Summary
Biotic interactions are potent, widespread causes of natural selection and divergent phenotypic evolution, and can lead to genetic differentiation with gene flow among wild populations (“isolation by ecology”) [1–4]. Biotic selection has been predicted to act on more genes than abiotic selection thereby driving greater adaptation [5]. However, difficulties in isolating the genome-wide effect of single biotic agents of selection have limited our ability to identify and quantify the number and type of genetic regions responding to biotic selection [6–9]. We identified geographically interspersed lakes in which threespine stickleback fish (Gasterosteus aculeatus) have repeatedly adapted to the presence/absence of a single member of the ecological community, prickly sculpin (Cottus asper), a fish that is both competitor and predator of stickleback [10]. Whole genome sequencing revealed that sculpin presence/absence accounted for the majority of genetic divergence among populations, more so than geography. The major axis of stickleback genomic variation within and between the two lake types was correlated with multiple traits, indicating parallel natural selection across a gradient of biotic environments. A large proportion of the genome - about 1.8%, encompassing more than 600 genes – differentiated stickleback from the two biotic environments. Divergence occurred in 141 discrete genomic clumps located mainly in regions of low recombination, suggesting that genes brought to lakes by the colonizing ancestral population often evolved together in linked blocks. Strong selection and a wealth of standing genetic variation explain how a single member of the biotic community can have such a rapid and profound evolutionary impact.
eTOC Blurb
Miller et al. analyze genetic differentiation in wild populations of threespine stickleback fish in response to a single agent of biotic selection, intraguild predation by prickly sculpin. Sculpin presence was correlated with parallel, widespread genetic divergence. Ecological interactions between these species had large evolutionary consequences.
Results and Discussion
Threespine stickleback were sampled in British Columbia (Canada) from seventeen geographically interspersed lakes of two types: “lakes with sculpin” (n=9) and “lakes without sculpin” (n=8) (Figure 1A). Sculpin fish are a piscivorous intraguild predator – preying upon stickleback and competing with it for food [10]. Sculpin presence/absence is associated with heritable phenotypic differences in stickleback defensive armor, body shape, and behavior [10,11]. Lakes formed about 10,000 years ago and were colonized from the sea by marine or anadromous stickleback (hereafter “marine”) [12]. Study lakes are similar in size and were chosen to minimize regional differences in lake characteristics (Figure 1B, Figure S1, Table S1). The result is an unusual natural system of populations having access to shared ancestral standing genetic variation that allowed for independent and repeated adaptation to environments differing primarily in the presence or absence of sculpin.
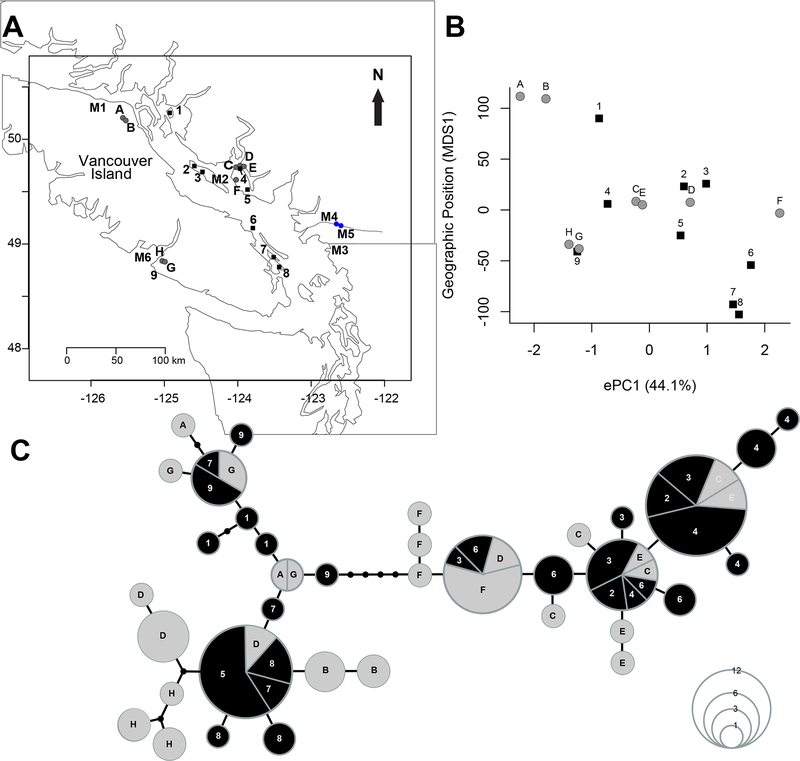
Figure 1. Sculpin Presence/Absence is primary difference among lakes:
(A) Location of study populations. Lakes in which stickleback co-occur with sculpin are indicated in dark gray (A-H) and lakes without sculpin are given in black (1–9). Sampling locations of marine stickleback are shown in light gray (M1–M6). Lake names are provided in Table S1. (B) The association between the first principal component of physical and chemical properties of the study lakes (ePC1) and the relative geographical position of lakes (MDS1), calculated as the principal axis from a multidimensional scaling of pairwise geographical distances among lakes. Lakes located in closer proximity to each other, as indicated by similarity in MDS1, have ePC1 values that are more similar than ePC1 values between more distant lakes. Environmental trait values are in Table S1. (C) Genealogical haplotype network based on a putatively neutral DNA sequence from the mitochondrial control region. Circle size indicates the total number of individuals with that haplotype (see lower right corner). Length of line segments indicate the number of mutations separating haplotypes. Haplotypes are often shared between stickleback from lakes with sculpin (circles, A-H) and with stickleback from lakes without sculpin (squares, 1–9). The haplotype network shows no evidence for monophyly of stickleback from lakes with sculpin or of stickleback from lakes without sculpin. See Figure S1 for more information on lake characteristics.
We tested the phylogenetic independence of stickleback from study lakes using a haplotype network based on putatively neutral sequences from the mitochondrial control region. Systematic structuring between population types would suggest that distinct monophyletic clades colonized lakes with and without sculpin, whereas shared haplotypes among lake types would be consistent with colonization of lakes by a common ancestral population. We found that most populations contained multiple haplotypes, many haplotypes were shared among several populations, and stickleback from lakes with and without sculpin did not cluster in the haplotype network (Figure 1C). This pattern agrees with the geological history of the region [12] and phylogenetic studies indicating that freshwater stickleback populations formed at the end of the last ice age by multiple colonization events from the sea and subsequently evolved independently [13].
Most Genomic Variation is Associated with Sculpin Presence/Absence
To detect parallel adaptive evolution in response to the presence/absence of sculpin, we sequenced a single representative stickleback from each freshwater population to an average coverage of 9X using Illumina 100 bp paired-end whole genome re-sequencing. Sequences were aligned to the stickleback reference genome [14,15]. Following filtering, we generated a dataset of 6.3 million SNPs from the seventeen freshwater genomes, corresponding to approximately one SNP every 73 bp.
