ABSTRACT
Recent genomic/transcriptomic studies have identified a novel peptide family whose members share the carboxyl terminal sequence –GSEFLamide. However, the presence/identity of the predicted isoforms of this peptide group have yet to be confirmed biochemically, and no physiological function has yet been ascribed to any member of this peptide family. To determine the extent to which GSEFLamides are conserved within the Arthropoda, we searched publicly accessible databases for genomic/transcriptomic evidence of their presence. GSEFLamides appear to be highly conserved within the Arthropoda, with the possible exception of the Insecta, in which sequence evidence was limited to the more basal orders. One crustacean in which GSEFLamides have been predicted using transcriptomics is the lobster, Homarus americanus. Expression of the previously published transcriptome-derived sequences was confirmed by reverse transcription (RT)-PCR of brain and eyestalk ganglia cDNAs; mass spectral analyses confirmed the presence of all six of the predicted GSEFLamide isoforms – IGSEFLamide, MGSEFLamide, AMGSEFLamide, VMGSEFLamide, ALGSEFLamide and AVGSEFLamide – in H. americanus brain extracts. AMGSEFLamide, of which there are multiple copies in the cloned transcripts, was the most abundant isoform detected in the brain. Because the GSEFLamides are present in the lobster nervous system, we hypothesized that they might function as neuromodulators, as is common for neuropeptides. We thus asked whether AMGSEFLamide modulates the rhythmic outputs of the cardiac ganglion and the stomatogastric ganglion. Physiological recordings showed that AMGSEFLamide potently modulates the motor patterns produced by both ganglia, suggesting that the GSEFLamides may serve as important and conserved modulators of rhythmic motor activity in arthropods.
KEY WORDS: Stomatogastric, Cardiac ganglion, Neuropeptide, Neurohormone
Summary: A recently identified arthropod neuropeptide family is highly conserved within the crustaceans; one family member (AMGSEFLamide), which is highly conserved within the decapods, potently modulates three motor-pattern-generating circuits.
INTRODUCTION
Flexibility in the neuronal outputs that control a wide range of movements, including rhythmic movement patterns (e.g. locomotion), is often imparted by the physiological effects of circulating neurohormones and/or locally released neuromodulators, the largest class of which is peptides (e.g. Dickinson et al., 2016; Nusbaum and Blitz, 2012; Nusbaum et al., 2017; Taghert and Nitabach, 2012). Recent advances in genomics/transcriptomics have enabled the identification of many new peptides in a variety of animals, including crustaceans (e.g. Christie, 2014a,b,c,d,e,f, 2015b, 2016a,b, 2017; Christie and Chi, 2015c; Christie and Pascual, 2016; Christie et al., 2008, 2010a, 2011, 2013, 2015, 2017b, 2018a,b; Dircksen et al., 2011; Gard et al., 2009; Ma et al., 2009, 2010; Nguyen et al., 2016; Suwansa-Ard et al., 2015; Toullec et al., 2013, 2017; Veenstra, 2015; Ventura et al., 2014; Yan et al., 2012); these peptides include additional members of previously known families, as well as new peptide groups (e.g. Christie, 2014f; Dircksen et al., 2011; Veenstra et al., 2012).
Among the peptide families recently identified in the Arthropoda are the GSEFLamides, a group characterized by the carboxyl (C)-terminal motif –GSEFLamide (e.g. Christie, 2014f; Veenstra et al., 2012). Previous genomic/transcriptomic analyses have identified GSEFLamides in at least one species of each arthropod subphylum (Bao et al., 2015; Christie, 2014a,c,f, 2015a,b,c; Christie and Chi, 2015a,b,c; Christie and Pascual, 2016; Christie et al., 2015, 2017a,b, 2018a,b; Nguyen et al., 2016; Toullec et al., 2017; Veenstra et al., 2012), suggesting that they are highly conserved in this taxon. However, published studies characterizing GSEFLamides are limited; consequently, the extent to which this family is conserved is not clear. Moreover, no biochemical or mass spectrometric confirmation of members of this peptide family has been published; at this point, the identity/presence of GSEFLamides is based on in silico predictions from genomic/transcriptomic data.
The present study is a multi-faceted examination of the GSEFLamide family, with a primary focus on the American lobster, Homarus americanus, a model species for investigating the modulation of rhythmic motor behaviours (e.g. Christie, 2011; Christie et al., 2010b; Dickinson et al., 2016; Nusbaum and Blitz, 2012; Nusbaum et al., 2017; Stein, 2009). Several transcriptomes (Christie et al., 2015, 2018c,d; Northcutt et al., 2016), including one specific for the eyestalk ganglia (Christie et al., 2017b), which is the locus of the neuroendocrine X-organ–sinus-gland (XO–SG) complex (e.g. Christie, 2011), are available for H. americanus. To date, GSEFLamides have been identified from both the lobster eyestalk ganglia assembly (Christie et al., 2017b) and a transcriptome generated from multiple nervous system regions (Christie et al., 2015).
Here, we used publicly accessible databases to determine the extent to which GSEFLamides are conserved within the Arthropoda. Additionally, we validated GSEFLamide sequences predicted from H. americanus transcriptomic data using reverse transcription (RT)-PCR. We then confirmed the identity/presence of the predicted lobster GSEFLamides using mass spectrometry. Finally, we asked whether the most abundant H. americanus GSEFLamide, AMGSEFLamide, modulates lobster pattern-generating networks.
MATERIALS AND METHODS
Animals
Lobsters, Homarus americanus Milne-Edwards 1837, were purchased from local (Brunswick, Maine, USA) seafood distributors. Animals used were adults of ∼500 g, and included both males and females. Lobsters were housed in recirculating natural seawater aquaria at 10–12°C and were fed chopped squid weekly.
In silico genome/transcriptome mining
Publicly accessible nucleotide sequence datasets were searched using a well-established workflow (e.g. Christie, 2014a,c,f, 2015a,b,c; Christie and Chi, 2015a,b,c; Christie et al., 2015, 2017a,b, 2018a,b) for evidence of GSEFLamide-encoding genes/transcripts in all extant arthropod subphyla, i.e. Chelicerata, Crustacea, Hexapoda and Myriapoda. In brief, the database option of the online tblastn program (National Center for Biotechnology Information, Bethesda, MD; http://blast.ncbi.nlm.nih.gov/Blast.cgi) was set to ‘nucleotide collection’, ‘whole-genome shotgun contigs’, ‘transcriptome shotgun assembly’ or ‘expressed sequence tags’ and restricted to data from one of a number of selected arthropod taxa (Table S1); a GSEFLamide precursor predicted from the H. americanus eyestalk ganglia, i.e. prepro-GSEFLamide variant 1 (Christie et al., 2017b), was used as the query sequence for these searches. To confirm that the identified nucleotide sequences encode putative GSEFLamide precursors, the hits returned by each BLAST search were translated using the Translate tool of ExPASy (http://web.expasy.org/translate/). They were then assessed for the signature motif that defines the GSEFLamide family, namely ‘XGSEFLG’ (where X represents one or more variable residues), bounded on both its amino (N)- and C-terminus by putative dibasic residues (unless it is the C-terminal- most portion of the deduced protein) that are likely to serve as prohormone convertase cleavage loci, e.g. KRAMGSEFLGKR, a sequence contained within the H. americanus precursor used as the query sequence. This analysis was a broad survey, designed to assess the likely presence/absence of GSEFLamides in the major taxa of the Arthropoda, rather than an all-encompassing search directed at identifying either all species within a given taxon for which there is evidence for GSEFLamides or all isoforms of the peptide family that likely exist within the phylum.
RT-PCR
Individual lobsters were anaesthetized by packing in ice for ∼30 min, after which the supraoesophageal ganglion (brain) and eyestalk ganglia were isolated via manual microdissection in chilled (8–10°C) physiological saline (composition in mmol l−1: 479.12 NaCl, 12.74 KCl, 13.67 CaCl2, 20.00 MgSO4, 3.91 Na2SO4, 11.45 Trizma base and 4.82 maleic acid; pH 7.45). Total RNAs were purified from individual brains or eyestalk ganglia pairs (n=3 independent samples from each tissue) as described previously (Christie et al., 2017b, 2018c) using TRIzol reagent (Life Technologies Corp., Carlsbad, CA, USA) and a Direct-zol RNA MiniPrep spin column system (Zymo Research, Irvine, CA, USA) with on-column DNase I treatment according to the manufacturer-supplied protocol. Before storage at −80°C, RNA quality and quantity were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). cDNAs were synthesized from ∼500 ng of total RNA with random pentadecamers (IDT, San Diego, CA, USA) and a SuperScript III First-Strand Synthesis System (Life Technologies Corp.). The full-length H. americanus (Homam)-prepro-GSEFLamide transcript was amplified from the respective cDNA sets using SapphireAmp Fast PCR Master Mix (Takara Bio USA, Inc., Mountain View, CA, USA) in a 20 μl reaction volume with 0.5 μl of cDNA and oligonucleotide primers designed using the first in-frame initiation codon in GenBank accession number GFDA01138849 (5′-ATGGTGAGGGGTTGGCCA-3′; an N-terminal partial sequence) and the stop codon in accession number GFDA01064392 (5′-TCAACCAAGGAACTCCGAACC-3′; a C-terminal partial sequence). To confirm the integrity of the cDNA templates, a 500-bp fragment of the H. americanus glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene was likewise amplified with oligonucleotide primers (sense: 5′-TCGGTCGTCTTGTCCTTC-3′; antisense: 5′-CAGTGACGGCATGAACAG-3′) designed to a previously deposited sequence (GenBank accession number FE043664). Thermocycler conditions consisted of: 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 56°C for 20 s, and 72°C for 90 s, with a final extension at 72°C for 5 min. The resulting products were visualized on a 1.5% agarose gel stained with SYBR Safe (Life Technologies Corp.). Aliquots from each reaction were then cloned into pCR2.1TOPO TA (Life Technologies Corp.) and sequenced at the Arizona State University DNA Core laboratory (Tempe, AZ, USA). Consensus sequences for the three cloned transcripts have been deposited in GenBank under accession numbers MH615811, MH615812 and MH615813.
Mass spectrometry
Sample preparation
Individual brains were placed in 58 μl of LCMS water (Fisherbrand; Life Technologies Corp.) in a 1.5 ml low retention tube. The sample was heated at 100°C for 5 min to deactivate proteolytic enzymes (Stemmler et al., 2013) and was cooled to room temperature or stored at −20°C. For tissue extraction, methanol (CH3OH; LCMS-grade; Fisherbrand; 128 μl) and glacial acetic acid (CH3CO2H; reagent grade; >99%; Sigma-Aldrich, St Louis, MO, USA; 14 μl) were added to the tube. The tissue was homogenized using a motor-driven tissue grinder equipped with a polypropylene pestle (Sigma-Aldrich). Following homogenization, the tissue was sonicated for 5 min and centrifuged at 14,000 g for 5 min. The supernatant was removed from the tissue and the remaining tissue pellet was re-suspended in 50 μl of extraction solvent (29% water; 64% methanol; 7% glacial acetic acid), sonicated for 5 min, and centrifuged at 14,000 g for 5 min; the supernatant was again removed. The supernatants were combined and filtered through either 10 or 30 kDa molecular mass cut-off (MWCO) filters (Amicon Ultra-0.5 ml; Millipore, Burlington, MA, USA) that had been prewashed with two 200 μl volumes of extraction solvent. The MWCO filters were centrifuged at 14,000 g for 20 min. The flow-through (∼350 μl) was collected for further processing or analysis. In preparation for liquid chromatography/mass spectrometry (LC/MS) analysis, the flow-through from the filters was either desalted and concentrated using Pierce™ C18 Spin Columns (Life Technologies Corp.) or subjected to liquid–liquid extraction. The liquid–liquid extraction was conducted by adding chloroform (NMR-grade 13CDCl3; Cambridge Isotope Laboratories, Tewksbury, MA, USA; 200 μl) and water (LCMS-grade; Fisherbrand; Life Technologies Corp.; 150 μl) to the flow-through solution, shaking for 1 min, allowing the solutions to separate for 1 min, and then removing and discarding the bottom organic layer. Additional chloroform (200 μl) was then added and the extraction was repeated. The top aqueous layer was then analyzed by LC/MS.
