide sequence homology of the 55 BGLF2-like proteins of herpesviruses (Supplementary File).
3.5. Bioinformatics analysis of the Deduced Amino Acid (AA) Sequence of EBV BGLF2
For AA sequence comparison, homology search, and conserved domain analysis, the AA sequence of BGLF2 was analyzed with protein BLAST (16), FASTA (17), and the Conserved Domains search tool (18), respectively. To compare BGLF2 with BGLF2-like proteins of the other herpesviruses (see Supplementary File), we analyzed the AA sequence homology and phylogenetic relationships using DNAstar 7.0. To predict transmembrane domain, the signal peptide sequence, glycosylation site, phosphorylation site, hydrophobic and hydrophilic regions, B-cell epitope, secondary structure, and the 3-dimensional (3D) structure of the BGLF2, TMHMM (19), SignalP-4.0 Server (20), NetNGlyc 1.0(21), NetPhos 2.0(22), Bioedit 7.0, DNAstar 7.0, PSIpred (23), and CPHmodels 3.2(24) were applied, respectively.
4. Results
4.1. PCR Amplification and Cloning of EBV BGLF2
To get BGLF2 gene, we performed PCR on the basis of the DNA template from the purified BAC DNA of the EBV Akata strain. As shown in Figure 1, no specific band was amplified from the negative control (Fig. 1, lane 1), while a target fragment of 1011 bp, which is consistent with the expected size was amplified from EBV BAC DNA (see Fig. 1, lane 2). The DNA fragment was then ligated into the eukaryotic expression vector pEYFP-C1, transformed into E. coli, and characterized by colony PCR (Fig. 1, lane 3 and lane 4). Subsequently, the identified pEYFP-BGLF2 plasmid was extracted from the positive colony (see Fig. 1, lane 5), which was further verified by restriction digestion analysis (Fig.1, lane 6 and lane 7) and DNA sequencing. Compared to that of the EBV Akata strain, the sequencing result indicated that there was no AA mutation existing in the clone (data not shown).
Figure 1.
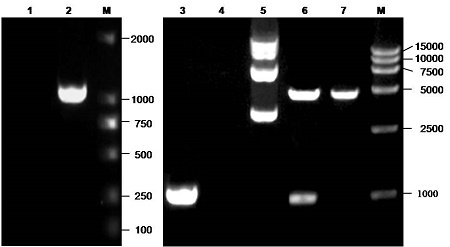
Cloning and verification of the plasmid pEYFP-BGLF2. Lane 1 to lane 4, PCR amplification product of no template control (lane 1), Akata-derived EBV-BAC DNA (lane 2), PCR product of pEYFP-BGLF2 (lane 3) or PCR product of the empty vector of pEYFP-C1 (lane 4) colony by primers BGLF2-F and BGLF2-R; lane 5, plasmid pEYFP-BGLF2; lane 6 and lane 7, restriction digestion product (approximately 4700 and 1018 bp, respectively) of the pEYFP-BGLF2 or restriction digestion product (approximately 4700 and 31 bp, respectively) of the pEYFP-C1 with EcoRI and BamHI. Lane M, DNA marker (TaKaRa). Samples were electrophoresed using a 1% agarose gel and stained with ethidium bromide.
4.2. Expression of EBV BGLF2 in Eukaryotic Cells
After sequence confirmation, the recombinant plasmid pEYFP-BGLF2 and the empty vector pEYFP-C1 were transfected into the expression host; the COS-7 cells. After 24 hours, the expression of EYFP-BGLF2 and EYFP were examined via Western blot using anti-YFP mAb. As a result a distinct band of approximately 66 kDa molecular weight, corresponding to the expected size of EYFP-tagged BGLF2 fusion protein (Fig. 2a), was detected. However, a band with only about 27 kDa was observed in the control COS-7 cells transfected with pEYFP-C1 (Fig. 2a). This result assured that the open reading frame of the BGLF2 was properly expressed in the transfected COS-7 Cells
Figure 2.
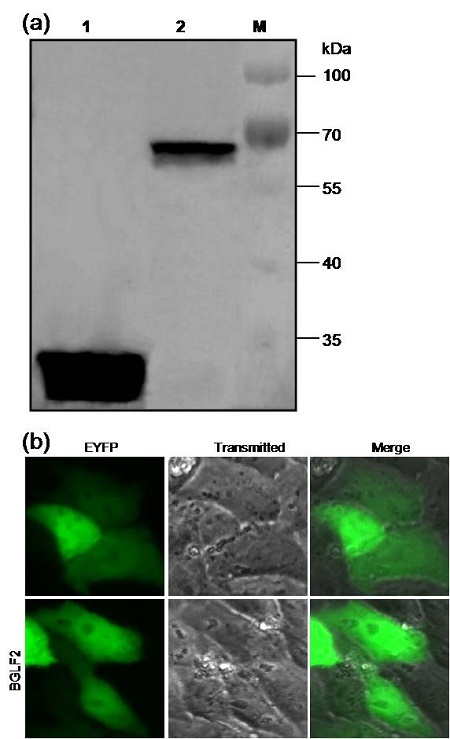
Protein expression and subcellular localization of the EBV BGLF2 in transfected cells. (a) Expression analysis of the EBV BGLF2 in transfected cells using anti-YFP mAb through Western blot. Lane 1, COS-7 cells transfected with plasmid pEYFP-C1; lane 2, COS-7 cells transfected with plasmid pEYFP-BGLF2; lane M, protein marker (Thermo Scientific PageRuler Prestained Protein Ladder). (b) The subcellular distribution of EYFP and EYFP-BGLF2. Each fluorescence image is representative of the vast majority of the observed cells.