A principal component analysis (PCA) of all nuclear SNPs, excluding those on the sex chromosome, revealed sculpin presence/absence to be the predominant factor organizing stickleback genome-wide variation. The first genomic principal component (gPC1) explained 11.6% of the genome-wide SNP variation and separated stickleback from the two lake types with only slight overlap (Figure 2A; F1,15=22.5, P<2.6e-4; Lakes with sculpin = 412.1 ±120.6, Lakes without sculpin = −366.3 ±108.7). The second genomic principal component (gPC2) accounted for 7.8% of SNP variation and was associated with geographical position of lakes (Figure 1B). The two lake types overlapped broadly in gPC2 and their means were not significantly different (F1,15=0.15, P=0.71; Lakes with sculpin = 42.8 ±191.5, Lakes without sculpin = −38.1± 105). We found a weak spatial autocorrelation in gPC1 (Moran’s I: I=0.08, P=0.08), but a significant spatial autocorrelation in gPC2 (Moran’s I: I=−0.141 P=0.02) (Figure S2). We retested the difference between the lake types in gPC1 after controlling for spatial effects using a distance-based Moran’s eigenvector map analysis (dbMEM) and again found a significant association (F4,12=9.2, P=0.001). Overall, we conclude that sculpin presence/absence had a large overall effect on genomic variation in stickleback, more so than geography in this sample of populations.
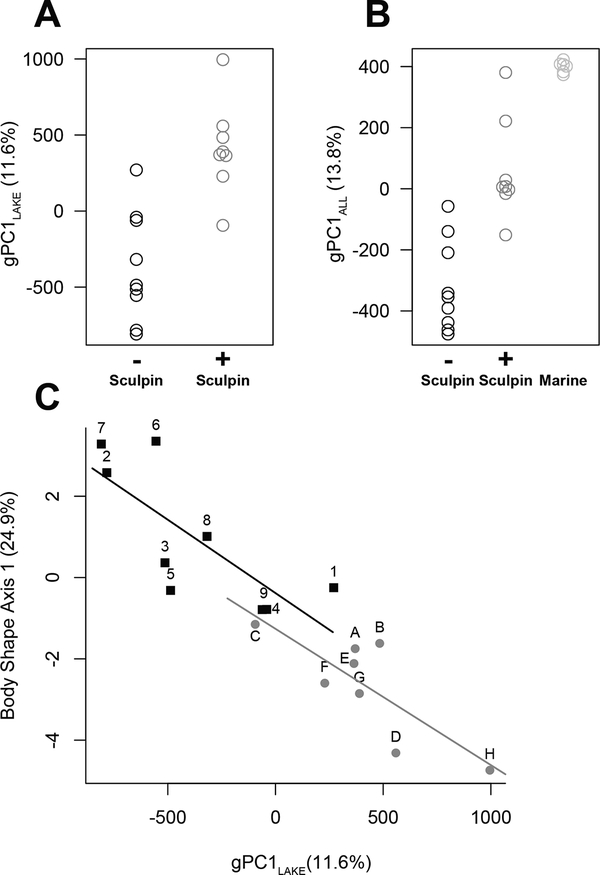
Figure 2. Genomic Variation is Associated with Sculpin Presence/Absence:
Separation of stickleback from lakes with and without sculpin along the first principal component of whole genome SNP variation. (A) First principal component values based on all seventeen lake populations (gPC1). (B) First principal component values based on an analysis that also includes marine (ancestral) stickleback sampled at six localities (gPC1). Each point represents a single individual from a population. Stickleback from lakes with sculpin resemble marine stickleback more closely than do stickleback from sculpin-absent lakes. The percent of SNP variation explained by each first principal component is shown in parentheses. (C) Comparison of the first principal component of all SNPs in all lake populations (gPC1LAKE) with the first linear discriminant axis of body shape (Body Shape Axis 1). Greater phenotypic differentiation both between lake types and within lake types is associated with more extreme genotypic differentiation along gPC1LAKE. See Figures S2 and S3.
We repeated these analyses including stickleback from six marine locations. The first genomic principal component explained 13.8% of the genomic variation and again differentiated stickleback from lakes with and without sculpin (Figure 2B; F6,16=18.9, P<2e-6). Stickleback sympatric with sculpin were more similar to the marine populations along the gPC1 than stickleback from sculpin-absent lakes (Figure 2B), indicating that populations from lakes with sculpin have retained mainly marine alleles at the SNPs having high loadings. Since the marine population represents the putative ancestral form, the alleles at SNPs evolving in parallel are most often in the derived state in sculpin-absent lakes compared with alleles in sculpin-present lakes. This agrees with previous phenotypic trait comparisons of the three groups [16].
Genomic and Phenotypic Differentiation are Associated
Position along the major axis of genomic variation among lakes (gPC1) was correlated with phenotypic differentiation, both within and between lake types in body shape (Figure 2C; Figure S2; lakes with sculpin r=0.80, P=0.02; lakes without sculpin r=0.77, P=0.02; overall r=0.91, P=4e-7), and bony armor (Figure S2). This pattern might reflect widespread pleiotropy, with suites of phenotypic traits mapping to the same parallel diverging genetic variants, but this seems unlikely given the diversity of traits and large number of genomic regions involved (see below). A more likely cause is correlated selection on multiple traits and underlying genes along a gradient of environmental differences, chiefly sculpin presence/absence.
Genomic Differentiation is Extensive
Widespread genomic regions differed between stickleback from lakes with and without sculpin (Figure 3A). To quantify absolute divergence between the two lake types, we calculated the raw variance between the two groups at each nucleotide (FSTNUM) and then took the average in 10,000 bp sliding windows across the genome. This divergence metric is the numerator of the FST statistic and has a maximum of 1 at bi-allelic SNPs and a value of 0 at invariant sites. Although lakes types are not strictly geographically structured subpopulations, we also calculated FST to quantify relative differentiation between stickleback from the two lake types. FST, allele frequency difference (AFD), and a modified version of the cluster separation score of [14] produced similar findings (Figure S3, Supplemental methods).
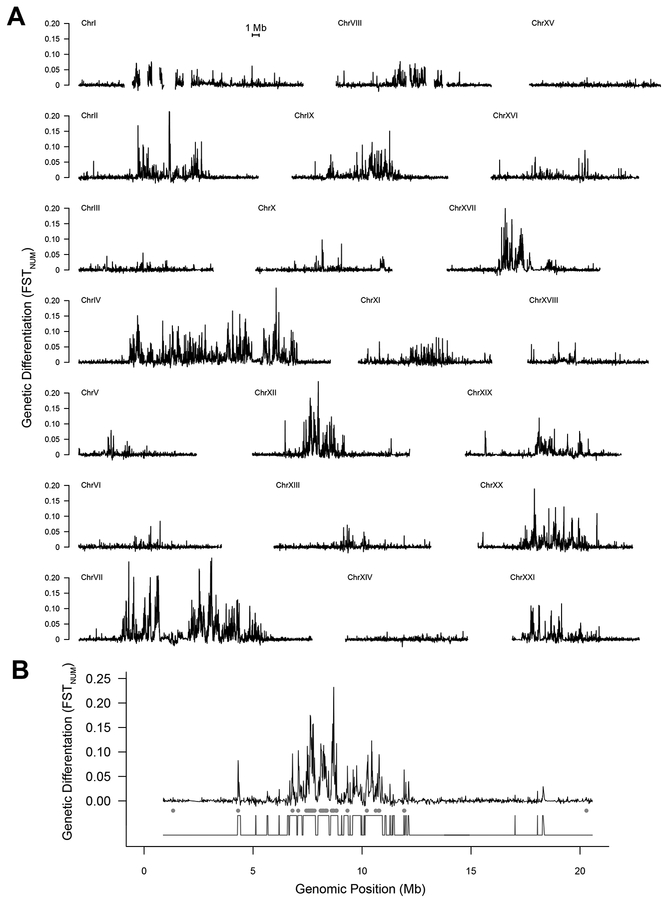
Figure 3. Extensive Genomic Differentiation Between Lake Types:
(A) Genomic differentiation between stickleback from lakes with and without sculpin. FSTNUM was measured in 10,000 bp sliding windows (step size 5,000 bp). All chromosomes are plotted on the same scale and ordered by size. (B) FSTNUM between stickleback from lakes with and without sculpin for an exemplary chromosome showing marked divergence (ChrXII). Estimated state changes (high compared to low differentiation) identified with the HMM are shown below. Outlier windows are indicated with gray circles. See Figures S3–S4 and Tables S2 and S3.