Chip-based nanoLC-QTOF tandem mass spectrometry (MS/MS) analysis
Mass spectrometric analysis was performed using a 6530 quadrupole time-of-flight (Q-TOF) mass analyzer (Agilent Technologies). Mass spectra (MS and MS/MS) were collected in positive ion mode; the ionization voltage ranged from 1850 to 1975 V and the ion source temperature was held at 350°C. Spectra were internally calibrated using dibutyl phthalate (C16H22O4) and hexakis(1H, 1H, 4H-hexafluorobutyloxy)phosphazine (HP-1221; C24H18O6N3P3F36) continuously infused and detected as [M+H]+. Collision-induced dissociation (CID)-MS/MS experiments were executed with precursor ions subjected to CID using nitrogen as the target gas. The collision energy (CE), in electron-volts (eV), used to dissociate a precursor ion of mass-to-charge ratio, m/z, was calculated using the equation: CE=[(3.7×m/z)/100]+2.5.
Chromatographic separation and nano-electrospray ionization (nanoESI) were performed using a 1260 Chip Cube system (Agilent Technologies) using a ProtID-chip with a 40 nl enrichment column and a 150 mm×75 µm analytical column (Agilent Technologies). The enrichment and analytical columns were packed with 300 Å, 5 µm particles with Zorbax 300SB-C18 stationary phase. The mobile phases were 0.1% formic acid/H2O (A) and 0.1% formic acid/2% water/acetonitrile (B). Samples (1–8 µl) were loaded on the enrichment column using 98:2 (A:B) at 4 µl min−1. Samples were analyzed with the 150 mm analytical column using a linear gradient of 98:2 (A:B) for 1 min to 65:35 (A:B) at 30 min, to 40:60 (A:B) at 33 min and 0:100 (A:B) at 40.0 min using a flow rate of 0.3 µl min−1.
LC/MS figures were generated by exporting Mass Hunter (Agilent Technologies) chromatograms or mass spectra as metafiles and importing these graphics into CorelDRAW X4 (Corel Corporation, Ottawa, Canada) for final figure production.
Electrophysiology
Experiments were conducted on three pattern-generating networks in the lobster nervous system: the cardiac ganglion (CG), which controls the rhythmic beating of the neurogenic heart (Cooke, 2002), and two networks that form part of the stomatogastric nervous system (STNS) and are located in the stomatogastric ganglion (STG). Specifically, we examined the effects of AMGSEFLamide on the gastric mill and pyloric neuronal circuits, which control the chewing movements of the gastric mill teeth and the filtering movements of the pyloric filter, respectively (e.g. Harris-Warrick et al., 1992; Selverston and Moulins, 1987). To isolate the CG and STNS, the heart and foregut were removed from cold-anesthetized lobsters and pinned out in cold (∼4°C) saline.
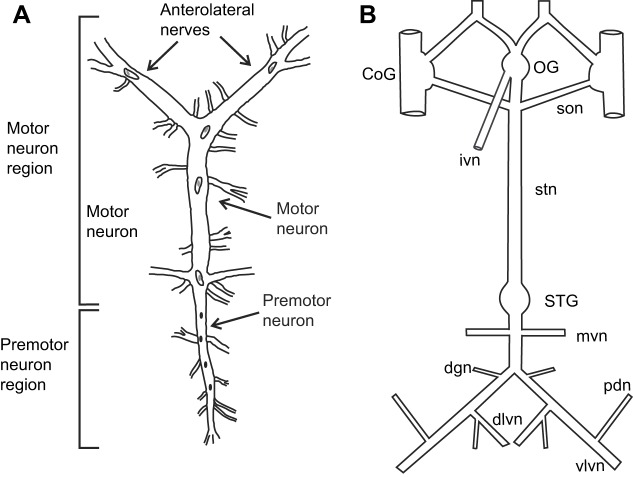
To isolate the CG, the heart was opened along the ventral axis, and the main trunk of the ganglion, together with lengths of the anterolateral and posterolateral nerves (Fig. 1A), was dissected from the inner dorsal surface of the heart. To isolate the STNS, the foregut was opened along the ventral side. The STNS (Fig. 1B), including the paired commissural ganglia (CoGs), the single oesophageal ganglion (OG) and the single STG, together with the connecting nerves and motor nerves, was removed by microdissection. Both neural tissues were pinned out in a Petri dish covered in Sylgard 184 (KR Anderson, Santa Clara, CA, USA). The connective sheath that overlies the STG was removed to ensure access to the ganglion by the peptide. Additionally, a section of the sheath surrounding the stomatogastric nerve (stn) was removed. The desheathed region of the stn was surrounded by a petroleum jelly well, allowing us to block the passage of action potentials through the stn by replacing the saline in the well with isotonic sucrose (750 mmol l−1); in most cases, the stn was subsequently transected within the well to ensure complete isolation of the STG from the rest of the nervous system. Although blocking the stn is reversible, cutting it is not; thus, AMGSEFLamide was applied to all preparations at multiple concentrations before the stn was blocked.
Fig. 1.
Schematic drawings of the Homarus americanus cardiac ganglion (CG) and stomatogastric nervous system (STNS). (A) CG; the cardiac motor pattern was recorded distally on the anterolateral nerves, which contain motor neuron axons. (B) STNS; activity of stomatogastric networks was recorded on motor nerves at the posterior end of the ganglion. CoG, commissural ganglion; dgn, dorsal gastric nerve; dlvn, dorsal lateral ventricular nerve; ivn, inferior ventricular nerve; mvn, medial ventricular nerve; OG, oesophageal ganglion; pdn, pyloric dilator nerve; son, superior oesophageal ganglion; STG, stomatogastric ganglion; stn, stomatogastric nerve; vlvn, ventral lateral ventricular nerve. Drawings are not to scale.
Petroleum jelly wells were built around small portions of the motor nerves in both preparations; bipolar stainless steel electrodes were used for recordings, with one electrode in the well and one nearby in the bath. Neuronal activity was amplified with a 1700 A-M Systems Differential AC amplifier (A-M Systems, Sequim, WA, USA) and a Brownlee Precision amplifier (Brownlee Instruments, San Jose, CA, USA). Activity was recorded onto a computer using a CED Micro 1401 digitizer and Spike2 version 7 (Cambridge Electronic Design, Cambridge, UK), with a sampling rate of 10 kHz.
Throughout the recordings, temperature was maintained at 10–12°C using an in-line Peltier temperature regulator (CL-100 bipolar temperature controller and SC-20 solution heater/cooler; Warner Instruments, Hamden, CT, USA). Physiological saline was superfused continuously across the ganglia at a flow rate of ∼5 ml min−1. AMGSEFLamide, custom synthesized by GenScript Corporation (Piscataway, NJ, USA), was introduced to the bath containing the ganglia via the perfusion system. The peptide was dissolved in deionized water at 10−3 mol l–1, stored in small aliquots at −20°C, and diluted in saline to the appropriate concentration just before use.
Data analysis
Physiological data were analyzed using the functions built into Spike2, as well as scripts available at http://stg.rutgers.edu/Resources.html. Data were analyzed statistically and graphed using Prism 7 (GraphPad Software, San Diego, CA, USA). To normalize for variation in baseline parameters, some values are presented as percent change from baseline. Only preparations that returned to baseline values during the saline wash following peptide application were considered in our data analysis.
For CG recordings, data were averaged over 50 bursts, with the control values taken shortly before the peptide was added to the bath, and the peptide values taken at the peak of the peptide effect, 5–10 min into peptide application. Because one data set (CG burst frequency at 10−9 mol l–1) was not normally distributed (Shapiro–Wilk normality test), the nonparametric Wilcoxon signed rank test was used to determine whether changes differed significantly from a hypothetical value of zero. To determine whether there were differences in effects of AMGSEFLamide when applied at different concentrations, responses at peptide concentrations that exerted significant effects (10−8, 10−7 and 10−6 mol l–1, all normally distributed) were compared using an ANOVA followed by Tukey's post hoc test for multiple comparisons.
For STNS recordings, measurements were averaged over ten bursts before and ten bursts at the peak of the peptide effect, or 5–10 min into peptide application, when the peak response generally took place. All parameters were normally distributed (Shapiro–Wilk normality test); thus, one-sample t-tests (two-tailed) were used to determine whether these changes differed significantly from zero. No comparisons were made between peptide concentrations because all changes were significant only at 10−6 mol l–1.
N-values for all experiments refer to individual animals. All error values in the text and error bars in the figures for physiological data represent standard deviations (s.d.).
RESULTS
GSEFLamides appear broadly conserved within the Arthropoda, but may have been lost in all but the most basal orders of the Insecta
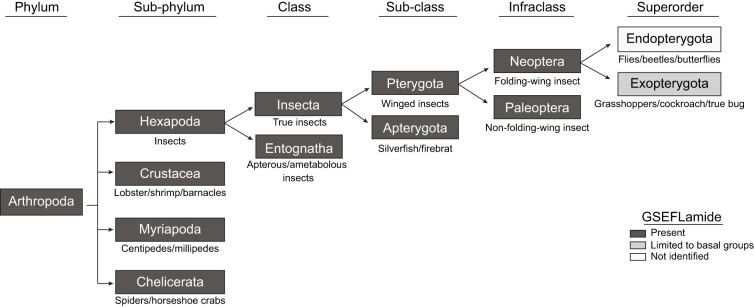
Previous genomic/transcriptomic studies have identified genes/transcripts encoding GSEFLamide precursors from at least one species from each arthropod subphylum, i.e. the Chelicerata, Myriapoda, Crustacea and Hexapoda. However, the full extent to which this peptide group is conserved within arthropods remains unclear. Thus, to broaden our understanding of GSEFLamide conservation within the Arthropoda, we conducted BLAST searches of multiple genomic/transcriptomic datasets for most classes/orders within each subphylum, looking for evidence of GSEFLamide-encoding genes/transcripts. Our BLAST searches support the hypothesis, suggested by previous analyses, that GSEFLamides are broadly conserved within the arthropods, with one or more potential losses in the more advanced taxa of the Hexapoda (Table S1). Specifically, evidence for the existence of GSEFLamides was found in most Chelicerate and Crustacean classes/orders for which large genomic/transcriptomic resources are publicly accessible, including many for which no data supporting the presence of the family previously existed, e.g. the Merostomata, a chelicerate class that includes only one extant order, the Xiphosura (horseshoe crabs), and the Thecostraca, a subclass of the crustacean class Maxillopoda that includes the order Cirripedia (barnacles). Although publicly accessible nucleotide sequences are limited, there is also evidence for broad GSEFLamide conservation in members of the Myriapoda, with in silico data now supporting the presence of GSEFLamides in at least two of its four classes: Symphyla [pseudocentipedes (Christie, 2015c)] and Chilopoda [true centipedes (this study)]. In contrast, the evidence we found for GSEFLamide-encoding genes/transcripts in hexapods was limited to orders within the ametabolous class Entognatha, i.e. the Collembola (springtails), Diplura (bristletails) and Protura [coneheads (Christie and Chi, 2015a; this study)], and the most basal orders within the Insecta, i.e. the ametabolous Zygentoma (silverfish), and the hemimetabolous Odonata (dragonflies and damselflies) and Ephemeroptera (mayflies). We found no evidence for the existence of GSEFLamides in the more advanced orders of hemimetabolous insects, e.g. the Blattodea (cockroaches and termites) and Orthoptera (grasshoppers, locusts and crickets), or in any holometabolous insect order, e.g. the Diptera (flies), Hymenoptera (ants, bees and wasps) and Lepidoptera (butterflies and moths), despite the sequence richness of the publicly deposited datasets for many of these taxa. These data suggest the possibility of a large-scale loss of the GSEFLamides early in the insect lineage. However, one report of a GSEFLamide-like precursor from Lygus hesperus (Christie et al., 2017a), a member of the order Hemiptera (true bugs), raises the possibility of multiple losses, or potentially gains, of the family in at least some higher insect orders (Fig. 2).
Fig. 2.