4.3. Subcellular Localization of BGLF2 in the Transfected Cells
It’s well known that determination of the subcellular localization is one way to evaluate the potential roles of some specific proteins. To further investigate the subcellular distribution of BGLF2 in transfected live cells, fluorescence microscopy was employed. The plasmid pEYFP-BGLF2 encoding BGLF2 fused to the C terminus of EYFP was transfected into COS-7 cells to study the subcellular localization of the BGLF2 in absence of other viral proteins. As shown in Fig. 2b, the control EYFP fluorescence was evenly distributed throughout the cytoplasm and the nucleus in the cells transfected with pEYFP-C1. Moreover, EYFP-BGLF2 also exhibited a similar subcellular distribution pattern with that of EYFP (Fig. 2b).
4.4 Bioinformatics Analysis of the EBV BGLF2 Nucleotide Sequence
The nucleotide sequence similarity search using NCBI BLASTN yielded 5 nucleotide sequences (accession Nos. DQ279927, JQ009376, HQ020558, AJ507799, and AY961628) with strong identity to the EBV Akata strain BGLF2 (up to 98.5, 99.8, 99.8, 99.9, and 100%, respectively) (see Table 1 and Fig. 3); these accessions were corresponded to the BGLF2 gene of the EBV AG876, HKNPC1, GD2, B95-8, and GD1 strains, respectively. Multiple alignments of EBV BGLF2 with 54 homologous reference herpesviruses revealed a remarkably high similarity of 82.2% and 54.5% with the members of subfamily Gammaherpesvirinae, that is Cercopithecine herpesvirus 15 (CeHV-15) and Callitrichine herpesvirus 3 (CalHV-3). However, a relatively low similarity was detected between EBV and other members of the subfamily Betaherpesvirinae and Alphaherpesvirinae (Table 1 and Fig. 3).
Table 1.
Multiple nucleic acid sequence and amino acid sequence alignments of EBV BGLF2 gene with its reference species .
| Virus name | AlHV-1 | AnHV-1 | AoHV-1 | AtHV-3 | BBHV | BoHV-1 | BoHV-4 | BoHV-5 | CalHV-3 | CavHV-2 | CeHV-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PNHa (%) | 35.3 | 23.2 | 25.1 | 36.8 | 22.8 | 24.4 | 38.7 | 25.4 | 54.5 | 21.9 | 22.7 |
| PAHb (%) | 32.5 | 13.4 | 14.0 | 36.7 | 16.7 | 15.2 | 38.6 | 16.4 | 60.4 | 16.5 | 15.2 |
| Virus name | CeHV-2 | CeHV-5 | CeHV-9 | CeHV-15 | CeHV-16 | CeHV-17 | CMCMV | EEHV | EHV-1 | EHV-2 | EHV-4 |
| PNH(%) | 22.9 | 23.8 | 20.2 | 82.2 | 23.7 | 34.8 | 21.4 | 22.1 | 22.3 | 39.4 | 22.7 |
| PAH(%) | 14.6 | 16.4 | 14.3 | 87.8 | 14.3 | 35.1 | 15.8 | 16.1 | 13.4 | 35.1 | 13.7 |
| Virus name | EHV-8 | EHV-9 | FeHV-1 | GaHV-3 | HHV-1 | HHV-3 | HHV-5 | HHV-6 | HHV-7 | HHV-8 | McHV-3 |
| PNH(%) | 22.1 | 21.7 | 22.4 | 22.1 | 23.1 | 22.8 | 22.5 | 20.8 | 21.5 | 35.2 | 22.8 |
| PAH(%) | 13.4 | 13.1 | 14.6 | 14.3 | 13.7 | 12.5 | 14.3 | 15.2 | 16.1 | 30.8 | 16.1 |
| Virus name | MFRV | MuHV-1 | MuHV-2 | MuHV-4 | MuHV-8 | OvHV-2 | PLHV-1 | PLHV-2 | PLHV-3 | PnHV-2 | RHVP |
| PNH(%) | 35.8 | 23.9 | 20.3 | 32.9 | 24.1 | 39.9 | 42.4 | 42.8 | 42.5 | 21.7 | 35.2 |
| PAH(%) | 34.8 | 15.2 | 14.9 | 29.4 | 16.1 | 36.0 | 42.3 | 42.6 | 42.6 | 14.3 | 28.7 |
| Virus name | SaHV-1 | SaHV-2 | SaHV-3 | TuHV-1 | WMHV | HHV-4 AG876 | HHV-4 HKNPC1 | HHV-4 GD2 | HHV-4 B95-8 | HHV-4 GD1 | |
| PNH(%) | 22.0 | 39.1 | 20.8 | 24.0 | 33.4 | 99.5 | 99.8 | 99.8 | 99.9 | 100 | |
| PAH(%) | 15.2 | 39.1 | 16.4 | 15.8 | 29.4 | 100 | 100 | 100 | 100 | 100 |
a PNH indicates the multiple nucleic acid sequence alignment of BGLF2 gene of EBV Akata strain with its homologous genes of 54 selected species (Table 1) by using the MEGALIGN program in LASERGENE (DNAStar 7.0) with Clustal V Method, and sequence distance was calculated using weight matrix Identity. Gaps had been introduced by the alignment program to maximize the homology.
b PAH indicates the multiple aa sequence alignment of BGLF2 of EBV Akata strain with its homologous proteins of 54 selected species (Table 1) by using the MEGALIGN program in LASERGENE (DNAStar 7.0) with Clustal V Method, and sequence distance was calculated using weight matrix PAM250. Gaps had been introduced by the alignment program to maximize the homology.