Using permutation tests, and controlling false discovery rate, we identified significant differences in 1,395 of 76,109 informative genomic windows (hereafter ‘outliers’), representing ~1.8% of the genome. Outlier windows frequently contained alleles not yet fixed between populations from the different lake types as only 689 SNPs (out of 6.3 million) had FSTNUM > 0.8 between stickleback from the two lake types. Outlier windows overlapped introns or exons of 609 (3.3%) of the 18,254 annotated genes that fell within sampled windows. Each outlier window contained an average of 0.88 genes (sd=0.79), with individual genes often spanning multiple windows. Genes in outlier windows were enriched for GO terms related to neuron development, synaptic transmission, heart morphogenesis and many other diverse biological functions (Table S3) suggesting that interactions with sculpin had a comprehensive effect on stickleback evolution.
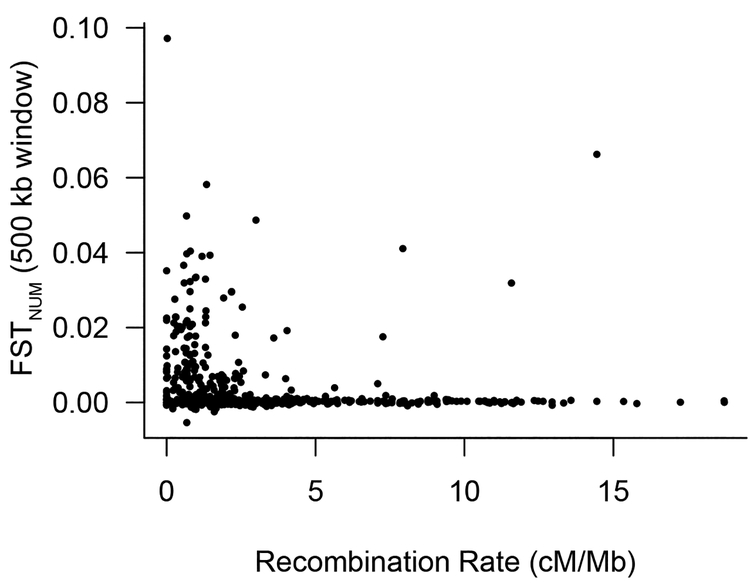
Outlier windows occurred on many chromosomes, with chromosomes IV, VII, XII, and XX displaying especially elevated levels of divergence (Figure 3A). This spatial distribution of outlier windows across chromosomes was highly non-random (χ2 =1753, df=20, P< 2.2e−16). Outlier widows occurred most frequently in chromosomal centers, which have relatively low recombination rates [17]. Across all windows, FSTNUM and local recombination rate were negatively correlated (Figure 4; r= −0.31, P=1.5e−15).
Figure 4. Genomic Differentiation Correlated with Recombination Rate:
The average value of FSTNUM calculated between lakes with and without sculpin in consecutive 500 kb windows are negatively correlated with average recombination rate (R=0.19, P<1.7e−5). Recombination rate data are from [17].
Influence of Abiotic Lake Environment
Lakes with and without sculpin overlapped along axes ePC1 and ePC2 describing abiotic environmental differences among lakes (Figure S1; ePC1: U=51, P=0.17; ePC2: U=40, P=0.74). However, lakes without sculpin had higher average pH (U=63, P=0.01) and greater average calcium concentrations (U=59, P=0.03) than lakes with sculpin (Table S1, Figure S1). Calcium concentration and pH were strongly correlated with each other (r=0.83, P=4e−5). Nevertheless, pH and calcium are unlikely to determine the presence or absence of sculpin. Sculpin occur in lakes with higher pH and calcium concentrations than are represented in our sample of non-sculpin lakes [18,19]. However, sculpin-absent study lakes were more variable in these parameters, in association with local differences in geological substrates: none of the study lakes with sculpin occurred on the high limestone substrates on which several of the sculpin-absent lakes occurred. We reassessed the relationship between gPC1LAKE and lake type while including pH and calcium concentration as covariates, and accounting for spatial effects. Stickleback from different lake types remained divergent along gPC1 (Type: t=2.9, P=0.016). No relationship was detected between gPC1LAKE and pH or calcium concentration (pH: t=0.26, P=0.80; CA: t=−0.91, P=0.39).
We investigated the influence of variation in pH and calcium separately on the stickleback genome, focusing only on lakes without sculpin, whose range of values in pH and calcium encompassed the narrower range of values in sampled lakes with sculpin (Figure S1). We calculated FSTNUM between stickleback sequences from sculpin-absent lakes grouped into low or high calcium concentration and low or high pH (Figure S4). The comparison of FSTNUM between stickleback in lakes with and without sculpin shared 23 outlier windows with the analysis of lakes with low and high calcium, and 12 outlier windows with the analysis of lakes with low and high pH.
We conclude that abiotic differences between study lakes in pH or calcium do not drive the majority the genomic changes differentiating stickleback between lakes with and without sculpin. To minimize the possibility of influence, the 35 shared outlier windows were excluded from subsequent analyses, leaving 1,298 outlier windows and 584 genes associated with the presence/absence of sculpin.
Estimating Boundaries of Divergent Regions
Outlier windows typically occurred consecutively in the genome, suggesting that genomic blocks of differentiated sequence might often have evolved together during adaptation to contrasting lake types [20]. To estimate the extent of these blocks, we used a two state Hidden Markov Model (HMM) to define boundaries between genomic regions of high and low divergence. This model collapsed the 1,298 outlier windows into 141 discrete outlier blocks across the genome (Figure 3B, Table S2), with a median width of 220 kb (range 10–1,470 kb). This number might still overestimate the actual number of independent genetic loci under selection during lake adaptation, since it is possible that linked blocks of standing genetic variation in the ancestral population included a mixture of windows with low and high divergence.
Broader Implications
Extensive genomic divergence of stickleback was associated with the presence/absence of a single biotic agent of selection, the prickly sculpin. Parallel differentiation of genomes between stickleback from the different lake types involved ~1.8% of the genome, overlapping 584 genes with a wide diversity of biological functions. Widespread adaptation is implicated because genetic drift is unlikely to cause repeated, parallel evolution in multiple evolving populations in association with a specific environmental feature [21]. These extensive changes underscore the rapid and profound effects of a seemingly simple biotic interaction on stickleback evolution.