Evolutionary lineage tree showing that the GSEFLamides are broadly, but perhaps not ubiquitously, conserved within the Arthropoda. Taxa for which there is genomic/transcriptomic evidence of GSEFLamide genes/transcripts are boxed in dark grey. For the hexapod infraclass Neoptera and superorder Exopterygota, evidence of GSEFLamides is limited to a single identification of a GSEFLamide precursor from the true bug Lygus hesperus (Christie et al., 2017a). No evidence was found for the existence of GSEFLamides in any member of the Endopterygota, suggesting a putative loss of this peptide family in holometabolous insects. Data from: Bao et al., 2015; Christie, 2014a,c,f, 2015a,b,c; Christie and Chi, 2015a,b,c; Christie and Pascual, 2016; Christie et al., 2015, 2017a,b, 2018a,b; Nguyen et al., 2016; Toullec et al., 2017; Veenstra et al., 2012; this study.
RT-PCR confirms the presence and identity of GSEFLamide-encoding transcripts in the H. americanus eyestalk ganglia and brain
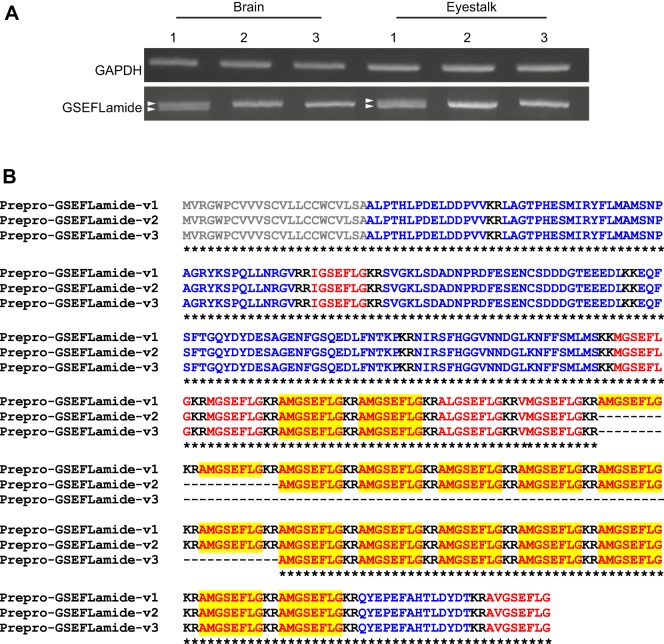
Two previous studies have provided transcriptomic evidence for GSEFLamides in neural tissues of the lobster, H. americanus (Christie et al., 2015, 2017b). In silico analyses of an eyestalk-ganglia-specific transcriptome identified two transcripts that separately encode the N- and C-termini of putative GSEFLamide preprohormones (Christie et al., 2017b). To generate a complete open reading frame (ORF) for the lobster GSEFLamide precursor(s), primers were designed based on the partial eyestalk ganglia nucleotide sequences GFDA01138849 and GFDA01064392, which correspond to the 5′ and 3′ terminal fragments of the putative transcript(s), respectively. These primers were used in RT-PCR to amplify products from multiple eyestalk ganglia and brain cDNAs (n=3/tissue). While amplimers were generated from each replicate, sizes varied, with multiple products generated in a subset of each cDNA template (Fig. 3A), suggesting potential variant amplification. Sanger sequencing revealed the presence of three GSEFLamide precursors, the largest of which encompasses a 1221 nucleotide (nt) ORF (accession number MH615811); it contains 338 base pairs (bp) of new sequence that bridges the gap between the two in silico identified fragments. The other two variants, accession numbers MH615812 and MH615813, are differentiated by 60 nt (781–840) and 240 nt (781–1020) deletions, respectively. Little variation was observed between sequences derived from the eyestalk ganglia and brain, with no evidence for tissue-specific variant expression. BLASTx analyses of the 1221 nt ORF revealed 77, 76 and 53% nucleotide identity with sequences from the crayfish Cherax quadricarinatus (accession number AWK57519; Nguyen et al., 2016), the hexapod Thermobia domestica (accession number ALG35950 GenBank direct submission) and the crab Scylla paramamosain (accession number ALQ28590; Bao et al., 2015), respectively, each annotated as encoding a GSEFLamide precursor.
Fig. 3.
Reverse transcription (RT)-PCR confirms the presence of multiple prepro-GSEFLamide splice variants in both the brain and eyestalk ganglia. (A) RT-PCR-based amplification of GSEFLamide precursors from brain and eyestalk ganglia cDNAs. Gene-specific primers designed to the 5′ and 3′ ends of the non-overlapping transcriptomic sequences GFDA01138849 and GFDA01064392 generated amplimers from all three biological replicates for each tissue. All amplimers were sub-cloned and sequence-verified. The varied amplimer sizes correspond to splice variants, with two variants (arrowheads) resolvable in a subset of tissue cDNAs. To control for cDNA quality, a 500 bp fragment of H. americanus GAPDH was amplified. Image corresponds to PCR products electrophoresed on 1.5% agarose gels stained with SYBR Safe. (B) Alignment of the three GSEFLamide precursors deduced from cloned sequences. Signal peptides are shown in grey; mono/dibasic cleavage loci are shown in black. GSEFLamide isoforms are shown in red; linker/precursor-related peptides are shown in blue. Copies of AMGSEFLamide are highlighted in yellow. In the line below each sequence grouping, an asterisk (*) indicates identical amino acid residues. Homam-prepro-GSEFLamide-v1, deduced from accession number MH615811; Homam-prepro-GSEFLamide-v2, deduced from accession number MH615812; Homam-prepro-GSEFLamide-v3, deduced from accession number MH615813.
Translation of the longest variant (MH615811) revealed a 406-amino acid preprohormone, Homam-prepro-GSEFLamide-v1 (Fig. 3B), with a signal peptide predicted to span the first 23 residues. Proteolytic processing of dibasic cleavage sites in the prohormone would yield 23 peptides with the –GSEFLamide motif, including 17 copies of AMGSEFLamide, two copies of MGSEFLamide, and one copy each of IGSEFLamide, VMGSEFLamide, ALGSEFLamide and AVGSEFLamide. The deletions observed in the other two variants would affect only the number of AMGSEFLamide copies (Fig. 3B), with 15 predicted in Homam-prepro-GSEFLamide-v2 (deduced from MH615812) and nine in Homam-prepro-GSEFLamide-v3 (deduced from MH615813).
Mass spectrometric confirmation of predicted GSEFLamide isoforms in the lobster brain
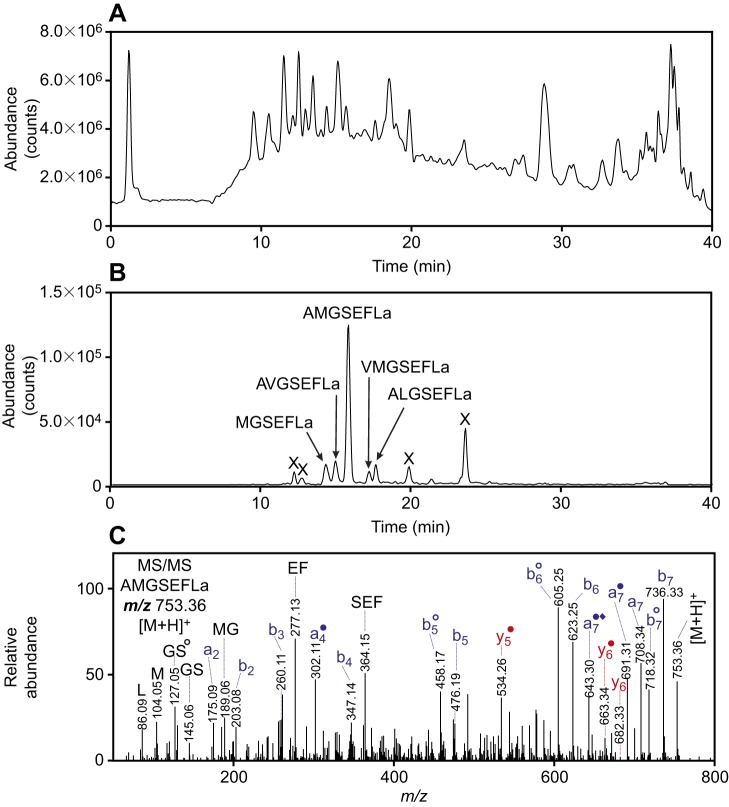
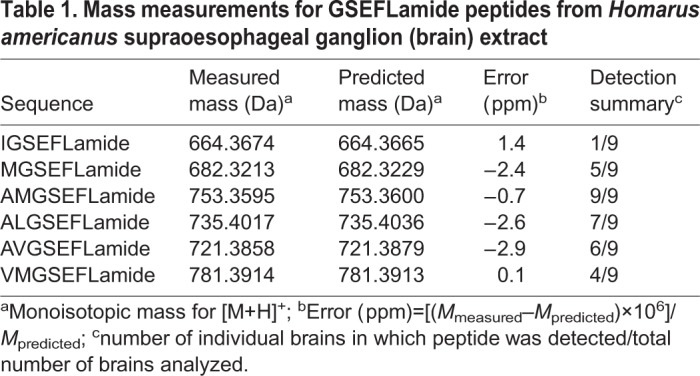
Peptides in the brains of individual lobsters (n=9) were extracted and analyzed using a chip-based nanoLC-QTOF-MS/MS instrument. A representative chromatogram for the extract from one animal is shown in Fig. 4A. We used exact mass measurements to search for signals reflecting the six GSEFLamides. The peptides, with no basic amino acid residues in their sequences, produced singly-protonated [M+H]+ ions; we used the signals at the predicted m/z values to locate the individual GSEFLamides in the chromatogram (shown as a summed extracted ion chromatogram in Fig. 4B). The [M+H]+ ion (m/z 753.36) from AMGSEFLamide yielded the most intense signal (Fig. 4B); four other GSEFLamides (MGSEFLamide, AVGSEFLamide, VMGSEFLamide and ALGSEFLamide) were detected with lower signal strength. The identification of these peptides was supported by exact mass measurements (Table 1) and by MS/MS spectra to confirm the amino acid sequence (Fig. 4C for AMGSEFLamide and Figs S1-S5). The peptide IGSEFLamide, which eluted between MGSEFLamide and AVGSEFLamide, yielded a weaker signal and was confirmed only by exact mass measurements.
Fig. 4.
Mass spectrometric identification of GSEFLamide isoforms from the brain. (A) Total ionization chromatogram for H. americanus supraoesophageal ganglion (brain) extract, showing signals from the many peptides detected in brain tissue extracts. (B) Summed extracted ion chromatograms for the [M+H]+ peaks from IGSEFLamide (m/z 664.3665), MGSEFLamide (m/z 682.3229), AVGSEFLamide (m/z 721.3880), AMGSEFLamide (m/z 753.3600), VMGSEFLamide (m/z 781.3914) and ALGSEFLamide (m/z 735.4036), showing signals specific to GSEFLamide peptides; the strongest signal originated from AMGSEFLamide; ‘X’ indicates that the signal does not originate from a GSEFLamide peptide; signals from IGSEFLamide were too weak to contribute to the chromatogram. (C) Tandem mass spectrometry (MS/MS) spectrum for the m/z 753.36 [M+H]+ ion from AMGSEFLamide at a collision energy of 30.4 eV. The assigned sequence was supported by a complete series of N-terminus-containing b-type product ions, many of which lost CO to produce a-type ions. C-terminus-containing y-type ions provided additional sequence support, as did the detection of internal product ions, including MG, GS, EF and SEF, and immonium ions, including peaks for L and M. Ions that have lost NH3 are shown with a filled circle; ions that have lost H2O are shown with an open circle; ions that have lost CH3SH from a methionine residue are shown with a filled diamond. Monoisotopic masses are displayed. Blue text indicates b- and a-type product ions; red text indicates y-type product ions.
Table 1.