Figure 3.
The multiple nucleic acid sequence alignments of the EBV BGLF2 gene against its reference species (supplementary file) by using the MEGALIGN program in LASERGENE (DNAStar 7.0) with Clustal V Method, and sequence distance calculated using weight matrix identity. Gaps had been introduced by the alignment program to maximize the homology. (The original file is available at www.ijbiotech.com).
4.5 Bioinformatics Analysis of the EBV BGLF2 Polypeptide Sequence
An AA sequence similarity search using NCBI BLASTP yielded 5 AA sequences (accession Nos. ABB89265, AFJ06878, AEM00649, CAD53441, and AAY41138) that completely matched with the target sequence of the EBV Akata BGLF2 (accession No. AFY97879; all reach to 100%), and these sequences corresponded to the BGLF2 protein of EBV AG876, HKNPC1, GD2, B95-8, and GD1 strains, respectively (Table 1 and Fig. 4). In addition, multiple sequence alignments of the BGLF2 with its homologs in the 54 references for the herpesviruses showed a high similarity of 60.4% or 87.8% between BGLF2 and its CalHV-3 or CeHV-15 counterparts of subfamily Gammaherpesvirinae. However, BGLF2 shared low similarity with the BGLF2-like proteins from the members of the Betaherpesvirinae and Alphaherpesvirinae subfamilies with values lower than 20% (Table 1 and Fig. 4). The phylogenetic analyses of the EBV and other herpesviruses were performed based on the sequences of BGLF2 and the BGLF2-like proteins of the 54 reference herpesviruses (Table 1). As the results showed, the proteins were preliminarily separated into different subfamilies, i.e. Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae (see Fig. 5), which is corresponded with the existing classification for the subfamily of herpesvirus. The general branching pattern is also in line with previously published phylogenetic analyses (25, 26). Furthermore, the EBV Akata, AG876, HKNPC1, GD2, B95-8, and GD1 strains were different from the other Gammaherpesviruses. They clustered together and formed a separate branch and then clustered with members of the subfamily Gammaherpesvirinae such as CeHV-15 and CalHV-3. Subsequently, they clustered with other members of the subfamily Gammaherpesvirinae (Fig. 5). Therefore, EBV might have a closer evolutionary relationship with the member of subfamily gammaherpesviruses.
Figure 4.
The multiple AA sequence alignments of the EBV BGLF2 gene against its reference species (supplementary file) by using the MEGALIGN program in LASERGENE (DNAStar 7.0) with Clustal V Method and sequence distance was calculated using the weight matrix PAM250. Gaps had been introduced by the alignment program to maximize the homology. (The original file is available at www.ijbiotech.com).
Figure 5.
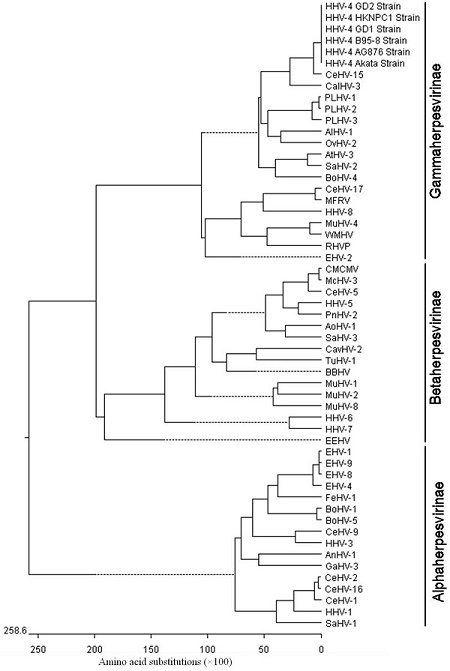
The evolutionary relationships of the EBV BGLF2 protein with its 54 reference herpesviruses from different species (see supplementary file). The phylogenetic tree of these proteins was generated using the Megalign program in LASERGENE (DNAStar 7.0) with the Clustal V method and the sequence distance was calculated using weight matrix PAM250. Gaps were introduced by the alignment program to maximize the homology.
The transmembrane domain and the signal polypeptide prediction showed no transmembrane domain (Fig. 6a) and the signal polypeptide cleavage site (Fig. 6b) in BGLF2. However, N-linked glycosylation site (Asn-X-Ser/Thr) prediction demonstrated that there are 3 potential N-glycosylation sites in the BGLF2 (Fig. 6c). Interestingly, 15 potential phosphorylation sites were observed in the BGLF2 (Fig. 6d) including 6 serine, 5 tyrosine, and 4 threonine residues.
Figure 6.