Study lakes are recent (< 10,000 generations), meaning that differentiation between lake types has repeatedly occurred in a remarkably short time period. On the other hand, genes in outlier windows are not evolutionarily independent, because most occur in contiguous blocks of differentiated genes whose alleles might have increased in frequency in unison during adaptation. We found that stickleback populations sympatric with sculpin have retained more marine genetic variants than populations in lakes without sculpin. Because marine stickleback co-occur with numerous other fish species, including several other sculpin species, recent release from selection by sculpin produced the direction of most genetic divergence between stickleback populations in the two lake types.
How can a single species from the ecological community rapidly produce differentiation across so many genomic regions? One possible explanation is that traits under selection might be highly polygenic. Stickleback from the two lake types differ in body shape, defensive armor, diet, and behavior [10,11], and variation in such traits is commonly controlled by many genetic loci [16,22,23]. Second, stickleback traits in lakes with sculpin might represent the cumulative outcome of a long series of reciprocal coevolutionary changes in the two species that unraveled in lakes where sculpin are absent. This hypothesis could be addressed in future studies investigating the evolutionary dynamics of sculpin.
A third potential explanation is that sculpin and stickleback are members of a larger network of interacting species. Stickleback may indirectly experience multifarious selection if the presence/absence of sculpin leads to changes in the strength of stickleback interactions with other species. The accumulated effects of “diffuse” or “indirect” selection might be as great or greater than direct selection by sculpin. For example, the lack of sculpin may allow stickleback to colonize the shallow benthic environment, changing stickleback diet and thereby leading to selection to alter behaviors and morphology to improve catching and handling performance on new food items and avoid predators.
Lastly, differentiation of a large portion of the genome might be a correlated response to selection on a smaller set of genes. Most identified differentiated genome regions are clustered in blocks of relatively old standing genetic variation brought to the lakes by colonists [20], with genes in the same block remaining in relatively high linkage disequilibrium during adaptation of a more limited set of loci [24]. In this way, neutral and even deleterious alleles could hitchhike along with linked loci under selection, overestimating the number of genes under selection [25]. It is likely that freshwater allele copies present as standing genetic variation in the colonizing marine population originated from gene flow with other nearby freshwater populations not long before lake colonization [26] and so were initially present as co-segregating blocks. By estimating where blocks of differentiated windows begin and end, we calculate that at most 141 genomic regions were involved in the process of adaptation by stickleback to the presence/absence of sculpin. This still represents a large number of genomic regions responding to biotic selection over a relatively short time span.
Although recombination would eventually break down linkage between genes in the marine population, regions of low recombination would aid the persistence of large blocks of freshwater alleles in the marine population [27]. Consistent with this, we found a negative correlation between local recombination rate and genetic divergence between stickleback from lakes with and without sculpin. We note that by using whole genome sequence data, our study is less likely to be biased towards detecting selection in regions of low recombination than reduced representation methods (e.g. RAD-seq).
How did selection bring so many genes or blocks of genes from low to high frequency in so few generations? Selection on many genomic regions would presumably generate a high substitution load requiring many selective deaths [28]. One possibility is that biotic interactions may lead to “soft” selection, in which the fitness of an individual is a function of the difference between its phenotype and the most fit phenotype in the population, rather than its difference from an optimum phenotype, reducing substitution load [29]. A change to a new optimum can occur quickly when initial allele frequency in a population is high [30], therefore a second possible answer is that adaptive genetic variants did not start at a low frequency. Colonizing populations were likely large, since colonization occurred during post-glacial rebound of coastal lands. At this time, increased gene flow may have maintained much higher levels of standing variation in the ancestral marine population compared to the present-day marine population. As a result, differentiated genes may have begun at relatively high frequency in the colonizing population, reducing substitution load.
This study provided evidence that ecological interactions between species – even non-intimate and symbiotic interactions – can have large evolutionary consequences. Other studies have found biotic selection to be associated with substantial genomic divergence [14,31–33], although examples remain few. It is tempting to suggest, based on our study and others, that biotic selection is likely to have more substantial effects on the genome and the phenotype than abiotic agents of selection. Testing this hypothesis will require future controlled studies on the genomic impact of different agents of selection.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sara Miller (sem332@cornell.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Adult threespine stickleback fish (Gasterosteus aculeaus) specimens were collected using minnow traps from lake and marine regions in Southwestern British Columbia (Table S1). Upon sampling, stickleback were euthanized with an overdose of buffered MS-222 anesthetic (Argent Chemical Laboratories, Redmond, WA) and stored in 95% ethanol.
METHOD DETAILS
Sampling Strategy
Our study design, based on [14], applied whole genome re-sequencing of a single representative (diploid) stickleback individual from multiple independent lakes, maximizing the number of populations sampled rather than the number of individuals within populations. The design minimizes false positives signatures of selection produced by the effects of demographic history. The approach is mainly adept at identifying genomic regions that evolved repeatedly from shared standing genetic variation present in the common ancestral population [13,34]. Standing genetic variation has been previously shown to be a source of adaptive alleles in stickleback [13]. The method is unlikely to detect genomic regions evolving by new mutations unless they produce a parallel signature at the same loci in different populations.
Hence our approach provides a conservative estimate of genes responding to biotic environments. Study lakes were initially identified either based on previous lake observations [11] or were found using Habitat Wizard (www.env.gov.bc.ca/habwiz), an online database maintained by the Provincial government of British Columbia. We searched for lakes with and without prickly sculpin and whose fish community otherwise contained only threespine stickleback and coastal cutthroat trout (Oncorhyncus clarkii clarkii). Cutthroat trout are piscivorous predators of stickleback and sculpin [35] and their presence was unavoidable as they are found in virtually every lake in this region (as are diving piscivorous birds during summer, especially loons (Gavia immer). Therefore, we emphasize that our comparisons are between lakes with and without sculpin, and not between lakes with and without predation. The presence or absence of sculpin was verified in all lakes using minnow traps.
Study lakes selected were geographically interspersed to minimize the contribution of regional differences in lake characteristics. All study lakes, except North Lake (a lake with sculpin) and Klein Lake (a lake without sculpin), were in separate watersheds, and all were inaccessible from the sea, ensuring no contemporary gene flow between populations. We therefore treat each lake as an independent replicate.
We sampled up to 25 adult threespine stickleback during the breeding season (May-June 2012–2014) from nine lakes with sculpin and eight lakes without sculpin (Figure 1A). In addition, we sampled marine stickleback at six localities (Figure 1A). Present day marine stickleback are thought to be phenotypically representative of the ancestral marines that colonized freshwater habitats after the end of the last ice age [12,36]. Modern Pacific Ocean marine populations are considered nearly panmictic, with high gene flow among nearby populations [13,14,34,37].
Mitochondrial Haplotype Network
We used mitochondrial DNA (mtDNA) to test for systematic structuring of populations from lakes with and without sculpin. We chose mtDNA rather than whole genome sequences to minimize the effect of parallel evolution of shared standing variation, which can give a false signal of monophyly [14,38]. Shared haplotypes among lake types would suggest a monophyletic origin, whereas haplotypes interspersed among populations would suggest multiple independent origins.