Mass measurements for GSEFLamide peptides from Homarusamericanus supraoesophageal ganglion (brain) extract
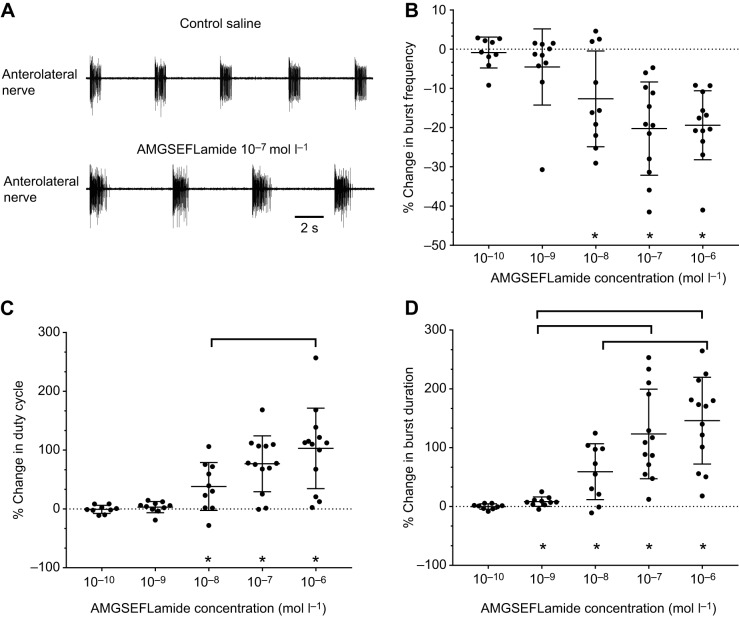
AMGSEFLamide modulates the motor output of the H. americanus cardiac ganglion
Because GSEFLamide transcripts are present in the eyestalk ganglia, as well as in other neuronal tissues in the lobster, we asked whether AMGSEFLamide, the most abundant GSEFLamide, might function as a neuromodulator. One neuronal tissue that is often subject to neuromodulation is the CG, which is located on the inner dorsal wall of the single-chambered heart of the lobster. The rhythmic contractions of the H. americanus heart are driven by bursts of action potentials generated in the nine neurons that comprise the CG (e.g. Cooke, 2002). All of the CG neurons fire at approximately the same time due to tight coupling, both electrical and chemical, among these neurons. The output of the CG is thus a single-phase pattern (e.g. Cooke, 2002). Bursts of action potentials in the motor neurons travel down the paired anterolateral and posterolateral nerves to the myocardium, where each burst induces a single cardiac contraction, the amplitude of which is a complex function of the burst cycle frequency and duty cycle (i.e. burst duration/cycle period; Williams et al., 2013). It is thus possible to characterize the pattern by quantifying cycle frequency, burst duration and duty cycle in any of the motor nerves.
We recorded from the anterolateral nerves in control saline and during superfusion with AMGSEFLamide at concentrations ranging from 10−10 to 10−6 mol l–1. AMGSEFLamide clearly modulated the rhythmic output of the CG (Fig. 5A): burst frequency decreased, while burst duration and duty cycle increased. The threshold for AMGSEFLamide effects was between 10−9 and 10−8 mol l–1, with significant changes in cycle frequency and duty cycle at concentrations of 10−8 mol l–1 and higher (Fig. 5B,C). Significant changes in burst duration were seen at an even lower concentration, i.e. 10−9 mol l–1, although the magnitude of these changes was relatively small (8.3±7.9%) (Fig. 5D). At higher peptide concentrations, the magnitude of the effect increased for both burst duration and duty cycle. In contrast, cycle frequency did not change significantly across peptide concentrations.
Fig. 5.
AMGSEFLamide modulates CG output. (A) Recordings of activity in the anterolateral nerve in control saline and during bath application of 10−7 mol l–1 AMGSEFLamide. AMGSEFLamide elicited an increase in burst duration and a decrease in cycle frequency. (B–D) Graphs depicting changes in CG cycle frequency (B), duty cycle of motor neuron bursts (C) and duration of motor neuron bursts (D) in response to bath application of AMGSEFLamide at concentrations ranging from 10−10 to 10−6 mol l–1. (B) Threshold concentration for changes in cycle frequency was ∼10−8 mol l–1; at concentrations of 10−8 mol l–1 and higher, the percent change in burst frequency decreased significantly (Wilcoxon signed rank test against a theoretical median of 0, two-tailed: 10−10 mol l–1, P=0.9102; 10−9 mol l–1, P=0.1309; 10−8 mol l–1, P=0.0273; 10−7 mol l–1, P=0.0005; 10−6 mol l–1, P=0.0005). However, changes in cycle frequency were not concentration dependent; all concentrations above threshold elicited similar effects (ANOVA, P=0.2345). (C) Threshold concentration for changes in duty cycle was ∼10−8 mol l–1, with 10−8 mol l–1 and higher concentrations eliciting significant changes in duty cycle (Wilcoxon signed rank test; theoretical median of 0, two-tailed: 10−10 mol l–1, P=0.7344; 10−9 mol l–1, P=0.1934; 10−8 mol l–1, P=0.0137; 10−7 mol l–1, P=0.0005; 10−6 mol l–1, P=0.0002). The response to AMGSEFLamide increased with increasing concentrations (ANOVA, P=0.0279), with a significant difference between responses to 10−8 and 10−6 mol l–1 (Tukey's multiple-comparison test, P=0.0211). (D) Threshold for changes in burst duration elicited by AMGSEFLamide was ∼10−9 mol l–1; concentrations of 10−9 mol l–1 and higher elicited significant changes in burst duration (Wilcoxon signed rank test; theoretical median of 0, two-tailed: 10−10 mol l–1, P=0.9102; 10−9 mol l–1, P=0.0098; 10−8 mol l–1, P=0.0098; 10−7 mol l–1, P=0.0002; 10−6 mol l–1, P=0.0002). Changes in burst duration were concentration dependent (ANOVA, P<0.0001), with burst duration increasing as peptide concentration increased. Responses to 10−9 mol l–1 AMGSEFLamide differed significantly from those to 10−7 and 10−6 mol l–1 peptide; responses to 10−8 mol l–1 also differed from those to 10−6 mol l–1 peptide (Tukey's multiple-comparison test; 10−9 mol l–1 versus 10−7 mol l–1, P=0.0003; 10−9 mol l–1 versus 10−6 mol l–1, P<0.0001; 10−8 mol l–1 versus 10−6 mol l–1, P=0.0080). An asterisk (*) indicates averages significantly different from zero. Square brackets indicate significant differences between responses to peptide concentrations. For all parameters (B–D): 10−10 mol l–1, n=9; 10−9 mol l–1, n=10; 10−8 mol l–1, n=10; 10−7 mol l–1, n=13; 10−6 mol l–1, n=13.
AMGSEFLamide modulates both the gastric mill and pyloric motor patterns produced by the lobster stomatogastric ganglion
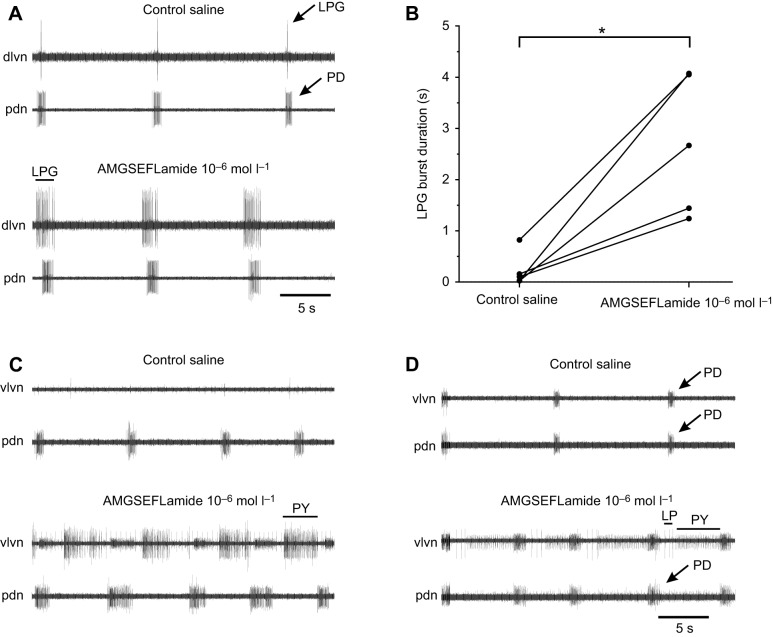
Two rhythmic motor programs are generated in the STG, the gastric mill pattern, a two-phase pattern that controls the protraction and retraction of the three teeth that make up the gastric mill, where food is shredded, and the pyloric pattern, a three-phase pattern that governs the movements of the pyloric filter (e.g. Harris-Warrick et al., 1992; Selverston and Moulins, 1987). Because both patterns are targets of the constitutive release of modulators from the CoGs and OG when the single input nerve to the STG (i.e. the stn) from these ganglia is intact, we examined the effects of AMGSEFLamide in the two networks both when the stn was intact and when action potentials travelling through the stn were eliminated.
When the stn was intact, a regular three-phase pyloric pattern was always recorded. Superfusion with AMGSEFLamide at concentrations ranging from 10−8 to 10−6 mol l–1 had only a minor effect on the pyloric pattern under these conditions. To quantify these changes, we analyzed the pyloric cycle frequency and the duration of bursts in the pyloric dilator (PD) neurons, which are members of the pacemaker group that drives the pattern. At 10−6 mol l–1, AMGSEFLamide induced a small increase in pyloric cycle frequency (∼10%; Fig. 6A). No significant change in PD burst duration was recorded at any of the concentrations used here (Fig. 6B).
Fig. 6.
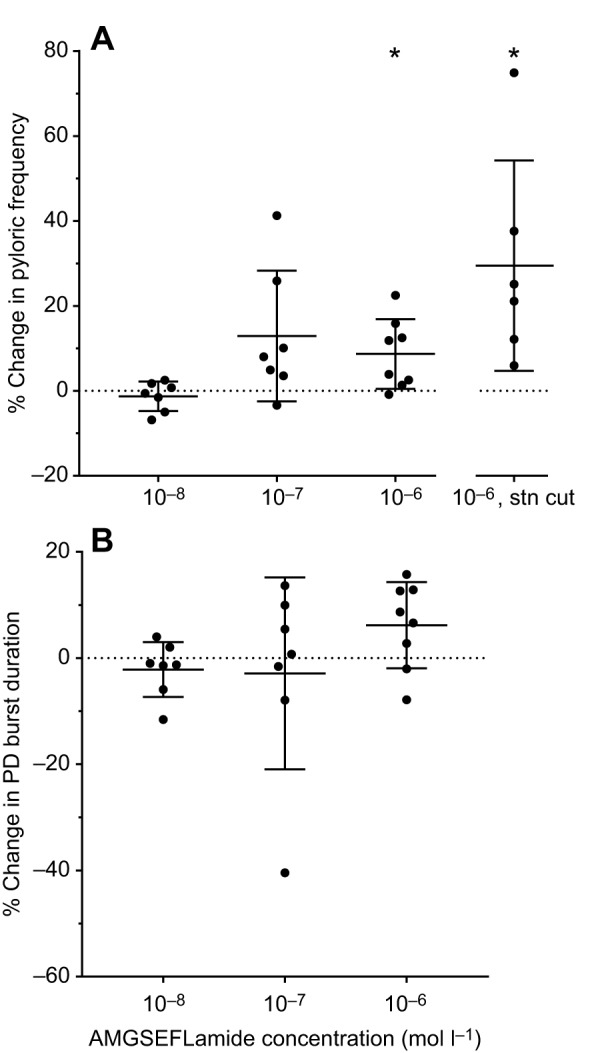
AMGSEFLamide modulated the pyloric motor network. (A) AMGSEFLamide elicited an increase in pyloric cycle frequency (percent change from frequency in control saline). When inputs from anterior ganglia were present, i.e. the stn was intact, threshold was ∼10−7–10−6 mol l–1. When the stn was blocked or cut (10−6 mol l–1, stn cut), 10−6 mol l–1 AMGSEFLamide elicited a significant increase in pyloric cycle frequency. An asterisk (*) indicates means significantly different from zero; single-sample t-test (two-tailed) against a hypothetical value of 0; 10−8 mol l–1: n=10, P=0.3668; 10−7 mol l–1: n=7, P=0.0684; 10−6 mol l–1: n=8, P=0.020; 10−6 mol l–1, stn cut: n=6, P=0.0333. (B) AMGSEFLamide did not alter the duration of pyloric dilator (PD) bursts [single-sample t-test (two-tailed) against a hypothetical value of 0: 10−8 mol l–1: n=7, P=0.3101; 10−7 mol l–1: n=7, P=0.6870; 10−6 mol l–1: n=8, P=0.0683].