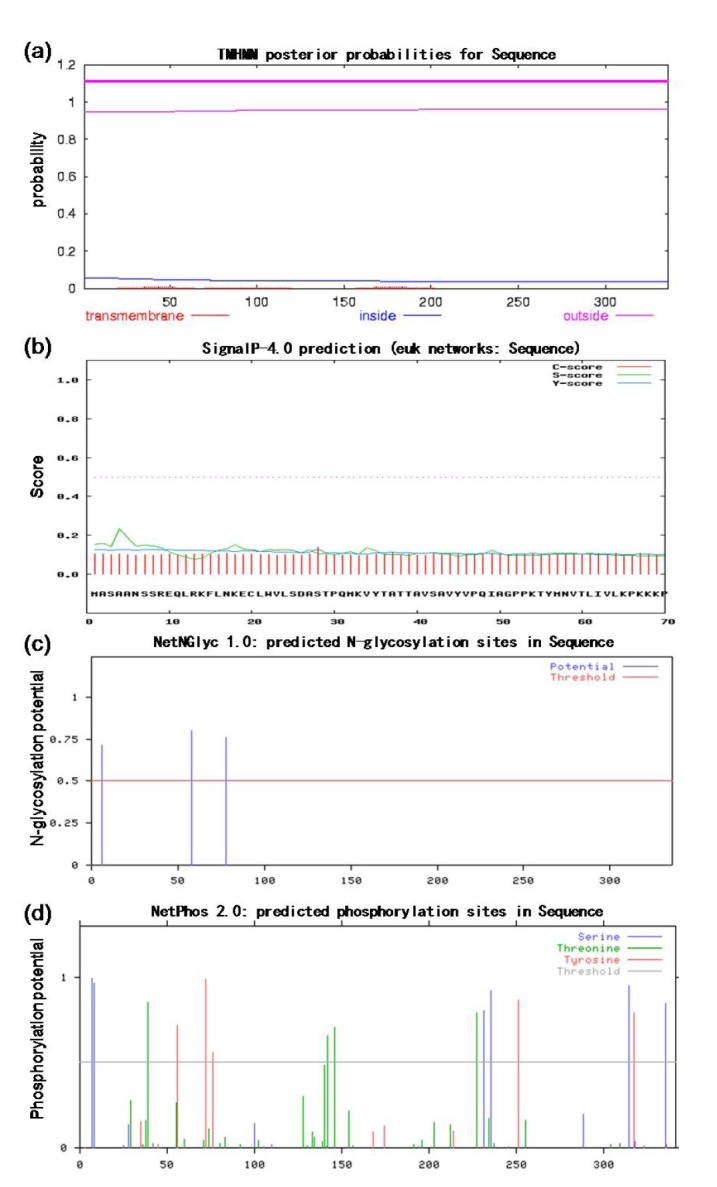
The prediction of transmembrane domain, signal peptide sequence, glycosylation site, and phosphorylation site of the EBV BGLF2 using TMHMM program (a), SignalP-4.0 (b), NetNGlyc 1.0 program (c), and NetPhos 2.0 program (d), respectively.
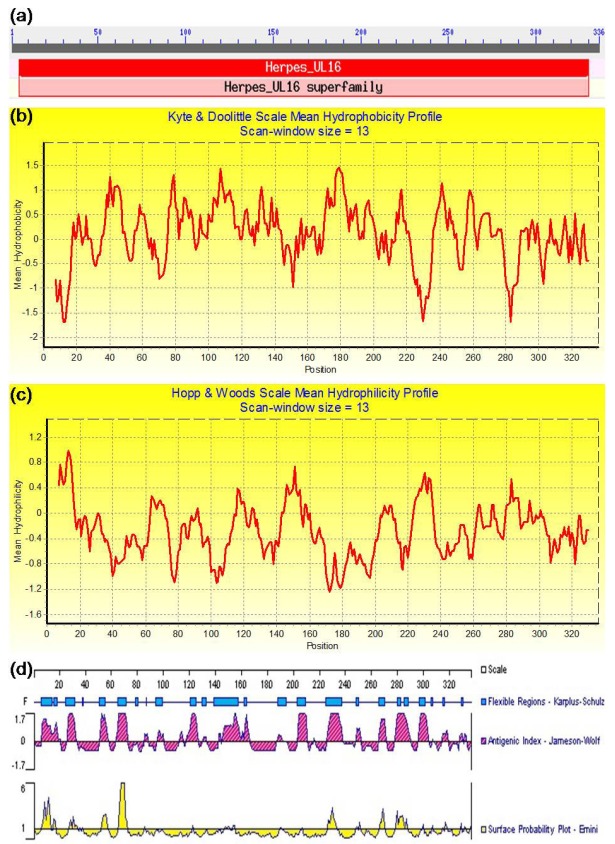
In addition, the conserved domain analysis indicated that BGLF2 contained an apparently conserved domain of the Herpes_UL16, which is closely related to the Herpes UL16 tegument protein family (Fig. 7a). The hydrophobicity analysis revealed several potential hydrophobic regions located at the AAs 17-26, 35-48, 56-63, 75-144, 155-164, 168-201, 212-221, 236-278, 289-300, and 314-327, respectively (Fig. 7b). Compared to the hydrophobic region, the proportionality of the hydrophilic region was smaller than the hydrophobic region (Fig. 7c). Furthermore, analysis of a potential B-cell epitope determinant demonstrated several potential B-cell epitopes in the BGLF2 situated in or adjacent to the AAs 7-19, 27-34, 52-57, 65-71, 121-126, 147-165, 204-211, 226-237, 266-272, 277-289, and 296-305, correspondingly (Fig. 7d). Moreover, the secondary structure analysis (Fig. 8a) suggested that the AA sequence of BGLF2 is composed of the random coils (58.63%), β-strand (25.30%), and α-helices (16.07%). Besides, the three dimensions (3D) structure prediction for the BGLF2 demonstrated that a known 3D structure model with a relatively high homology with the BGLF2 was searched (Fig. 8b), which was predominantly consisted by 81 H-bond, 7 α-helix, 2 β-strand, and 16 β-turn.