We Sanger sequenced a 352 bp stretch of the neutrally evolving mitochondrial control region (D-Loop) using the standard primers L-Pro-F [39] and TDK-D [40]. Sequences were aligned and visually checked in CodonCode Aligner (v6.0.2). A ML-phylogeny of haplotypes was inferred in RAxML (v8.0.0) [41] using the GTRCAT model of sequence evolution with rate heterogeneity among sites. The sequence alignment and phylogeny were then used to construct a haplotype genealogy with FITCHI [42]. The final analysis was based on 3–15 stickleback individuals per lake (112 sequences in total) and a total of 25 SNPs.
Morphometrics
Body Shape Measurements
Stickleback samples for body shape analysis were stained with alizarin red [43] and photographed on the left with a Nikon D300 camera. Twenty landmarks outlining the shape of the fish and the insertion points of spines and fins (Figure S2) [44] were placed using tpsDig 2.3 [45]. Landmarks were centered, scaled, and rotated using the shapes R package [46]. Shape differences among lakes were visualized using a linear discriminant function analysis (LDA), with population as the classification variable.
Description of Body Shape Landmarks
Dorsal insertion of the caudal peduncle
Posterior midpoint of the caudal peduncle
Ventral insertion of the caudal penducle
Posterior insertion of the anal fin at the first soft ray
Anterior insertion of the anal fin at the first soft ray
Hinge of the pelvic spine
Dorsal insertion of the pectoral fin
Ventral insertion of the pectoral fin
Anterior edge of the ectocoracoid bone
Anterior extent of the preopercle
Posteroventral extent of the maxilla
Anterior extent of the premaxilla
Naris
Anterior orbit in line with the midpoint of the eye
Posterior orbit in line with the midpoint of the eye
Posterior extent of the supraoccipital
Insertion point of the first dorsal spine
Insertion point of the second dorsal spines
Anterior insertion of the dorsal fin at the first soft ray
Posterior insertion of the dorsal fin at the first soft ray
Armor Trait Measurements
The following traits were measured on stained stickleback specimens: length of the first dorsal spine, length of second dorsal spine, length of the pelvic spine, width of pelvic girdle, and number of lateral plates. All traits, with the exception of lateral plates, were size corrected following the methods in [11]. Armor differences among populations were visualized using a principal component analysis of log-transformed values.
Armor Trait Measurements
The following traits were measured on stained stickleback specimens: length of the first dorsal spine, length of second dorsal spine, length of the pelvic spine, width of pelvic girdle, and number of lateral plates. All traits, with the exception of lateral plates, were size corrected following the methods in [11]. Armor differences among populations were visualized using a principal component analysis of log-transformed values.
Whole Genome Re-sequencing
We chose a single representative fish from each lake by performing a PCA on size scaled and rotated morphological landmarks (see above) for up to 25 individuals per population. The fish nearest to the mean of PC1 and PC2 was selected for sequencing, whether male or female. Genomic DNA was extracted from fin clips using a standard phenol/chloroform method. Paired-end whole genome libraries were prepared for each fish using the Illumina TruSeq sample kit (Illumina, San Diego CA), with a target insert size of 500 bp and quantified using High-Sensitivity Bioanalyzer chips (Agilent Technologies, Inc). Libraries were sequenced using the Illumina HiSeq2000 (100 bp paired-end reads) at the University of British Columbia and at Génome Québec.
Bioinformatics Pipeline
Raw sequence reads were aligned to the stickleback reference genome (gasAcu1 2006 assembly) using BWA (v0.7.13) [47]. Single Nucleotide Polymorphisms (SNPs) were identified using the HaplotypeCaller tool in GATK (v3.6) in conjunction with Picard (v2.8.2) (http://broadinstitute.github.io/picard) following the GATK 3.6 best practices recommendations [48]. Low quality SNPs and insertion/deletions were removed with VCFtools (v0.1.14) [49] using the filters --minGQ 100, --min-meanDP 6, --remove-indels, and --max-missing-count 12 (for full dataset) or 8 (freshwater stickleback only). The seventeen freshwater genomes generated a final dataset, after filtering, of 6.3 million SNPs. Overall lower sequence coverage for marine samples (mean 5X) resulted in a dataset of 2.5 million SNPs for all 23 genomes, following filtering. This correspondsto approximately one SNP every 73 bp for the freshwater dataset, and one SNP every 184 bp for the full dataset.
Genome coordinates were converted to the improved stickleback reference genome assembly [15] using R scripts provided by the authors. The stickleback sex chromosome is chromosome XIX and was excluded from subsequent analyses.
Genomic Divergence Among Lakes
The overall patterns of divergence among our samples were established using a PCA of SNP genotype values. We then determined if the presence of sculpin was correlated with the main axis of genetic differentiation among stickleback lake populations.
A VCF file was loaded into R using the VariantAnnotation package [50]. SNPs occurring in masked regions of the genome, as identified in the RepeatMasker track in the USCS stickleback genome table browser (sticklebrowser.stanford.edu), were removed using custom R scripts. In each individual, SNPs were given a numerical value relative to the reference sequence (REF/REF = 0, ALT/ALT = 1; REF/ALT = 0.5). Missing values were filled in using the average value of that SNP across all samples. The PCA of the covariance matrix among all pairs of SNPs was calculated using the ‘svd’ function in the pcaMethods package [51].
We tested for spatial autocorrelation in the values of genome-wide principal components using Moran’s I test in R’s Ape package (v5) [52]. Distance-based Moran’s eigenvector map analysis (dbMEM) was then used to control for the effect of geographical autocorrelation [53]. This method was implemented using R’s quanteco package [54].
To measure genome-wide differentiation between stickleback from the two lake types, we calculated the raw variance between groups, in sliding windows across the genome using a modified version of vcftools (https://github.com/dalloliogm/vcftools_custom/). We refer to this value as FSTNUM because it is equivalent to the numerator of Weir-Cocherham FST [55]. FSTNUM, is a measure of absolute divergence, and has a maximum of 1 at bi-allelic SNPs and a value of 0 at invariant sites. FSTNUM in a window is the sum of its value over all nucleotide bases divided by the number of sequenced bases in each window. The statistical significance of FSTNUM values was assessed using a permutation test. Within each window, individual fish were randomly reassigned to the two groups, keeping the number of fish in each group the same. This process was repeated 10,000 times and a FSTNUM value between groups was calculated for each permutation. The P-value is the proportion of times in which the permutated value exceeded FSTNUM calculated from the real data. Outlier windows were determined using a significance level corresponding to a genome-wide false discovery rate of 0.05 using the qvalue R package [56]. Outlier windows fulfilling this significance criterion are not only highly divergent but also show parallel allele frequency differences between stickleback in lakes with and without sculpin. A χ2 goodness of fit test was performed to determine whether the number of outlier windows per chromosome was different from the number expected based on the number of nucleotides per chromosome.
Candidate Genes
The number of genes within outlier windows were counted using the biomaRt R package [57]. Pseudogenes and ncRNAs were excluded by sorting genes based upon the “gene_biotype” attribute. We classified “outlier genes” as those genes within 2000-bp upstream or downstream of outlier windows.