When the stn is blocked or cut, eliminating all modulatory inputs from the CoGs and OG, the pyloric pattern in H. americanus is severely disrupted; under these conditions, only the PD neurons continue to fire regular bursts of action potentials (Mizrahi et al., 2001). Additionally, in our experiments, the lateral posterior gastric (LPG) neuron fired a small number (1–3) of action potentials during each PD burst in most preparations (five of seven); the LPG neuron never fired with gastric-like timing when the stn was cut or blocked. This activity in the LPG neuron was likely driven by weak electrical coupling between the PD and LPG neurons in this species. Under these conditions, modulatory effects of AMGSEFLamide were evident. Regardless of whether or not the LPG neurons were firing before application of AMGSEFLamide, they were strongly activated in all preparations (n=7; Fig. 7A). In the presence of 10−6 mol l–1 AMGSEFLamide, the duration of LPG bursts increased significantly (paired t-test; P=0.0116), from less than 1 s to a mean of 2.47±1.25 s (Fig. 7B). Interestingly, firing in the LPG neurons, which started after that in the PD neurons in control saline, usually began to precede the onset of PD neuron bursting during superfusion with 10−6 mol l–1 AMGSEFLamide (Fig. 7A). Moreover, in two of seven preparations, LPG bursts became long enough that each LPG burst was accompanied by two PD neuron bursts in the presence of the peptide.
Fig. 7.
The pyloric pattern was activated by AMGSEFLamide when inputs from the CoGs and OG were blocked. (A) Recording of activity in the lateral posterior gastric (LPG; dlvn) and PD (pdn) neurons in control saline and after ∼5 min superfusion with 10−7 mol l–1 AMGSEFLamide. During AMGSEFLamide superfusion, the LPG neuron was strongly activated. Burst duration increased; bursts were longer than those in PD; onset of LPG bursts preceded the first spikes recorded in PD. (B) Graph showing the increase in LPG burst duration across preparations. An asterisk (*) indicates significant difference, paired two-tailed t-test, P=0.0116, n=5. (C) In most cases, pyloric (PY) neurons were also activated by AMGSEFLamide. (D) In a few preparations, AMGSEFLamide activated the complete three-phase core pyloric pattern, with activity in PD, LP and PY neurons. Arrows and lines indicate spikes from: LP, lateral pyloric; LPG, lateral posterior gastric; PD, pyloric dilator; PY, pyloric.
In addition to the effects of the peptide on the LPG firing pattern, pyloric cycle frequency increased significantly in the presence of AMGSEFLamide when input via the stn was eliminated (Fig. 6A). In most lobsters (six of seven), superfusion with AMGSEFLamide at 10−6 mol l–1 also activated the pyloric (PY) neurons (Fig. 7C). In a subset of these lobsters (two of six), the full triphasic pyloric core pattern, with coordinated bursting in the PD, PY and lateral pyloric (LP) neurons, was restored when the STG was superfused with 10−6 mol l–1 AMGSEFLamide (Fig. 7D).
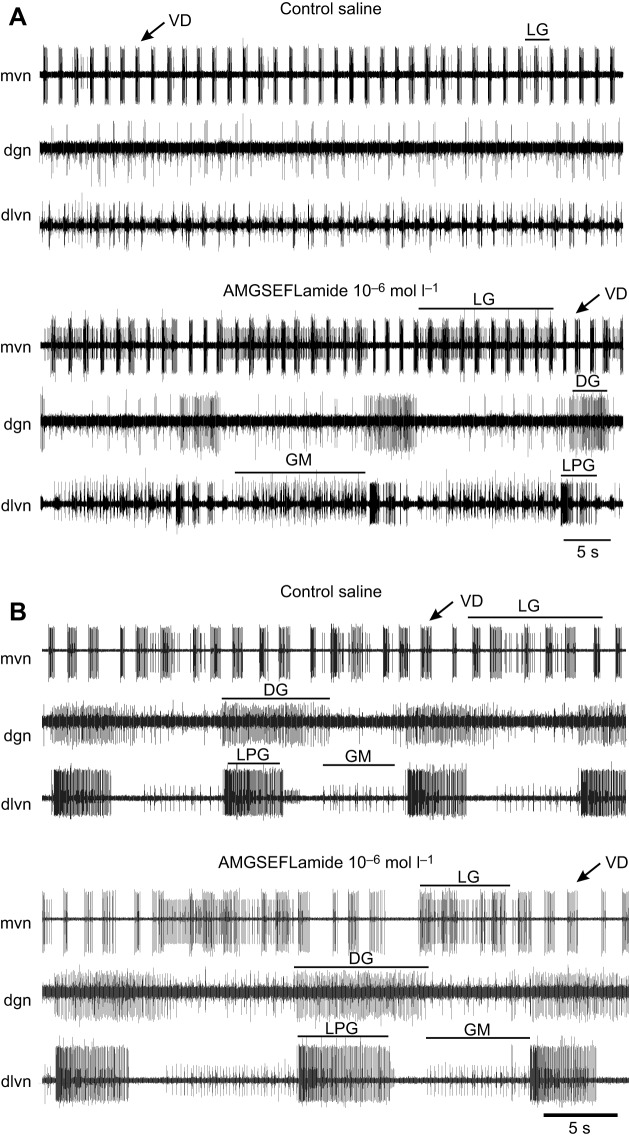
In contrast to the pyloric pattern, in which the effects of AMGSEFLamide were evident primarily when inputs from the anterior ganglia were absent, AMGSEFLamide modulated the gastric mill pattern only when the stn was intact; when the stn was blocked/cut, no gastric activity was activated. Although the LPG neuron, which generally fired with the gastric mill pattern when the stn was intact, was activated under these conditions, it fired only in pyloric time.
When the stn was intact, but an ongoing gastric mill pattern was absent, AMGSEFLamide superfusion activated the gastric pattern (10−6 mol l–1; Fig. 8A); ongoing gastric mill patterns were enhanced by AMGSEFLamide application (10−6 mol l–1; Fig. 8B). The resulting pattern included coordinated bursting in the lateral gastric (LG) and gastric mill (GM) neurons in antiphase with bursting in the dorsal gastric (DG) and LPG neurons (Fig. 8). To characterize the effects of AMGSEFLamide on the gastric mill pattern and to determine the extent to which it was dependent on peptide concentration, we quantified the effects of the peptide on the cycle period of the gastric mill pattern and on burst duration in the LG neuron, which was consistently activated by AMGSEFLamide (Fig. 9A,B). Effects on both parameters had thresholds near 10−6 mol l–1; no significant changes were recorded when the peptide was superfused at 10−7 mol l–1, but the changes in both parameters (Fig. 9A,B) were significantly greater than zero in the presence of 10−6 mol l–1 AMGSEFLamide.
Fig. 8.
AMGSEFLamide activated the gastric mill pattern when the stn was intact. Representative recordings from preparations with no ongoing pattern and with ongoing gastric mill patterns. (A) When the gastric mill pattern was not active in control saline, superfusion with 10−6 mol l–1 AMGSEFLamide activated a complete gastric pattern, with coordinated bursting in DG and LPG neurons alternating with bursts in LG and GM neurons. (B) In preparations in which the gastric mill pattern was active in control saline, the pattern was enhanced in 10−6 mol l–1 AMGSEFLamide. Burst durations in LG, DG, GM and LPG neurons increased, and spike frequency increased in LG and GM neurons. DG, dorsal gastric neuron; dgn, dorsal gastric nerve; dlvn, dorsal lateral ventricular nerve; GM, gastric mill neuron; LG, lateral gastric neuron; LPG, lateral posterior gastric neuron; mvn, medial ventricular nerve; VD, ventricular dilator neuron.
Fig. 9.
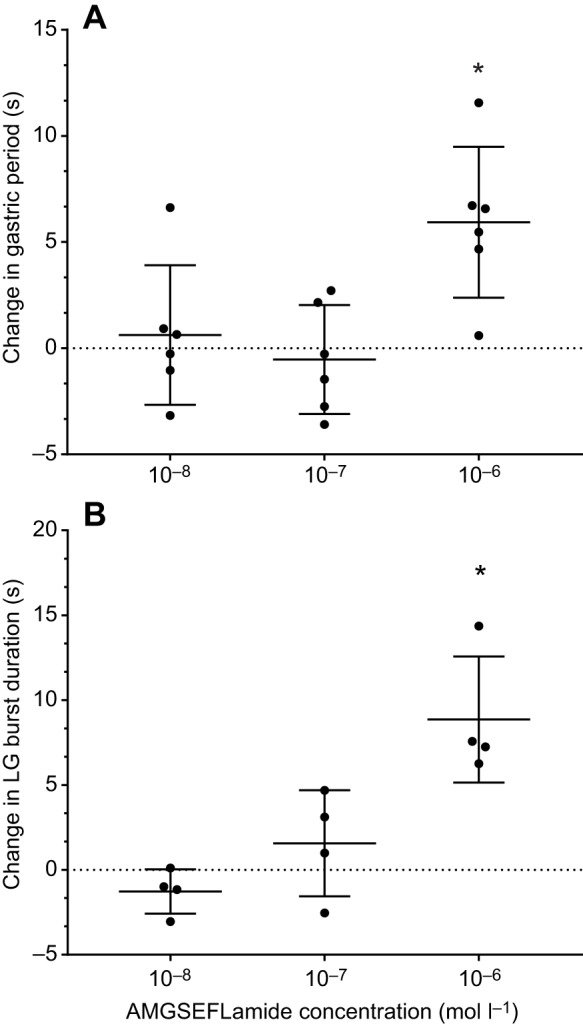
AMGSEFLamide activated the gastric mill pattern with a threshold of ∼10−6 mol l–1 when the stn was intact. (A) Bath application of AMGSEFLamide at 10−6 mol l–1, but not at 10−7 or 10−8 mol l–1, significantly increased the gastric mill cycle period [single-sample t-test (two-tailed) against hypothetical value of 0: 10−8 mol l–1: n=6, P=0.6646; 10−7 mol l–1: n=6, P=0.6312; 10−6 mol l–1: n=6, P=0.00946]. (B) Bath application of AMGSEFLamide at 10−6 mol l–1, but not at 10−7 or 10−8 mol l–1, significantly increased the duration of bursts in the LG neuron [single-sample t-test (two-tailed) against a hypothetical value of 0: 10−8 mol l–1: n=4, P=0.1469; 10−7 mol l–1: n=4, P=0.3913; 10−6 mol l–1: n=4, P=0.0175]. An asterisk (*) indicates means significantly different from zero.
DISCUSSION
Conservation of the GSEFLamide family in the Arthropoda
Recent advances in both the technology used for high-throughput nucleotide sequencing and in the computational algorithms used to assemble the resulting data have led to a rapid expansion in the genomic/transcriptomic datasets that are publicly accessible for many organisms, including arthropods. These publicly accessible genomes/transcriptomes have been used extensively to expand our knowledge of peptidergic signalling systems in arthropods, including the identification of several previously undescribed peptide families, e.g. the GSEFLamides (Veenstra et al., 2012). Prior genomic/transcriptomic analyses suggested that the GSEFLamides are broadly conserved within the Arthropoda, with evidence for their existence being found in one or more species from each subphylum, i.e. Chelicerata (Christie, 2015a; Christie and Chi, 2015b; Veenstra et al., 2012), Myriapoda (Christie, 2015c), Crustacea (Bao et al., 2015; Christie, 2014a,c,f, 2015b; Christie and Chi, 2015c; Christie and Pascual, 2016; Christie et al., 2015, 2017b, 2018a,b; Nguyen et al., 2016; Toullec et al., 2017) and Hexapoda (Christie and Chi, 2015a; Christie et al., 2017a). However, the number of studies that have reported putative GSEFLamide-encoding genes/transcripts is limited, especially for the Myriapoda and Hexapoda; thus, precisely how conserved this peptide family is within the phylum/subphyla remains unresolved. Here, we conducted a survey of publicly accessible genomic/transcriptomic datasets to broaden our understanding of GSEFLamide conservation in the Arthropoda. These analyses confirmed the hypothesis that this peptide family is broadly conserved within the phylum; they also suggest the existence of one or more losses of GSEFLamides in the more advanced taxa of the Hexapoda (Fig. 2), i.e. a potential loss at the split between the pterygotan (winged insects) infraclasses Paleoptera and Neoptera, and/or a loss (or possibly a gain) at the split between the superorders Exopterygota (hemimetabolous species) and Endopterygota (holometabolous species). Specifically, we found genomic/transcriptomic evidence for GSEFLamides in multiple species from multiple orders within the Paleoptera, e.g. the Ephemeroptera (mayflies) and Odonata (dragonflies and damselflies), while evidence for the peptide group in the Neoptera and more derived taxa is limited to a single prior report of a GSEFLamide-like peptide-encoding sequence from the plant bug L. hesperus (Christie et al., 2017a). As more sequence data are deposited for the Arthropoda, losses and/or gains of GSEFLamides can be further quantified, and further inferences can be drawn concerning possible functional roles of the peptide family in members of this phylum.