Figure 7.
The conserved domain, hydrophobicity, hydrophilicity, and antigenic analyses of the EBV BGLF2. (a) The conserved domain analysis of the BGLF2 using NCBI Conserved Domains search tool. (b) The hydrophobicity or (c) hydrophilicity profile was determined using the values of Kyte and Doolittle (Kyte and Doolittle, 1982) or Hopp and Woods (Hopp and Woods, 1981), respectively, with a 13-amino-acid window. The peaks pointing up represent the most hydrophobic (b) and hydrophilic (c) regions, respectively. (d) Antigenic analysis of the BGLF2 was carried out through determination of its primary structure using the PROTEAN software of the DNAStar based on its flexibility, antigenic index, and surface probability.
Figure 8.
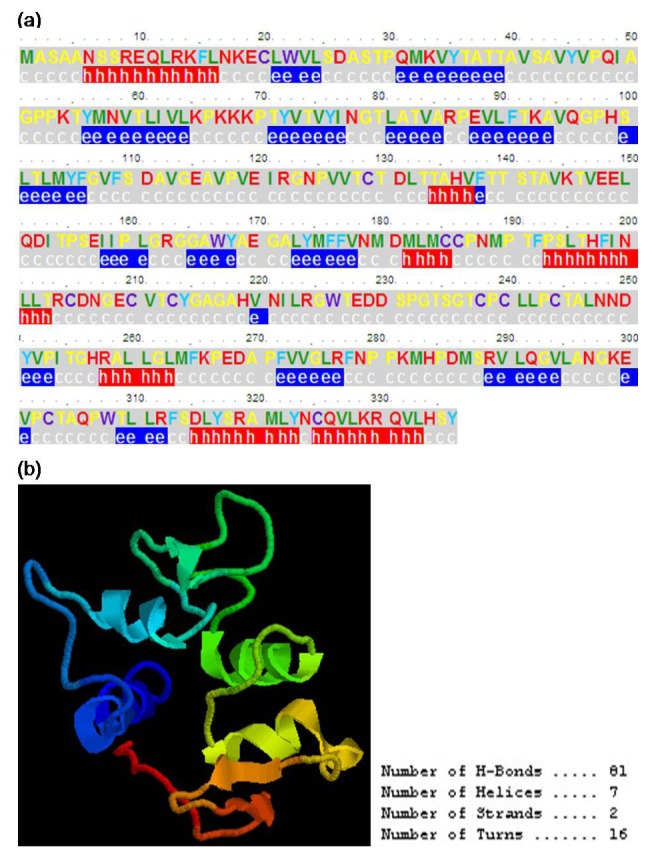
The secondary structure and three dimensional (3D) structure prediction of the EBV BGLF2. (a) The secondary structure of the BGLF2. The secondary structure was predicted by using the PSIpred program and the letters h, e, and c indicate alpha Helix, extended (beta strand) and Coil, respectively. (b) The 3D structure of the BGLF2. The 3D structure was predicted by employing the protein modeling server database CPHmodels 3.2 Server. The number of H-Bond, Helice, Strand and Turn included in this model were 81, 7, 2, and 16, respectively.
5. Discussion
The BGLF2 gene of the EBV Akata strain was confirmed by PCR amplification, cloning, and sequencing. In addition, in an attempt to further exploring the biological functions of the BGLF2, an expression plasmid pEYFP-BGLF2 that drives expression of the BGLF2 protein fused with EYFP-tag was constructed in this investigation, since the GFP (green fluorescent protein) or its mutant such as EYFP has been employed as a probe to detect protein expressed in the living cells in the previous work (27, 28). After characterization of the EBV BGLF2 expression in the eukaryotic cells by Western blot, the molecular properties of the BGLF2 was analyzed by several bioinformatics tools.
Since viruses are generally conserved and only a few genes go through mutation in the process of evolution, viral evolution thus can be assessed at the molecular level. Our analysis uncovered that the nucleotide sequence similarity of the EBV BGLF2 of the Akata strain to that of AG876, HKNPC1, GD2, B95-8, and GD1 strains was up to 99.5% to 100%, and the AA sequence similarities were all up to 100% (Table 1), respectively, clearly indicated that GD1 is the most closely related strain to that of the Akata strain. These results also manifested a close relationship among these studied EBV strains.
It is well known that the mutation of the virus may be caused by environmental pressure. Thus, a limited extent of gene mutation among different strains may render the variance of EBV virulence. Carrying our multiple nucleotide and AA sequence alignments of the BGLF2 and the BGLF2-like proteins, it was shown that BGLF2 is conserved throughout all the herpesvirus families and has greater similarity with the members of subfamily Gammaherpesvirinae, especially CalHV-3 or CeHV-15 (54.5% and 60.4% or 82.2% and 87.8%, respectively). Therefore, BGLF2 has high homology with the subfamily of Gammaherpesvirinae, but not Alphaherpesvirinae or Betaherpesvirinae. Accordingly, phylogenetic analysis (Fig. 5) obviously indicated that EBV belongs to the subfamily Gammaherpesvirinae, in line with the conclusions of a previous report (25).