Calculating Genomic Divergence Among Lakes using Other Metrics
FST
We also calculated FST in all genomic windows, a common measure of relative differentiation [55] that ignores invariant sites. Individuals from freshwater populations were grouped by whether their lakes contained sculpin. These two groups are not true subpopulations either spatially or via gene flow, which is absent between all lakes. We calculated FST between these groups in 10,000 bp sliding widows with a step size of 5,000 bp using VCFtools (v0.1.14) [49]. Outlier windows were identified using a permutation test, following the procedures described above for CS’. FST was highly correlated with FSTNUM and CS’ (Figure S3).
CS’
To measure absolute genomic differentiation between stickleback from the two lake types, we used a modified version of the cluster separation score (CSS) from [14] in 10,000 bp sliding windows (step size 5,000 bp) across the genome. CSS is based on pairwise sequence divergence between individuals at variable sites within a window between individuals belonging to two different groups. Pairwise divergence is used to calculate the first two principal axes from a multi-dimensional scaling (MDS). The CSS score is the average pairwise Euclidean distance between individuals from different groups minus average distance between individuals within groups. CSS mainly describes divergence on axes of co-varying sequence variation among sites within a window. High CSS within a window indicates a comparatively large number of SNPs diverging in parallel between groups of lake.
We made two modifications of Jones et al. [14] method. First, we used the first two principal components of SNP variation within each window instead of MDS axes. Second, we scaled the score for each window by dividing it by the total number of called bases (variant and invariant) in the window. These modifications increased the speed of computation and convert CSS to a per-base metric of sequence divergence between groups, which we refer to as CS’.
A PCA was conducted within each 10,000 bp sliding widow (step size 5,000 bp) on the numerical genotype scores (0, 0.5, and 1) at each SNP. After applying the data quality filters listed in the bioinformatics pipeline, a total of 69,215 windows containing 18,232 genes were available from the lake samples. We retained the first two principal components in each window and calculated the pairwise Euclidean distance D between individual fish from the two lake types. CS’ is then calculated as:
The numerator of CS’ corrects a typographical error in the formula from [14]. D is the Euclidean distance between two fish, i and j are individual fish belonging to different groups, and s and n are the number of stickleback individuals from sculpin lakes and non-sculpin lakes. To convert to a per-base measure, we divided by N, the number of variant and invariant bases sequenced within a window, yielding CS’. Windows containing fewer than 250 called bases (out of 10,000) or containing fewer SNPs than the total number of fish were dropped. CS’ is occasionally negative, which occurs when the average pairwise distance between fish in different lake types is less than the average pairwise distance between fish of the same lake type.
Permutation tests were used to assess the statistical significance of CS’ values. Within each window, individual fish were randomly reassigned to the two groups, keeping the number of fish in each group the same. This was repeated 10,000 times and a CS’ score was calculated for each permutation. The P-value is the proportion of times in which the permutated value exceeded the CS’ score calculated from the real data. Outlier windows were determined using a significance level corresponding to a genome-wide false discovery rate of 0.05 using the qvalue R package [56]. Outlier windows fulfilling this significance criterion are not only highly divergent but also show parallel allele frequency differences between stickleback in lakes with and without sculpin. A χ2 goodness of fit test was performed to determine whether the number of outlier windows per chromosome was different from the number expected based on the number of nucleotides per chromosome.
CS’ was more sensitive at identifying outlier windows than the other metrics (Table S2). Using CS’ we identified significant differences in 1,645 of 69,215 informative windows (hereafter, outliers), representing ~2.4% of the genome. Median CS’ was 0.0117 in outlier windows compared to 0.0001 in other windows. Outlier windows overlapped introns or exons of 684 of the 18,254 annotated genes in sampled windows (3.7%). There was an average of 0.84 genes (sd=0.80) in each outlier window. Outlier windows were non-randomly distributed across the genome (X2 =1986, df=20, P< 2.2e−16). CS’ and local recombination rate were negatively correlated (R= −0.206, P=3e−6).
Allele Frequency Difference
We also calculated Allele Frequency Difference (AFD) between groups within window. AFD is a commonly used metric of assessing genetic differentiation between populations [58]. Allele frequency was calculated between the two groups at each bi-allelic SNP using VCFtools [49]. The allele frequency difference was calculated for each SNP using the formula:
where SC is the frequency of the reference allele in the sculpin-lake group and NS is the frequency of reference allele in the non-sculpin lake group. The average AFD was calculated in 10,000 bp sliding widows with a step size of 5,000 bp using custom scripts in R. AFD was highly correlated with FSTNUM and CS’ (Figure S4).
Abiotic Lake Characteristics
Physical and chemical properties were compared among lakes to identify environmental differences that might be correlated with sculpin presence/absence. Information on the total surface area, perimeter, and mean depth of each study lake was obtained from Habitat Wizard (www.env.gov.bc.ca/habwiz). Lake elevation and shortest straight-line distance from the lake to the ocean was determined using Google maps (www.maps.google.com). To quantify the spatial distribution of study lakes, we calculated pairwise great circle distances among lakes (in km). Pairwise distances were then summarized using major axes from a multidimensional scaling analysis.
We collected water samples from the seventeen freshwater lakes to test for systematic differences in water chemistry between sculpin and non-sculpin lakes. Water samples were stored at 4°C for several months prior to testing. Sodium (Na) and Calcium (Ca) concentrations were assessed by averaging two independent measurements per lake water sample using flame atomic absorption spectroscopy (machine model: Spectra AA-220FS, Varian Inc.). In the same samples, we measured Soluble Reactive Phosphorus (SRP) with a spectrophotometer (machine model: SpectraMAX 340pc, Marshall Scientific) and pH. Readings from water samples taken at different time points from the same lake (i.e., in different years, or in different months between May and August within the same year) revealed little variation, indicating relatively high temporal stability in overall water chemistry within a lake. Whenever several water samples per lake were available from different dates, we report the mean of all measurements. All nine variables were log transformed to improve normality before performing a principal components analysis (PCA) of the correlation matrix using the pcaMethods package in R [51].
Influence of Abiotic Environment on Genetic Divergence
Physical and chemical properties of lakes (Table S1) were weakly associated with sculpin presence/absence. To assess this association, nine measured abiotic environmental variables were log transformed then summarized using principal component analysis. The first major axis of environmental variation mainly reflected increased sodium and calcium concentration and decreased distance to the ocean. The second principle component axis (ePC2) mainly reflected increased distance of lakes to the ocean and decreased lake area and perimeter. Lakes with and without sculpin broadly overlapped along ePC1 and ePC2 (Figure S1) and their means were not detectably different (ePC1: U=51, P=0.17; ePC2: U=40, P=0.74).
Lakes with sculpin had higher pH and greater calcium concentrations than lakes without sculpin. We investigated the influence of variation in pH and calcium separately on the stickleback genome, focusing only on lakes without sculpin, whose range of values in pH and calcium was broad and encompassed the narrower range of values in sampled lakes with sculpin (Figure S1). We calculated FSTNUM between stickleback sequences from sculpin-absent lakes grouped by low (n=5) and high (n=4) calcium concentration (Figure S4). After false discovery rate correction, there were no statistically significant outlier windows, but this may be caused by the lower power from a reduced number of lakes in this analysis. Instead we used a relaxed significance threshold that considered the 1188 windows corresponding to the lowest 0.05% of uncorrected P-values (P=0.0082) to be outliers. We repeated this analysis between sequences from non-sculpin lakes grouped by low (n=4) and high (n=5) pH, which yielded 759 outlier windows under the relaxed criterion (Figure S4). Similar results were obtained when considering the top 1% of P-values as outliers.