GSEFLamide isoform diversity appears highly conserved within decapods
Based on the precursor proteins deduced from the full-length clones described earlier, six isoforms of GSEFLamide are predicted to exist in the H. americanus nervous system, i.e. IGSEFLamide, MGSEFLamide, AMGSEFLamide, ALGSEFLamide, VMGSEFLamide and AVGSEFLamide; mass spectrometry confirmed the existence of all six peptides in extracts of the lobster brain. Comparisons of the complement of H. americanus GSEFLamides with those predicted for other decapod species, which span both suborders of the Decapoda, i.e. the Dendrobranchiata (Litopenaeus vannamei) and the Pleocyemata, including members of four infraorders, i.e. Astacidea (Astacus astacus, Procambarus clarkii and C. quadricarinatus), Caridea (Palaemon varians), Achelata (Jasus edwardsii) and Brachyura (Carcinus maenas, Eriocheir sinensis and Cancer borealis), suggest that a high level of isoform conservation likely exists within the decapods. Specifically, MGSEFLamide and AMGSEFLamide (both encoded as multiple copies in most species) are likely ubiquitously conserved within the order, with IGSEFLamide (present as a single copy) conserved in all but the penaeid and caridean shrimp, where a single copy of LGSEFLamide is substituted. The other lobster isoforms show varying levels of conservation among the species thus far examined. The high level of sequence conservation within the Decapoda for at least some GSEFLamide isoforms suggests that they may serve conserved functional roles in members of this order.
Physiological effects of AMGSEFLamide on the neural circuits of the cardiac and stomatogastric ganglia
Although GSEFLamide transcripts are present in both the brain and the eyestalk ganglia of the lobster, and all six native peptide isoforms have been identified in brain tissue, the presence or absence of these peptides in the CG and the STG has not yet been determined. Similarly, it remains to be determined whether any of these peptides are present in the pericardial organ, a major neuroendocrine organ that is known to release modulators into the heart (e.g. Christie, 2011). Nonetheless, the presence of GSEFLamide transcripts in the eyestalk ganglia suggests the possibility that these peptides may be released as circulating neurohormones from the XO–SG complex, a neuroendocrine system located in the eyestalk (e.g. Christie, 2011). The widespread phylogenetic conservation of the GSEFLamides, together with the identification of the nervous system as the site of synthesis of these peptides in a number of species (e.g. Bao et al., 2015; Christie et al., 2015; Christie and Pascual, 2016; Christie et al., 2017b), suggests that they may be neuroactive. Because of their locations, both the CG and the STG are fully exposed to virtually all hormones present in the hemolymph; the CG lies within the lumen of the heart, and the STG lies within one of the five major anterior arteries, i.e. the ophthalmic artery. AMGSEFLamide is present in the highest copy number on the deduced lobster GSEFLamide precursors (Fig. 3B) and is likely to be the most prevalent isoform in the lobster nervous system and/or hemolymph. Mass spectrometric evidence suggests that this is the case at least in the brain, where it was the most commonly detected GSEFLamide isoform. Thus, we chose to focus on this isoform of GSEFLamide.
Our data suggest that AMGSEFLamide modulates the cardiac neural circuit at concentrations as low as 10−8–10−9 mol l–1. Moreover, a number of the effects of this peptide were concentration dependent, with increasing effects as peptide concentration increased. For the CG, these changes could have profound implications for the functioning of the heart as a whole. Although the experiments reported here were conducted in isolated CGs, the amplitude of contractions is at least in part a function of the bursting pattern generated by the CG. The relationship between motor output and contraction amplitude can be characterized by the neuromuscular transform (Brezina et al., 2000; Brezina and Weiss, 2000; Williams et al., 2013). In the absence of peripheral modulation, the amplitudes of contractions in an isolated lobster heart are a function of the cycle frequency and duty cycle of bursts of action potentials in the CG motor neurons (Williams et al., 2013). Thus, we can predict to some extent the functional implications of the central modulatory effects of AMGSEFLamide, bearing in mind that the peptide could also have peripheral effects. While the cycle frequency, which averaged about 0.36 Hz in control saline, was decreased by an average of ∼13–22%, the differences across concentrations were not significant. Based on previously measured neuromuscular transforms, the effects of this change in frequency alone would likely elicit only modest changes in contraction amplitude, ranging from about a 10% increase to a 4% decrease; the amplitude of these changes would be dependent on the duty cycle (Williams et al., 2013). In the experiments presented here, the baseline duty cycle varied considerably, ranging from 0.07 to 0.24, suggesting that baseline contraction amplitudes might also vary considerably. However, since the increase in duty cycle resulting from AMGSEFLamide superfusion was concentration dependent, the increases in duty cycle recorded here would result in changes in contraction amplitude as a function of peptide concentration. Such changes would be most pronounced if the peptide were released locally as well as hormonally. It would thus be of particular interest to determine whether the GSEFLamides are present within the CG.
A number of neuromodulators have also been shown to alter the amplitude of cardiac contractions, and thus overall cardiac function, by effects exerted at the periphery (Dickinson et al., 2015a; Fort et al., 2007a,b; Stevens et al., 2009; Wilkens et al., 2005). For example, the peptide proctolin, in addition to central effects, exerts effects both on the cardiac muscle itself and at the neuromuscular junction (Wilkens et al., 2005). The peptides myosuppressin and two native FLRFamide-related peptides (GYSDRNYLRFamide and SGRNFLRFamide) have been shown to modulate both the output of the CG and the periphery in the lobster, although whether the peripheral effects are at the neuromuscular junction or within the muscle has not yet been determined (Dickinson et al., 2015a; Stevens et al., 2009). In the crab Callinectes sapidus, crustacean cardioactive peptide (CCAP) and a native FMRFamide-related peptide likewise exert both central and peripheral effects (Fort et al., 2007a,b). Because the GSEFLamides are likely to be present as circulating hormones, they would reach peripheral cardiac sites, where they, like the peptides mentioned above, could exert effects if appropriate receptors are present.
Like the neural circuit in the CG, the pyloric network in the STG is constitutively active in intact animals (e.g. Clemens et al., 1998a,b; Harris-Warrick et al., 1992; Rezer and Moulins, 1983; Selverston and Moulins, 1987; Yarger and Stein, 2015); it is also virtually always active in the isolated STNS when the stn, which carries inputs from the more anterior ganglia (CoGs and OG) to the STG, is intact (e.g. Harris-Warrick et al., 1992; Selverston and Moulins, 1987). In contrast, the gastric mill pattern is only active periodically. Such differences may play a role in determining the effects of modulators on the two patterns, since many modulators have state-dependent effects (Brezina et al., 2003; Nagy and Dickinson, 1983; Nusbaum and Marder, 1989; Sakurai and Katz, 2003). Nonetheless, both patterns are the targets of numerous neuromodulators in a range of decapod crustaceans, although the number of modulators whose effects on the gastric mill pattern in H. americanus have been examined is relatively small (e.g. Dickinson et al., 2015b; Kwiatkowski et al., 2013; Rehm et al., 2008).
When the stn was intact and the pyloric pattern was active, the effects of AMGSEFLamide were minimal. The peptide, even at concentrations of 10−6 mol l–1, which would be expected from local release, did not elicit major alterations of the core pyloric pattern, although cycle frequency did increase slightly (∼10%). When the stn was blocked, bursts of action potentials were recorded only in the PD neurons. Additionally, a few (2–4) action potentials in the LPG neuron were fired during PD bursts. These action potentials in the LPG neuron appear to be triggered by weak electrical coupling between the PD and LPG neurons; such coupling is prominent in the crab C. borealis (Shruti et al., 2014; Weimann et al., 1991), but is less known in the lobster. Under these conditions, the activity of the LPG neuron was markedly increased and, in many instances, the onset of the LPG burst preceded that of the PD burst. Without intracellular recordings, it is not possible to determine the mechanism that underlies this change in timing; it could reflect an increase in the bursting ability of the LPG neuron, such that it is able to trigger bursts independently, or it could reflect changes in the coupling or in the amplitude of the subthreshold depolarizations in one or both neurons. At the same time, AMGSEFLamide induced firing in the PY and occasionally the LP neurons, such that a more complete pyloric pattern could be induced even in the absence of other modulatory inputs.
The gastric mill pattern was weakly active in most of the STNSs that we recorded from (five of seven). In those in which the pattern was not active, AMGSEFLamide induced activity in the network; in those in which the pattern was ongoing, the peptide enhanced the activity of the network, leading to longer bursts and a slower gastric frequency. However, it did not induce a gastric mill pattern in the isolated STG, indicating that other modulators are required to put the network into a state in which it is responsive to AMGSEFLamide. What those modulators are and how they act is not known. Very few modulators are able to activate the gastric pattern in an isolated STG. In the lobster, the neuropeptide red pigment concentrating hormone (RPCH) activated bursting with timing appropriate for the gastric mill pattern in the DG neuron (Rehm et al., 2008), although whether the rest of the gastric mill pattern is activated by RPCH in this condition is not known. Even in the crab STNS, which has been studied extensively, only pyrokinins have been shown to activate the gastric mill pattern in an isolated STG (Rodriguez et al., 2013; Saideman et al., 2007a,b).
Although GSEFLamide concentrations in lobster hemolymph have not been measured, hemolymph concentrations of other peptides have been measured in both insects and crustaceans, with values ranging from sub-nanomolar to sub-micromolar. For example, hemolymph concentrations of an FMRFamide-related peptide are ∼5×10−9–8×10−9 mol l–1 in the locust (Robb and Evans, 1990) and 10−9–2×10−8 mol l–1 in the blood-sucking bug Rhodnius prolixus (Elia et al., 1993), while concentrations as high as 3×10−8–4×10−8 mol l–1 have been reported for ecdysis-triggering hormone and vitellogenin-inhibiting hormone in insects and shrimp (Fastner et al., 2007; Kang et al., 2014; Z̆itn̆an et al., 1999). The concentrations at which AMGSEFLamide exerted effects on the CG in the experiments reported here are within these ranges, suggesting that GSEFLamides could function as both hormonal and locally released modulators. In contrast, the thresholds for effects of AMGSEFLamide on the pyloric and gastric mill patterns were between 10−7 and 10−6 mol l–1, suggesting that they would be able to exert effects on the networks located in the STG only if the peptide is locally released. At present, we do not know the distribution of any of the GSEFLamides; this information is critical to understanding whether or not AMGSEFLamide is physiologically important in the STG. However, because both the CG and the STG are very small tissues, the total concentration of peptides in these ganglia, if they are present, would also be very low. This, coupled with the fact that these peptides do not contain basic amino acids, which enhance ionization and facilitate mass spectrometric detection, suggests that they would be difficult to detect using mass spectrometry as was done to validate the structures of these peptides in the H. americanus brain. Immunohistochemistry would be a more promising approach, but must await the development of an antibody directed against the GSEFLamides.
In the present study, we focused on one of the six isoforms of GSEFLamide that are encoded by the GSEFLamide transcript and are present in the brain of the lobster. It will be of interest to determine whether all of the isoforms exert similar effects on the three networks examined here. Although it seems likely that any differences in the responses to isoforms will be similar across networks, differences in the responses to peptide isoforms across networks within the lobster were recently reported for the pyrokinin peptides (Dickinson et al., 2015b,c). Although the native lobster pyrokinins were not known at the time of that study, the effects of five crustacean pyrokinins were examined. All five exerted identical effects on the gastric mill pattern; none had any effect on the pyloric pattern. Surprisingly, one of the isoforms clearly activated the cardiac pattern, as seen in whole heart contractions, while the others had no effect on cardiac contractions. This suggested the possibility that there are multiple receptors for the pyrokinins, with different levels of specificity, or that they act on different second messenger systems, again with different levels of specificity. Although similar phenomena have not been widely reported, the existence of a family of similar GSEFLamides is another opportunity to ask whether such differences may be more widespread than previously thought.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.S.D., E.A.S., A.E.C.; Methodology: P.S.D., E.S.D., E.A.S., J.J.H., A.E.C.; Software: E.S.D.; Validation: P.S.D., E.S.D., E.R.O., C.D.R., M.E.S., E.A.S., J.J.H., A.E.C.; Formal analysis: P.S.D., E.S.D., E.A.S., J.J.H., A.E.C.; Investigation: P.S.D., E.S.D., E.R.O., C.D.R., M.E.S., E.A.S., J.J.H., A.E.C.; Resources: P.S.D., E.A.S., J.J.H., A.E.C.; Data curation: P.S.D., E.A.S., J.J.H., A.E.C.; Writing - original draft: P.S.D., E.A.S., J.J.H., A.E.C.; Writing - review & editing: P.S.D., E.S.D., E.R.O., C.D.R., M.E.S., E.A.S., J.J.H., A.E.C.; Visualization: P.S.D., E.S.D., E.A.S., J.J.H., A.E.C.; Supervision: P.S.D., E.A.S., J.J.H., A.E.C.; Project administration: P.S.D., E.A.S., A.E.C.; Funding acquisition: P.S.D., J.J.H., A.E.C.