Conserved domain analysis demonstrated that EBV BGLF2 encompasses a conserved domain Herpes_UL16 (Fig. 7a) which encodes herpesvirus tegument protein UL16. Therefore, EBV BGLF2 may belong to the Herpes_UL16 superfamily. HSV-1 UL16, a member of the Herpes_UL16 superfamily that is conserved among all the herpesviruses, may be involved in the viral DNA packaging, virion assembly, budding, and egress by providing an interaction with the membrane-bound UL11 protein, UL21 protein and glycoprotein E (29). Another UL16 homologue is HCMV UL94 which is a nucleo-cytoplasmic shuttling protein that may play important role in the virion assembly and budding (30, 31). Besides, tegument protein ORF33 of the murine gammaherpesvirus 68 (MHV-68) is also a member of Herpes_UL16, which is required for the viral lytic replication, virion morphogenesis, and egress (32). Indeed, the subcellular localization study of EBV BGLF2 suggests that in the transfected cell EBV BGLF2 is predominantly dispersed throughout the cytoplasm and the nucleus which is similar to the subcellular localization of the MHV-68 ORF33 and confirms again the previous report (33). However, the future study needs to be carried out to determine whether BGLF2 possess similar functions with its homologues. It is reported that almost every protein is labeled with a correct subcellular localization signal, and various subcellular localizations of a specific viral protein may play diverse roles in the course of infection (34). Since BGLF2 is a capsid binding protein, in the future works, of course, it is crucial to identify the functions of the potential subcellular localization signal (including nuclear localization signal and nuclear export signal) of the BGLF2 in the process to virus replication, especially in the encapsidation process or viral assembly.
Protein phosphorylation is one of the most normal and essential types of protein modifications and certain aspects of the cell process, for instance, signal transduction, proliferation, differentiation, and metabolism, all of which are prone to be regulated by the activities of certain protein kinases and protein phosphatases upon pivotal target proteins. Applying phosphorylation site prediction we have shown that there are 15 potential phosphorylation sites in the EBV BGLF2, including 6 serine, 5 tyrosine, and 4 threonine residues. It is well known that tyrosine phosphorylation is associated with the modification of the protein translocation from the cytoplasm to the nucleus during the course of productive viral infection (35). Phosphorylation is also implicated in the replication of many herpesviruses (36). Indeed, modification by the phosphorylation has been described for several herpesvirus tegument proteins such as the process of tegument dissociation from the incoming nucleocapsids likely occurs as a result of phosphorylation by cellular and viral kinases (32). Thus, phosphorylation of EBV BGLF2 may also play an important role during EBV infection, perhaps in modulating its proper localization (37, 38), stable capsid structure assembling (39), or playing a potential regulatory role in the affinity for DNA (40), as well as virion morphogenesis, and egress (32).
In addition, sequence analysis indicated no potential transmembrane domain and signal peptide in EBV BGLF2, but it contained three N-linked glycosylation sites (Asn-X-Ser/Thr). In conformity with this prediction, pseudorabies virus UL16 was predicted to contain one potential N-linked glycosylation signal, but N-glycosidase F treatment experiment demonstrated that it is not N-glycosylated (41). Therefore, more works are required to establish whether BGLF2 is N-glycosylated.
The hydrophobicity and hydrophilicity analyses demonstrated that in BGLF2 the proportion of hydrophobic region was larger than the hydrophilic region, which might represent the internal and surface regions of the protein, respectively. Furthermore, the B-cell epitopes of BGLF2 were predicted using DNAStar PROTEAN programs based on its flexibility, antigenic index, and surface probability by the determination of its primary structure (42-44). The results indicated that the improved knowledge of the antigenic and structural features of the BGLF2 resulting from this work might generate suitable approaches for developing new antibodies and immunoassays for the application in the clinical diagnosis of EBV. Besides, the secondary structure prediction revealed that BGLF2 contained 7 potential α-helix regions and several possible random Coil and β-strand. Moreover, its 3D conformation also found to consist of the 7 potential α-helix and two potential β-strands (linked by β-turn) and a number of hydrogen bonds. Therefore, we speculated the component of these structures of BGLF2 might be engaged in its self-association or the interactions with other components of the nucleocapsid (45-47), tegument (46-48), envelope (29), or the host proteins (49).
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81772179, 31400150, and 31200120); the Natural Science Foundation of Guangdong Province (2015A030313473); the Training Program for Outstanding Young Teachers in Universities of Guangdong Province (YQ2015132); the Medical Scientific Research Foundation of Guangdong Province, China (A2017055 and B2012165); the Science and Technology Plan Projects of Guangzhou City, China (201607010088); the Scientific Research Projects in Colleges and Universities of Guangzhou (1201430024, 1201610025 and 1201610024); Innovation and entrepreneurship training program in colleges and universities in Guangzhou (2017224104); High-Level Universities Academic Backbone Development Program of Guangzhou Medical University; Nanshan Scholar of Guangzhou Medical University; Thousand Hundred Ten Projects of Guangzhou Medical University, Guangdong; and Laboratory Opening Program for College Students of Guangzhou Medical University (2018). We thank Dr. Teru Kanda for the generous gift of AK-BAC (EBV Akata strain-derived BAC).
Footnotes
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Supplementary Files
This manuscript includes supplementary file accessible on http://www.ijbiotech.com.