Defining Boundaries of Divergent Regions
We used a two state Hidden Markov Model (HMM) implemented in the R package depmixS4 [59], to estimate the number of and size of contiguous blocks of highly divergent windows between stickleback from lakes with and without sculpin. The model assumed two underlying states of genomic divergence of windows, low and high, each with a distinct mean level of divergence and a normal probability distribution of emissions estimated from the data. Transitions between states, whose probabilities are also estimated from the data, were used to demarcate blocks of divergent windows. A dependent mixture model was fitted to log FSTNUM values of contiguous (rather than sliding) 10,000 bp windows using the parameters ‘nstates=2’ and ‘instart=runif(2)’. Other settings were program defaults. Each chromosome was analyzed separately.
Correlation of FSTNUM with Recombination rate
FSTNUM values were averaged in non-overlapping 500 kb genomic windows, using the FSTNUM scores previously calculated between lakes with and without sculpin in 10,000-bp windows. Chromosomal positions were converted to the genome assembly positions in [17] using custom R scripts. Based on pedigree-derived crossover (recombination) rate estimates for the stickleback genome from [16], we calculated average recombination rates for the same genome-wide 500 kb sliding windows as above. For cases in which sliding windows overlapped regions with multiple recombination rate estimates, we weighted the estimates by the degree of overlap with a window, and then calculated the average of the recombination rates. A conventional correlation analysis was used to test the association between FSTNUM and recombination rate variation across the genome.
Gene Ontology (GO) Enrichment in Outlier Windows
We searched for enriched Gene Ontology (GO) terms among genes identified in outlier windows. To be conservative, we restricted our analysis to the 456 genes identified as outlier by three different metrics, FSTNUM, FST, and CS’. Genes within outlier windows were identified using the biomaRt R package [57]. Pseudogenes and ncRNAs were excluded by sorting genes based upon the “gene_biotype” attribute. We classified “outlier genes” as those genes within 2000-bp upstream or downstream of outlier windows. Outlier genes were searched for enriched GO terms using the ‘weight01’ algorithm in the TopGO R package [60], restricting the search to GO terms in the category of biological processes. TopGO takes the GO hierarchy into account when identifying enriched GO terms resulting in fewer false positives than other methods. Candidate enriched GO terms were identified using Fisher’s exact test (Table S3).
QUANTIFICATION AND STATISTICAL ANALYSIS
All analyses were conducted in R [61]. Statistical tests are described in Method Details (above).
Supplementary Material
Highlights.
Biotic selection correlated with rapid, parallel genetic differentiation
Stickleback from the two biotic environments differed in more than 580 genes
Identified 140 discrete genomic regions potentially under biotic selection
Genetic differentiation correlated with variation in morphology
Acknowledgments
Funding for this project was provided by a fellowship to S.M. from the University of British Columbia, grants from the Swiss National Science Foundation and the Janggen-Pöhn Foundation to M.R. and by grants to D.S. from the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs program. Michael Sackville and Sandra Fehsenfeld assisted with water sample chemistry assessments. Thanks to Dan Bolnick, Sean Rogers, and Monica Yau for providing samples. Telma Laurentino and Walter Salzburger facilitated Sanger sequencing. Whole genome re-sequencing was provided by McGill University and Génome Québec Innovation Centre. Data is deposited in the NCBI’s short read archive in project PRJNA387728.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
All data files and custom R scripts used to create these analyses are available at the Dryad data repository (doi:10.5061/dryad.nb37pn3). Sequence data used in this paper are available at the NCBI short read archive in project PRJNA387728.
References
- 1.Kingsolver JG, and Hoekstra HE (2001). The strength of phenotypic selection in natural populations. Am. Nat 157, 245–61. [DOI] [PubMed] [Google Scholar]
- 2.Rieseberg LH, Widmer A, Arntz AM, and Burke JM (2002). Directional selection is the primary cause of phenotypic diversification. P. Natl. Acad. Sci. USA 99, 12242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson JN (2013). Relentless Evolution. University of Chicago Press. [Google Scholar]
- 4.Shafer ABA, Wolf JBW. (2004). Widespread evidence for incipient ecological speciation: a meta-analysis of isolation-by-ecology. Ecol. Lett 16, 940–50. [DOI] [PubMed] [Google Scholar]
- 5.Strauss SY, Irwin RE (2010). Ecological and Evolutionary Consequences of Multispecies Plant-Animal Interactions. Annu. Rev. Ecol. Evol. Syst 35, 435–66. [Google Scholar]
- 6.Stapley J, Reger J, Feulner PGD, Smadja C, Galindo J, Ekblom R, Bennison C, Ball AD, Beckerman AP, and Slate J (2010). Adaptation genomics: the next generation. Trends Ecol. Evol 25, 705–12. [DOI] [PubMed] [Google Scholar]
- 7.Savolainen O, Lascoux M, and Merilä J (2013). Ecological genomics of local adaptation. Nat. Rev. Genet 14, 807–20. [DOI] [PubMed] [Google Scholar]
- 8.Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez- Barrio A, Promerová M, Rubin CJ, Wang C, Zamani N, et al. (2017). Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518, 371–375. [DOI] [PubMed] [Google Scholar]
- 9.Bento G, Routtu J, Fields PD, Bourgeois Y, Pasquier Du L, and Ebert D (2017). The genetic basis of resistance and matching-allele interactions of a host-parasite system: The Daphnia magna-Pasteuria ramosa model. PLoS Genet. 13, e1006596–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingram T, Svanback R, Kraft N, and Kratina P (2012). Intraguild predation drives evolutionary niche shift in threespine stickleback. Evolution 66, 1819–1832. [DOI] [PubMed] [Google Scholar]
- 11.Miller SE, Metcalf D, and Schluter D (2015). Intraguild predation leads to genetically based character shifts in the threespine stickleback. Evolution 69, 3194–203. [DOI] [PubMed] [Google Scholar]
- 12.Bell MA, and Foster SA (1994). In: The Evolutionary Biology of the Threespine Stickleback, Bell MA and Foster SA, eds. (New York: Oxford University Press; ). [Google Scholar]
- 13.Jones FC, Chan YF, Schmutz J, Grimwood J, and Brady SD (2012). A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr. Biol 22, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. (2012). The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glazer AM, Killingbeck EE, Mitros T, Rokhsar DS, and Miller CT (2015). Genome Assembly Improvement and Mapping Convergently Evolved Skeletal Traits in Sticklebacks with Genotyping-by-Sequencing. G3-Genes Genom. Genet 5, 1463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers SM, Tamkee P, Summers B, Balabahadra S, Marks M, Kingsley DM, and Schluter D (2012). Genetic signature of adaptive peak shift in threespine stickleback. Evolution 66, 2439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roesti M, Moser D, and Berner D (2013). Recombination in the threespine stickleback genome-patterns and consequences. Mol. Ecol 22, 3014–27. [DOI] [PubMed] [Google Scholar]
- 18.Saiki MK, and Martin BA (2001). Survey of fishes and environmental conditions in Abbotts Lagoon, Point Reyes National Seashore, California. Calif. Fish Game 87, 123–138. [Google Scholar]
- 19.Roch M, Nordin RN, Austin A, McKean CJP, Deniseger J, Kathman RD, McCarter JA and Clark MJR (1985). The effects of heavy metal contamination on the aquatic biota of Buttle lake and the Campbell river drainage (Canada). Arch. Environ. Contam. Toxicol 14, 347–62. [DOI] [PubMed] [Google Scholar]
- 20.Terekhanova NV, Logacheva MD, Penin AA, Neretina TV, Barmintseva AE, Bazykin GA, Kondrashov AS and Mugue NS (2014). Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback Gasterosteus aculeatus. PLoS Genet. 10, e1004696–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endler JA (1986). Natural Selection in the Wild. Princeton University Press. [Google Scholar]
- 22.Arnegard ME, McGee MD, Matthews B, Marchinko KB, Conte GL, Kabir S, Bedford N, Bergek S, Chan YF, and Jones FC (2014). Genetics of ecological divergence during speciation. Nature 511, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peichel CL, and Marques DA (2017). The genetic and molecular architecture of phenotypic diversity in sticklebacks. Philos T. R. Soc. B 372, 20150486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassham S, Catchen J, Lescak E, Hippen von FA, and Cresko WA (2018). Repeated Selection of Alternatively Adapted Haplotypes Creates Sweeping Genomic Remodeling in Stickleback. Genetics, 3006102017–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Excoffier L, and Ray N (2008). Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol 23, 347–51. [DOI] [PubMed] [Google Scholar]
- 26.Schluter D, and Conte GL (2009). Genetics and ecological speciation. P. Natl. Acad. Sci. USA 106 (Suppl 1), 9955–9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartl DL, and Clark AG (2007). Principles of Population Genetics. Sinauer Associates Incorporated.