Funding
Funding was provided by the National Science Foundation (IOS-1353023, IOS-1354567 and CHE-1126657), an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P20GM103423), U.S. Department of Agriculture (USDA) base CRIS funding (Project #2020-22620-022-00D), and the Cades Foundation. Mention of trade names/commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation/endorsement by the USDA. The USDA is an equal opportunity provider/employer. Deposited in PMC for release afer 12 months.
Data availability
Consensus sequences for the three cloned transcripts have been deposited in GenBank under accession numbers MH615811, MH615812 and MH615813.
Supplementary information
Supplementary information available online at http://jeb.biologists.org/lookup/doi/10.1242/jeb.194092.supplemental
References
- Bao C., Yang Y., Huang H. and Ye H. (2015). Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic analysis and expression profiles during vitellogenesis. Sci. Rep. 5, 17055 10.1038/srep17055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezina V. and Weiss K. R. (2000). The neuromuscular transform constrains the production of functional rhythmic behaviors. J. Neurophysiol. 83, 232-259. 10.1152/jn.2000.83.1.232 [DOI] [PubMed] [Google Scholar]
- Brezina V., Orekhova I. V. and Weiss K. R. (2000). The neuromuscular transform: the dynamic, nonlinear link between motor neuron firing patterns and muscle contraction in rhythmic behaviors. J. Neurophysiol. 83, 207-231. 10.1152/jn.2000.83.1.207 [DOI] [PubMed] [Google Scholar]
- Brezina V., Orekhova I. V. and Weiss K. R. (2003). Neuromuscular modulation in Aplysia. I. Dynamic model. J. Neurophysiol. 90, 2592-2612. 10.1152/jn.01091.2002 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2011). Crustacean neuroendocrine systems and their signaling agents. Cell Tissue Res. 345, 41-67. 10.1007/s00441-011-1183-9 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2014a). Expansion of the Litopenaeus vannamei and Penaeus monodon peptidomes using transcriptome shotgun assembly sequence data. Gen. Comp. Endocrinol. 206, 235-254. 10.1016/j.ygcen.2014.04.015 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2014b). Identification of the first neuropeptides from the Amphipoda (Arthropoda, Crustacea). Gen. Comp. Endocrinol. 206, 96-110. 10.1016/j.ygcen.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2014c). In silico characterization of the peptidome of the sea louse Caligus rogercresseyi (Crustacea, Copepoda). Gen. Comp. Endocrinol. 204, 248-260. 10.1016/j.ygcen.2014.05.031 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2014d). Peptide discovery in the ectoparasitic crustacean Argulus siamensis: identification of the first neuropeptides from a member of the Branchiura. Gen. Comp. Endocrinol. 204, 114-125. 10.1016/j.ygcen.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2014e). Prediction of the first neuropeptides from a member of the Remipedia (Arthropoda, Crustacea). Gen. Comp. Endocrinol. 201, 74-86. 10.1016/j.ygcen.2014.01.017 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2014f). Prediction of the peptidomes of Tigriopus californicus and Lepeophtheirus salmonis (Copepoda; Crustacea). Gen. Comp. Endocrinol. 201, 87-106. 10.1016/j.ygcen.2014.02.015 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2015a). In silico characterization of the neuropeptidome of the Western black widow spider Latrodectus hesperus. Gen. Comp. Endocrinol. 210, 63-80. 10.1016/j.ygcen.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2015b). Neuropeptide discovery in Eucyclops serrulatus (Crustacea, Copepoda): in silico prediction of the first peptidome for a member of the Cyclopoida. Gen. Comp. Endocrinol. 211, 92-105. 10.1016/j.ygcen.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2015c). Neuropeptide discovery in Symphylella vulgaris (Myriapoda, Symphyla): In silico prediction of the first myriapod peptidome. Gen. Comp. Endocrinol. 223, 73-86. 10.1016/j.ygcen.2015.09.021 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2016a). Expansion of the neuropeptidome of the globally invasive marine crab Carcinus maenas. Gen. Comp. Endocrinol. 235, 150-169. 10.1016/j.ygcen.2016.05.013 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2016b). Prediction of Scylla olivacea (Crustacea; Brachyura) peptide hormones using publicly accessible transcriptome shotgun assembly (TSA) sequences. Gen. Comp. Endocrinol. 230-231, 1-16. 10.1016/j.ygcen.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Christie A. E. (2017). Neuropeptide discovery in Proasellus cavaticus: prediction of the first large-scale peptidome for a member of the Isopoda using a publicly accessible transcriptome. Peptides 97, 29-45. 10.1016/j.peptides.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Christie A. E. and Chi M. (2015a). Identification of the first neuropeptides from the enigmatic hexapod order Protura. Gen. Comp. Endocrinol. 224, 18-37. 10.1016/j.ygcen.2015.05.015 [DOI] [PubMed] [Google Scholar]
- Christie A. E. and Chi M. (2015b). Neuropeptide discovery in the Araneae (Arthropoda, Chelicerata, Arachnida): elucidation of true spider peptidomes using that of the Western black widow as a reference. Gen. Comp. Endocrinol. 213, 90-109. 10.1016/j.ygcen.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Christie A. E. and Chi M. (2015c). Prediction of the neuropeptidomes of members of the Astacidea (Crustacea, Decapoda) using publicly accessible transcriptome shotgun assembly (TSA) sequence data. Gen. Comp. Endocrinol. 224, 38-60. 10.1016/j.ygcen.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Christie A. E. and Pascual M. G. (2016). Peptidergic signaling in the crab Cancer borealis: tapping the power of transcriptomics for neuropeptidome expansion. Gen. Comp. Endocrinol. 237, 53-67. 10.1016/j.ygcen.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Cashman C. R., Brennan H. R., Ma M., Sousa G. L., Li L., Stemmler E. A. and Dickinson P. S. (2008). Identification of putative crustacean neuropeptides using in silico analyses of publicly accessible expressed sequence tags. Gen. Comp. Endocrinol. 156, 246-264. 10.1016/j.ygcen.2008.01.018 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Durkin C. S., Hartline N., Ohno P. and Lenz P. H. (2010a). Bioinformatic analyses of the publicly accessible crustacean expressed sequence tags (ESTs) reveal numerous novel neuropeptide-encoding precursor proteins, including ones from members of several little studied taxa. Gen. Comp. Endocrinol. 167, 164-178. 10.1016/j.ygcen.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Stemmler E. A. and Dickinson P. S. (2010b). Crustacean neuropeptides. Cell. Mol. Life Sci. 67, 4135-4169. 10.1007/s00018-010-0482-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A. E., McCoole M. D., Harmon S. M., Baer K. N. and Lenz P. H. (2011). Genomic analyses of the Daphnia pulex peptidome. Gen. Comp. Endocrinol. 171, 131-150. 10.1016/j.ygcen.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Roncalli V., Wu L.-S., Ganote C. L., Doak T. and Lenz P. H. (2013). Peptidergic signaling in Calanus finmarchicus (Crustacea, Copepoda): in silico identification of putative peptide hormones and their receptors using a de novo assembled transcriptome. Gen. Comp. Endocrinol. 187, 117-135. 10.1016/j.ygcen.2013.03.018 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Chi M., Lameyer T. J., Pascual M. G., Shea D. N., Stanhope M. E., Schulz D. J. and Dickinson P. S. (2015). Neuropeptidergic signaling in the American lobster Homarus americanus: new insights from high-throughput nucleotide sequencing. PLoS ONE 10, e0145964 10.1371/journal.pone.0145964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A. E., Hull J. J., Richer J. A., Geib S. M. and Tassone E. E. (2017a). Prediction of a peptidome for the Western tarnished plant bug Lygus hesperus. Gen. Comp. Endocrinol. 243, 22-38. 10.1016/j.ygcen.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Roncalli V., Cieslak M. C., Pascual M. G., Yu A., Lameyer T. J., Stanhope M. E. and Dickinson P. S. (2017b). Prediction of a neuropeptidome for the eyestalk ganglia of the lobster Homarus americanus using a tissue-specific de novo assembled transcriptome. Gen. Comp. Endocrinol. 243, 96-119. 10.1016/j.ygcen.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A. E., Cieslak M. C., Roncalli V., Lenz P. H., Major K. M. and Poynton H. C. (2018a). Prediction of a peptidome for the ecotoxicological model Hyalella azteca (Crustacea; Amphipoda) using a de novo assembled transcriptome. Mar. Genomics 38, 67-88. 10.1016/j.margen.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Pascual M. G. and Yu A. (2018b). Peptidergic signaling in the tadpole shrimp Triops newberryi: a potential model for investigating the roles played by peptide paracrines/hormones in adaptation to environmental change. Mar. Genomics 39, 45-63. 10.1016/j.margen.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Yu A., Pascual M. G., Roncalli V., Cieslak M. C., Warner A. N., Lameyer T. J., Stanhope M. E., Dickinson P. S. and Hull J. J. (2018c). Circadian signaling in Homarus americanus: region-specific de novo assembled transcriptomes show that both the brain and eyestalk ganglia possess the molecular components of a putative clock system. Mar. Genomics 40, 25-44. 10.1016/j.margen.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Christie A. E., Yu A., Roncalli V., Pascual M. G., Cieslak M. C., Warner A. N., Lameyer T. J., Stanhope M. E., Dickinson P. S. and Hull J. J. (2018d). Molecular evidence for an intrinsic circadian pacemaker in the cardiac ganglion of the American lobster, Homarus americanus-Is diel cycling of heartbeat frequency controlled by a peripheral clock system? Mar. Genomics 41, 19-30. 10.1016/j.margen.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Clemens S., Combes D., Meyrand P. and Simmers J. (1998a). Long-term expression of two interacting motor pattern-generating networks in the stomatogastric system of freely behaving lobster. J. Neurophysiol. 79, 1396-1408. 10.1152/jn.1998.79.3.1396 [DOI] [PubMed] [Google Scholar]
- Clemens S., Massabuau J.-C., Legeay A., Meyrand P. and Simmers J. (1998b). In vivo modulation of interacting central pattern generators in lobster stomatogastric ganglion: influence of feeding and partial pressure of oxygen. J. Neurosci. 18, 2788-2799. 10.1523/JNEUROSCI.18-07-02788.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke I. M. (2002). Reliable, responsive pacemaking and pattern generation with minimal cell numbers: the crustacean cardiac ganglion. Biol. Bull. 202, 108-136. 10.2307/1543649 [DOI] [PubMed] [Google Scholar]
- Dickinson P. S., Calkins A. and Stevens J. S. (2015a). Related neuropeptides use different balances of unitary mechanisms to modulate the cardiac neuromuscular system in the American lobster, Homarus americanus. J. Neurophysiol. 113, 856-870. 10.1152/jn.00585.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P. S., Kurland S. C., Qu X., Parker B. O., Sreekrishnan A., Kwiatkowski M. A., Williams A. H., Ysasi A. B. and Christie A. E. (2015b). Distinct or shared actions of peptide family isoforms: II. Multiple pyrokinins exert similar effects in the lobster stomatogastric nervous system. J. Exp. Biol. 218, 2905-2917. 10.1242/jeb.124818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P. S., Sreekrishnan A., Kwiatkowski M. A. and Christie A. E. (2015c). Distinct or shared actions of peptide family isoforms: I. Peptide-specific actions of pyrokinins in the lobster cardiac neuromuscular system. J. Exp. Biol. 218, 2892-2904. 10.1242/jeb.124800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P. S., Qu X. and Stanhope M. E. (2016). Neuropeptide modulation of pattern-generating systems in crustaceans: comparative studies and approaches. Curr. Opin. Neurobiol. 41, 149-157. 10.1016/j.conb.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dircksen H., Neupert S., Predel R., Verleyen P., Huybrechts J., Strauss J., Hauser F., Stafflinger E., Schneider M., Pauwels K. et al. (2011). Genomics, transcriptomics, and peptidomics of Daphnia pulex neuropeptides and protein hormones. J. Proteome Res. 10, 4478-4504. 10.1021/pr200284e [DOI] [PubMed] [Google Scholar]