References
- 1.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, et al. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A. 2004;101(46):16286-91. doi: 10.1073/pnas.0407320101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen MR, Hsu TY, Lin SW, Chen JY, Yang CS. Cloning and characterization of cDNA clones corresponding to transcripts from the BamHI G region of the Epstein-Barr virus genome and expression of BGLF2. J Gen Virol. 1991;72 (Pt 12):3047-55. doi: 10.1099/0022-1317-72-12-3047 [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Cohen JI. Epstein-Barr Virus (EBV) Tegument Protein BGLF2 Promotes EBV Reactivation through Activation of the p38 Mitogen-Activated Protein Kinase. J Virol. 2015;90(2):1129-38. doi: 10.1128/JVI.01410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanda T, Shibata S, Saito S, Murata T, Isomura H, Yoshiyama H, et al. Unexpected instability of family of repeats (FR), the critical cis-acting sequence required for EBV latent infection, in EBV-BAC systems. PLoS One. 2011;6(11):e27758 doi: 10.1371/journal.pone.0027758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyashita T, Takami A, Takagi R. Molecular cloning and characterization of the 5’-flanking regulatory region of the carbonic anhydrase nacrein gene of the pearl oyster Pinctada fucata and its expression. Biochem Genet. 2012;50(9-10):673-83. doi: 10.1007/s10528-012-9510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li ML, Jiang S, Mo CC, Zeng ZC, Li XW, Chen CK, et al. Identification of molecular determinants for the nuclear import of pseudorabies virus UL31. Arch Biochem Biophys. 2015;587:12-7. doi: 10.1016/j.abb.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Cai MS, Jiang S, Li XW, Zeng ZC, Li ML. Characterization of the nuclear import mechanisms of HSV-1 UL31. Biol Chem. 2016;397(6):555-61. doi: 10.1515/hsz-2015-0299. [DOI] [PubMed] [Google Scholar]
- 8.Cai MS, Huang ZB, Liao ZM, Chen T, Wang P, Jiang S, et al. Characterization of the subcellular localization and nuclear import molecular mechanisms of herpes simplex virus 1 UL2. Biol Chem. 2017;398(4):509-17. doi: 10.1515/hsz-2016-0268. [DOI] [PubMed] [Google Scholar]
- 9.Li ML, Cui W, Mo CC, Wang JL, Zhao ZY, Cai MS. Cloning, expression, purification, antiserum preparation and its characteristics of the truncated UL6 protein of herpes simplex virus 1. Mol Biol Rep. 2014;41(9):5997-6002. doi: 10.1007/s11033-014-3477-y. [DOI] [PubMed] [Google Scholar]
- 10.Li ML, Li Z, Li WT, Wang BY, Ma CQ, Chen JH, et al. Preparation and characterization of an antiserum against truncated UL54 protein of pseudorabies virus. Acta Virol. 2012;56(4):315-22. DOI: 10.4149/av_2012_04_315. [DOI] [PubMed] [Google Scholar]
- 11.Cai MS, Jiang S, Mo CC, Wang JL, Huang JL, Zeng ZC, et al. Preparation and identification of an antiserum against recombinant UL31 protein of pseudorabies virus. Acta Virol. 2015;59(3):295-9. DOI: 10.4149/av_2015_03_295. [DOI] [PubMed] [Google Scholar]
- 12.Cai MS, Li ML, Wang KZ, Wang S, Lu Q, Yan J, et al. The herpes simplex virus 1-encoded envelope glycoprotein B activates NF-kappaB through the Toll-like receptor 2 and MyD88/TRAF6-dependent signaling pathway. PLoS One. 2013;8(1):e54586 doi: 10.1371/journal.pone.0054586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li ML, Jiang S, Wang JL, Mo CC, Zeng ZC, Yang YJ, et al. Characterization of the nuclear import and export signals of pseudorabies virus UL31. Arch Virol. 2015;160(10):2591-4. doi: 10.1007/s00705-015-2527-7.26195191 [Google Scholar]
- 14.Cai MS, Chen DX, Zeng ZC, Yang H, Jiang S, Li XW, et al. Characterization of the nuclear import signal of herpes simplex virus 1 UL31. Arch Virol. 2016;161(9):2379-85. doi: 10.1007/s00705-016-2910-z. [DOI] [PubMed] [Google Scholar]
- 15.Cai MS, Jiang S, Zeng ZC, Li XW, Mo CC, Yang YJ, et al. Probing the nuclear import signal and nuclear transport molecular determinants of PRV ICP22. Cell Biosci. 2016;6:3 doi: 10.1186/s13578-016-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.NCBI BLAST website. 1997 [cited 2016 Feb 12]; Available from: URL: http://www.ncbi.nlm.nih.gov/BLAST/.
- 17.FASTA website. 2016 [cited 2016 Feb 12]; Available from: URL: http://www.ebi.ac.uk/Tools/sss/fasta/.
- 18.Conserved Domains search tool website. 2005 [cited 2016 Feb 12]; Available from: URL: http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
- 19.TMHMM website. 2001 [cited 2016 Feb 12]; Available from: URL: http://www.cbs.dtu.dk/services/TMHMM/.
- 20.SignalP-4.0 Server website. 1997 [cited 2016 Feb 12]; Available from: URL: http://www.cbs.dtu.dk/services/SignalP/.
- 21.NetNGlyc 1.0 website. 2004 [cited 2016 Feb 12]; Available from: URL: http://www.cbs.dtu.dk/services/NetNGlyc/.
- 22.NetPhos 2.0 website. 1999 [cited 2016 Feb 12]; Available from: URL: http://www.cbs.dtu.dk/services/NetPhos/.
- 23.PSIpred website. 1999 [cited 2016 Feb 12]; Available from: URL: http://bioinf.cs.ucl.ac.uk/psipred/.
- 24.CPHmodels 3.2 website. 2010 [cited 2016 Feb 12]; Available from: URL: http://www.cbs.dtu.dk/services/CPHmodels/.