- 28.Kondrashov AS (1995). Contamination of the genome by very slightly deleterious mutations: why have we not died 100 times over? J. Theor. Biol 175, 583–94. [DOI] [PubMed] [Google Scholar]
- 29.Charlesworth B (2013). Why we are not dead one hundred times over. Evolution 67, 3354–61. [DOI] [PubMed] [Google Scholar]
- 30.Stephan W (2016). Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol. Ecol 25, 79–88. [DOI] [PubMed] [Google Scholar]
- 31.Turner TL, Bourne EC, Wettberg Von EJ, Hu TT, and Nuzhdin SV (2010). Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature 42, 260–3. [DOI] [PubMed] [Google Scholar]
- 32.Soria-Carrasco V, Gompert Z, Comeault AA Farkas TE, Parchman TL, Johnston JS, Buerkle CA, Feder JL, Bast J, Schwander T, and Egan SP (2014). Stick insect genomes reveal natural selection’s role in parallel speciation. Science 344, 738–742. [DOI] [PubMed] [Google Scholar]
- 33.Gou X, Wang Z, Li N, Qiu F, Xu Z, Yan D, Yang S, Jia J, Kong X, Wei Z, et al. (2014). Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24, 1308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roesti M, Gavrilets S, Hendry AP, Salzburger W, and Berner D (2014). The genomic signature of parallel adaptation from shared genetic variation. Mol. Ecol 23, 3944–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McPhail JD (2007) The freshwater fishes of British Columbia. Univ of Alberta Press. [Google Scholar]
- 36.Taylor EB, and McPhail JD (1999). Evolutionary history of an adaptive radiation in species pairs of threespine sticklebacks (Gasterosteus): insights from mitochondrial DNA. Biol. J. Linn. Soc. Lond 66, 271–291. [Google Scholar]
- 37.Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, and Cresko WA (2010). Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD Tags. PLoS Genet 6, e1000862–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, and Kingsley DM (2005). Widespread Parallel Evolution in Sticklebacks by Repeated Fixation of Ectodysplasin Alleles. Science 307, 1928–33. [DOI] [PubMed] [Google Scholar]
- 39.Meyer A, Morrissey JM, and Schartl M (1994). Recurrent origin of a sexually selected trait in Xiphophorus fishes inferred from a molecular phylogeny. Nature 368, 539–542. [DOI] [PubMed] [Google Scholar]
- 40.Lee W-J, Conroy J, Howell WH, and Kocher TD (1995). Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol 41, 54–66. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matschiner M (2016). Fitchi: haplotype genealogy graphs based on the Fitch algorithm. Bioinformatics 32, 1250–2. [DOI] [PubMed] [Google Scholar]
- 43.Peichel CL, Nereng KS, Ohgi KA, and Cole B (2001). The genetic architecture of divergence between threespine stickleback species. Nature 414, 901–5. [DOI] [PubMed] [Google Scholar]
- 44.Walker JA (1997). Ecological morphology of lacustrine threespine stickleback Gasterosteus aculeatus L. (Gasterosteidae) body shape. Biol. J. Linn. Soc. Lond 61, 3–50. [Google Scholar]
- 45.Rohlf FJ (2005). tpsDig, digitize landmarks and outlines, version 2.3 Department of Ecology and Evolution, State University of New York at Stony Brook. [Google Scholar]
- 46.Dryden I (2013). Shapes package. Vienna, Austria: R Foundation for Statistical Computing. Contributed package. Available at http://www.R-project.org. [Google Scholar]
- 47.Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013). From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST and McVean G (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obenchain V, Lawrence M, Carey V, Gogarten S, Shannon P, and Morgan M (2014). VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics 30, 2076–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stacklies W, Redestig H, Scholz M, Walther D, and Selbig J (2007). pcaMethods:a bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–7. [DOI] [PubMed] [Google Scholar]
- 52.Paradis E, Claude J, and Strimmer K (2004). APE: analyses of phylogeneics and evolution in R language. Bioinformatics 20, 289–290. [DOI] [PubMed] [Google Scholar]
- 53.Legendre P, Fortin MJ, Borcard D (2015). Should the Mantel test be used in spatial analysis? Methods Ecol. Evol 6,1239–47. [Google Scholar]
- 54.Anderson GB (2015). Quanteco: Quantitative Ecology in R R package version 0.1.2.
- 55.Weir BS, and Cockerham CC (1984). Estimating F statistics for the analysis of population structure. Evolution 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- 56.Storey JD, Bass AJ, Dabney A, and Robinson D (2015). qvalue: Q-value estimation for false discovery rate control. R package version 2.10.0. http://github.com/jdstorey/qvalue
- 57.Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4, 1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keinan A, Mullikin JC, Patterson N, and Reich D (2007). Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. genet 39, 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visser I, and Speekenbrink M (2010). DepmixS4: An R-package for fitting mixture models on mixed multivariate data with Markov dependencies. (Version 09).
- 60.Alexa A, & Rahnenfuhrer J (2010). topGO: enrichment analysis for gene ontology. R package version, 2(0). [Google Scholar]
- 61.R Core Team (2017). R: a language and environment for statistical computing (R Foundation for Statistical Computing).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.