- Elia A. J., Tebrugge V. A. and Orchard I. (1993). The pulsatile appearance of FMRF amide-related peptides in the haemolymph and loss of FMFRamide-like immunoreactivity from neurohaemal areas of Rhodnius prolixus following a blood meal. J. Insect Physiol. 39, 459-469. 10.1016/0022-1910(93)90077-5 [DOI] [Google Scholar]
- Fastner S., Predel R., Kahnt J., Schachtner J. and Wegener C. (2007). A simple purification protocol for the detection of peptide hormones in the hemolymph of individual insects by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 21, 23-28. 10.1002/rcm.2800 [DOI] [PubMed] [Google Scholar]
- Fort T. J., Brezina V. and Miller M. W. (2007a). Regulation of the crab heartbeat by FMRFamide-like peptides: multiple interacting effects on center and periphery. J. Neurophysiol. 98, 2887-2902. 10.1152/jn.00558.2007 [DOI] [PubMed] [Google Scholar]
- Fort T. J., Garcia-Crescioni K., Agricola H. J., Brezina V. and Miller M. W. (2007b). Regulation of the crab heartbeat by crustacean cardioactive peptide (CCAP): central and peripheral actions. J. Neurophysiol. 97, 3407-3420. 10.1152/jn.00939.2006 [DOI] [PubMed] [Google Scholar]
- Gard A. L., Lenz P. H., Shaw J. R. and Christie A. E. (2009). Identification of putative peptide paracrines/hormones in the water flea Daphnia pulex (Crustacea; Branchiopoda; Cladocera) using transcriptomics and immunohistochemistry. Gen. Comp. Endocrinol. 160, 271-287. 10.1016/j.ygcen.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Marder E., Selverston A. I. and Moulins M. (1992). Dynamic Biological Networks: the Stomatogastric Nervous System. MIT Press. [Google Scholar]
- Kang B. J., Okutsu T., Tsutsui N., Shinji J., Bae S. H. and Wilder M. N. (2014). Dynamics of vitellogenin and vitellogenesis-inhibiting hormone levels in adult and subadult whiteleg shrimp, Litopenaeus vannamei: relation to molting and eyestalk ablation. Biol. Reprod. 90, 12 10.1095/biolreprod.113.112243 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski M. A., Gabranski E. R., Huber K. E., Chapline M. C., Christie A. E. and Dickinson P. S. (2013). Coordination of distinct but interacting rhythmic motor programs by a modulatory projection neuron using different co-transmitters in different ganglia. J. Exp. Biol. 216, 1827-1836. 10.1242/jeb.082503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Bors E. K., Dickinson E. S., Kwiatkowski M. A., Sousa G. L., Henry R. P., Smith C. M., Towle D. W., Christie A. E. and Li L. (2009). Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen. Comp. Endocrinol. 161, 320-334. 10.1016/j.ygcen.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Gard A. L., Xiang F., Wang J., Davoodian N., Lenz P. H., Malecha S. R., Christie A. E. and Li L. (2010). Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei. Peptides 31, 27-43. 10.1016/j.peptides.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi A., Dickinson P. S., Kloppenburg P., Fénelon V., Baro D. J., Harris-Warrick R. M., Meyrand P. and Simmers J. (2001). Long-term maintenance of channel distribution in a central pattern generator neuron by neuromodulatory inputs revealed by decentralization in organ culture. J. Neurosci. 21, 7331-7339. 10.1523/JNEUROSCI.21-18-07331.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F. and Dickinson P. S. (1983). Control of a central pattern generator by an identified modulatory interneurone in crustacea. I. Modulation of the pyloric motor output. J. Exp. Biol. 105, 33-58. [DOI] [PubMed] [Google Scholar]
- Nguyen T. V., Cummins S. F., Elizur A. and Ventura T. (2016). Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci. Rep. 6, 38658 10.1038/srep38658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt A. J., Lett K. M., Garcia V. B., Diester C. M., Lane B. J., Marder E. and Schulz D. J. (2016). Deep sequencing of transcriptomes from the nervous systems of two decapod crustaceans to characterize genes important for neural circuit function and modulation. BMC Genomics 17, 868 10.1186/s12864-016-3215-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum M. P. and Blitz D. M. (2012). Neuropeptide modulation of microcircuits. Curr. Opin. Neurobiol. 22, 592-601. 10.1016/j.conb.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum M. P. and Marder E. (1989). A modulatory proctolin-containing neuron (MPN). II. State-dependent modulation of rhythmic motor activity. J. Neurosci. 9, 1600-1607. 10.1523/JNEUROSCI.09-05-01600.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum M. P., Blitz D. M. and Marder E. (2017). Functional consequences of neuropeptide and small-molecule co-transmission. Nat. Rev. Neurosci. 18, 389-403. 10.1038/nrn.2017.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm K. J., Deeg K. E. and Marder E. (2008). Developmental regulation of neuromodulator function in the stomatogastric ganglion of the lobster, Homarus americanus. J. Neurosci. 28, 9828-9839. 10.1523/JNEUROSCI.2328-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezer E. and Moulins M. (1983). Expression of the crustacean pyloric pattern generator in the intact animal. J. Comp. Physiol. 153, 17-28. 10.1007/BF00610338 [DOI] [Google Scholar]
- Robb S. and Evans P. D. (1990). FMRFamide-like peptides in the locust: distribution, partial characterization and bioactivity. J. Exp. Biol. 149, 335-360. [DOI] [PubMed] [Google Scholar]
- Rodriguez J. C., Blitz D. M. and Nusbaum M. P. (2013). Convergent rhythm generation from divergent cellular mechanisms. J. Neurosci. 33, 18047-18064. 10.1007/s10827-015-0587-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saideman S. R., Blitz D. M. and Nusbaum M. P. (2007a). Convergent motor patterns from divergent circuits. J. Neurosci. 27, 6664-6674. 10.1523/JNEUROSCI.0315-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saideman S. R., Ma M. M., Kutz-Naber K. K., Cook A., Torfs P., Schoofs L., Li L. J and Nusbaum M. P. (2007b). Modulation of rhythmic motor activity by pyrokinin peptides. J. Neurophysiol. 97, 579-595. 10.1152/jn.00772.2006 [DOI] [PubMed] [Google Scholar]
- Sakurai A. and Katz P. S. (2003). Spike timing-dependent serotonergic neuromodulation of synaptic strength intrinsic to a central pattern generator circuit. J. Neurosci. 23, 10745-10755. 10.1523/JNEUROSCI.23-34-10745.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selverston A. I. and Moulins M. (1987). The Crustacean Stomatogastric System: a Model for the Study of Central Nervous Systems. Springer. [Google Scholar]
- Shruti S., Schulz D. J., Lett K. M. and Marder E. (2014). Electrical coupling and innexin expression in the stomatogastric ganglion of the crab Cancer borealis. J. Neurophysiol. 112, 2946-2958. 10.1152/jn.00536.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein W. (2009). Modulation of stomatogastric rhythms. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 195, 989-1009. 10.1007/s00359-009-0483-y [DOI] [PubMed] [Google Scholar]
- Stemmler E. A., Barton E. E., Esonu O. K., Polasky D. A., Onderko L. L., Bergeron A. B., Christie A. E. and Dickinson P. S. (2013). C-terminal methylation of truncated neuropeptides: An enzyme-assisted extraction artifact involving methanol. Peptides 46, 108-125. 10.1016/j.peptides.2013.05.008 [DOI] [PubMed] [Google Scholar]
- Stevens J. S., Cashman C. R., Smith C. M., Beale K. M., Towle D. W., Christie A. E. and Dickinson P. S. (2009). The peptide hormone pQDLDHVFLRFamide (crustacean myosuppressin) modulates the Homarus americanus cardiac neuromuscular system at multiple sites. J. Exp. Biol. 212, 3961-3976. 10.1242/jeb.035741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwansa-Ard S., Thongbuakaew T., Wang T., Zhao M., Elizur A., Hanna P. J., Sretarugsa P., Cummins S. F. and Sobhon P. (2015). In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of eyestalk, central nervous system and ovary. PLoS ONE 10, e0123848 10.1371/journal.pone.0123848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert P. H. and Nitabach M. N. (2012). Peptide neuromodulation in invertebrate model systems. Neuron 76, 82-97. 10.1016/j.neuron.2012.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toullec J.-Y., Corre E., Bernay B., Thorne M. A. S., Cascella K., Ollivaux C., Henry J. and Clark M. S. (2013). Transcriptome and peptidome characterisation of the main neuropeptides and peptidic hormones of a euphausiid: the ice krill, Euphausia crystallorophias. PLoS ONE 8, e71609 10.1371/journal.pone.0071609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toullec J.-Y., Corre E., Mandon P., Gonzalez-Aravena M., Ollivaux C. and Lee C. Y. (2017). Characterization of the neuropeptidome of a Southern Ocean decapod, the Antarctic shrimp Chorismus antarcticus: focusing on a new decapod ITP-like peptide belonging to the CHH peptide family. Gen. Comp. Endocrinol. 252, 60-78. 10.1016/j.ygcen.2017.07.015 [DOI] [PubMed] [Google Scholar]
- Veenstra J. A. (2015). The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen. Comp. Endocrinol. 224, 84-95. 10.1016/j.ygcen.2015.06.013 [DOI] [PubMed] [Google Scholar]
- Veenstra J. A., Rombauts S. and Grbić M. (2012). In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem. Mol. Biol. 42, 277-295. 10.1016/j.ibmb.2011.12.009 [DOI] [PubMed] [Google Scholar]
- Ventura T., Cummins S. F., Fitzgibbon Q., Battaglene S. and Elizur A. (2014). Analysis of the central nervous system transcriptome of the eastern rock lobster Sagmariasus verreauxi reveals its putative neuropeptidome. PLoS ONE 9, e97323 10.1371/journal.pone.0097323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimann J. M., Meyrand P. and Marder E. (1991). Neurons that form multiple pattern generators: Identification and multiple activity patterns of gastric pyloric neurons in the crab stomatogastric system. J. Neurophysiol. 65, 111-122. 10.1152/jn.1991.65.1.111 [DOI] [PubMed] [Google Scholar]
- Wilkens J. L., Shinozaki T., Yazawa T. and ter Keurs H. (2005). Sites and modes of action of proctolin and the FLP F-2 on lobster cardiac muscle. J. Exp. Biol. 208, 737-747. 10.1242/jeb.01430 [DOI] [PubMed] [Google Scholar]
- Williams A. H., Calkins A., O'Leary T., Symonds R., Marder E. and Dickinson P. S. (2013). The neuromuscular transform of the lobster cardiac system explains the opposing effects of a neuromodulator on muscle output. J. Neurosci. 33, 16565-16575. 10.1523/JNEUROSCI.2903-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X.-C., Chen Z.-F., Sun J., Matsumura K., Wu R. S. and Qian P.-Y. (2012). Transcriptomic analysis of neuropeptides and peptide hormones in the barnacle Balanus amphitrite: evidence of roles in larval settlement. PLoS ONE 7, e46513 10.1371/journal.pone.0046513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarger A. M. and Stein W. (2015). Sources and range of long-term variability of rhythmic motor patterns in vivo. J. Exp. Biol. 218, 3950-3961. 10.1242/jeb.126581 [DOI] [PubMed] [Google Scholar]
- Z̆itn̆an D., Ross L. S., Z̆itn̆an I., Hermesman J. L., Gill S. S. and Adams M. E. (1999). Steroid induction of a peptide hormone gene leads to orchestration of a defined behavioral sequence. Neuron 23, 523-535. 10.1016/S0896-6273(00)80805-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.