- 25.Ehlers B, Spiess K, Leendertz F, Peeters M, Boesch C, Gatherer D, et al. Lymphocryptovirus phylogeny and the origins of Epstein-Barr virus. J Gen Virol. 2010;91(Pt 3):630-42. doi: 10.1099/vir.0.017251-0. [DOI] [PubMed] [Google Scholar]
- 26.Shi P, Du Y, Yang H, Huang H, Zhang X, Wang Y, et al. Molecular characterization of a new alkaline-tolerant xylanase from Humicola insolens Y1. Biomed Res Int. 2015;2015:149504 doi: 10.1155/2015/149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li ML, Cui W, Mo CC, Wang JL, Zhao ZY, Cai MS. Cloning, expression, purification, antiserum preparation and its characteristics of the truncated UL6 protein of herpes simplex virus 1. Mol Biol Rep. 2014;41(9):5997-6002. doi: 10.1007/s11033-014-3477-y. [DOI] [PubMed] [Google Scholar]
- 28.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802-5. [DOI] [PubMed] [Google Scholar]
- 29.Owen DJ, Crump CM, Graham SC. Tegument Assembly and Secondary Envelopment of Alphaherpesviruses. Viruses. 2015;7(9):5084-114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Cui Z, Zhang Z, Wei H, Zhou Y, Wang M, et al. The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology. 2009;388(1):68-77. doi: 10.1016/j.virol.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Phillips SL, Bresnahan WA. The human cytomegalovirus (HCMV) tegument protein UL94 is essential for secondary envelopment of HCMV virions. J Virol. 2012;86(5):2523-32. doi: 10.1128/JVI.06548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, Wang L, Peng L, Zhou ZH, Deng H. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J Virol. 2009;83(20):10582-95. doi: 10.1128/JVI.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai MS, Liao ZM, Chen T, Wang P, Zou XM, Wang YF, et al. Characterization of the subcellular localization of Epstein-Barr virus encoded proteins in live cells. Oncotarget. 2017; 8(41), 70006-34. doi: 10.18632/oncotarget.19549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng ZP. An overview on predicting the subcellular location of a protein. In Silico Biol. 2002;2(3):291-303. doi: 10.1002/0470857897. [PubMed] [Google Scholar]
- 35.Pomeranz LE, Blaho JA. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73(8):6769-81. doi: 10.1126/science.8303295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geiss BJ, Tavis JE, Metzger LM, Leib DA, Morrison LA. Temporal regulation of herpes simplex virus type 2 VP22 expression and phosphorylation. J Virol. 2001;75(22):10721-9. doi: 10.1128/JVI.75.22.10721-10729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knopf KW, Kaerner HC. Virus-specific basic phosphoproteins associated with herpes simplex virus type a (HSV-1) particles and the chromatin of HSV-1-infected cells. J Gen Virol. 1980;46(2):405-14. doi: 10.1099/0022-1317-46-2-405. [DOI] [PubMed] [Google Scholar]
- 38.Vales-Gomez M, Winterhalter A, Roda-Navarro P, Zimmermann A, Boyle L, Hengel H, et al. The human cytomegalovirus glycoprotein UL16 traffics through the plasma membrane and the nuclear envelope. Cell Microbiol. 2006;8(4):581-90. doi: 10.1111/j.1462-5822.2005.00645.x. [DOI] [PubMed] [Google Scholar]
- 39.Henson BW, Perkins EM, Cothran JE, Desai P. Self-assembly of Epstein-Barr virus capsids. J Virol. 2009;83(8):3877-90. doi: 10.1128/JVI.01733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilcox KW, Kohn A, Sklyanskaya E, Roizman B. Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J Virol. 1980;33(1):167-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klupp BG, Bottcher S, Granzow H, Kopp M, Mettenleiter TC. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J Virol. 2005;79(3):1510-22. doi: 10.1128/JVI.79.3.1510-1522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai MS, Zhao ZY, Cui W, Yang L, Zhu JY, Chen YL, et al. Molecular properties of the Epstein-Barr virus BFRF3 gene. Virol Sin. 2013;28(6):368-72. doi: 10.1007/s12250-013-3351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li ML, Cui W, Zhao ZY, Mo CC, Wang JL, Chen YL, et al. Molecular cloning and characterization of pseudorabies virus EP0 gene. Indian J Biochem Biophys. 2014;51(2):100-14. [PubMed] [Google Scholar]
- 44.Cai MS, Deng SX, Li ML. Comparison of the immune responses in BALB/c mice following immunization with DNA-based and live attenuated vaccines delivered via different routes. Vaccine. 2013;31(9):1353-6. doi: 10.1016/j.vaccine.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Ohta A, Yamauchi Y, Muto Y, Kimura H, Nishiyama Y. Herpes simplex virus type 1 UL14 tegument protein regulates intracellular compartmentalization of major tegument protein VP16. Virol J. 2011;8:365 doi: 10.1186/1743-422X-8-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meckes DG, Jr., Wills JW. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J Virol. 2007;81(23):13028-36. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshima S, Daikoku T, Shibata S, Yamada H, Goshima F, Nishiyama Y. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch Virol. 1998;143(5):863-80. [DOI] [PubMed] [Google Scholar]
- 48.Benach J, Wang L, Chen Y, Ho CK, Lee S, Seetharaman J, et al. Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68. J Biol Chem. 2007;282(43):31534-41. doi: 10.1074/jbc.M705637200. [DOI] [PubMed] [Google Scholar]
- 49.Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277(5330):1310-3. [DOI] [PubMed] [Google Scholar]