Abstract
Mouse double minute (Mdm) genes span an evolutionary timeframe from the ancient eukaryotic placozoa Trichoplax adhaerens to Homo sapiens, implying a significant and possibly conserved cellular role throughout history. Maintenance of DNA integrity and response to DNA damage involve many key regulatory pathways, including precise control over the tumour suppressor protein p53. In most vertebrates, degradation of p53 through proteasomal targeting is primarily mediated by heterodimers of Mdm2 and the Mdm2-related protein Mdm4 (also known as MdmX). Both Mdm2 and Mdm4 have p53-binding regions, acidic domains, zinc fingers, and C-terminal RING domains that are conserved throughout evolution. Vertebrates typically have both Mdm2 and Mdm4 genes, while analyses of sequenced genomes of invertebrate species have identified single Mdm genes, suggesting that a duplication event occurred prior to emergence of jawless vertebrates about 550–440 million years ago. The functional relationship between Mdm and p53 in T. adhaerens, an organism that has existed for 1 billion years, implies that these two proteins have evolved together to maintain a conserved and regulated function.
Keywords: Mdm2, Mdm4, evolution, p53
The discovery of double minute genes
The Mdm2 gene, along with Mdm1, was first identified in a screen for amplified genes in transformed BALB/c mouse 3T3DM cells to isolate factors associated with the appearance of double minutes—small, acentromeric, extrachromosomal nuclear bodies (Cahilly-Snyder et al., 1987), hence the name ‘mouse double minute’. Mdm3, co-amplified in 3T3DM cells, was subsequently identified using chromosome walking from Mdm2, but because of its lack of tumourigenicity, was not further investigated (Fakharzadeh et al., 1991). Mdm1 was eventually identified to be a microtubule-binding protein important in regulating centriole duplication (Snyder et al., 1988; Van de Mark et al., 2015), while Mdm2 became prominent with the discoveries that Mdm2 associates with, inhibits, and mediates the degradation of the tumour suppressor p53 (Momand et al., 1992; Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997), which by then was already recognized to play significant roles in cancer. The paralogue Mdm4 (also called MdmX) was identified in a cDNA screen for p53 binding partners and was named for its homology with Mdm2 (Shvarts et al., 1996). Subsequent biochemical and genetic data defined the roles of Mdm2 and Mdm4 in the inhibition of the tumour suppressive functions of p53, as well as p53-independent mechanisms in regulating genomic stability. Based on the similarities between their major functions, sequences, and structures, Mdm2 and Mdm4 became known as the Mdm2 family. Despite sharing similar names, Mdm1 and Mdm3 were excluded from the family due to vast differences in sequence and function.
Following the discovery in mice, human orthologues to Mdm2 and Mdm4 were isolated and have been distinguished throughout the literature by ‘HDM2’ or ‘HDM4’—human double minute 2 or 4. As this review attempts to explore the evolution of Mdm2 and Mdm4, we shall use the generic ‘Mdm2’ or ‘Mdm4’ and differentiate among the species with adjectives, like ‘murine Mdm2’ and ‘human Mdm2’.
Functions of Mdm2 and Mdm4
As a proto-oncogene, human Mdm2 is found amplified in 7% of cancers, with higher incidences in soft tissue tumours, osteosarcomas, and oesophageal carcinomas (Momand et al., 1998). Human Mdm2 transcript and protein levels are also found elevated in other malignancies even without gene amplification, in particular testicular and intracranial germ cell tumours, glioblastomas and astrocytomas, various forms of leukaemias, and breast and colorectal carcinomas (Rayburn et al., 2005).
The most well-studied role of Mdm2 is to bind, inhibit, and ubiquitinate the guardian of the genome p53, mediating its degradation by the 26S proteasome (Momand et al., 1992; Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997; Hock and Vousden, 2014). In response to cellular stress, such as oxidative damage (Liu and Xu, 2011), radiation damage (Fei and El-Deiry, 2003), or hypoxia (Sermeus and Michiels, 2011), p53 is activated by the respective damage sensors, becomes transcriptionally active, and accumulates. Mdm2, being a transcriptional target of p53 (Barak et al., 1993), is upregulated and regulates p53 levels through its E3 ligase activity in ubiquitinating p53, mediating its degradation. Mdm2 also auto-ubiquitinates, mediating its own proteasomal degradation (Ranaweera and Yang, 2013). In non-stressed cells, this feedback loop maintains a low level of Mdm2 and p53: the half-lives of the two proteins in unstressed cells are <20 min (Stommel and Wahl, 2004). The presence of stress signals the post-translational modification of either protein, inhibiting the interaction between p53 and Mdm2, thus preventing the degradation of p53. This allows the accumulation of p53 to facilitate its downstream functions, including cell cycle arrest or apoptosis (Zilfou and Lowe, 2009).
Although Mdm2 primarily functions as an E3 ligase mediating the transfer of ubiquitin to p53 or another Mdm2, its paralogue Mdm4 functions differently in the negative regulation of p53, despite being highly homologous. Like Mdm2, Mdm4 can bind to and negatively regulate p53 (Shvarts et al., 1996). However, Mdm4 is unable to function intrinsically as an E3 ubiquitin ligase, yet is important for the effective degradation of p53—Mdm4 stabilises Mdm2, and the ubiquitination of p53 is stimulated and enhanced by the formation of the Mdm2/Mdm4 heterodimer (Gu et al., 2002; Linares et al., 2003; Linke et al., 2008; Okamoto et al., 2009). Unlike Mdm2, Mdm4 is not a canonical p53 target but is ubiquitously expressed in the cytoplasm. In addition, Mdm4 lacks nuclear localization or export sequences, and is dependent on Mdm2 for nuclear translocation for p53 suppression (Migliorini et al., 2002). Mdm4 is also ubiquitinated by Mdm2, and this degradation of Mdm4 allows for p53 activity in response to cytotoxic stress (Pereg et al., 2005; Jin et al., 2006).
Human Mdm4 overexpression is implicated in a number of tumours, such as leukaemias, retinoblastomas, testicular cancers, and melanomas (Laurie et al., 2006; Han et al., 2007; Koster et al., 2011; Gembarska et al., 2012). Given the high prevalence of Mdm2 and/or Mdm4 overexpression in a large proportion of cancers with wild-type p53, targeting Mdm2 and Mdm4 represents a major strategy in cancer therapeutics, emphasizing a greater need to deepen our understanding of the p53–Mdm2–Mdm4 pathways.
The evolution of the domains of Mdm2 and Mdm4
Vertebrates carry both paralogues Mdm2 and Mdm4, but all invertebrates characterized thus far only harbour a single Mdm. The presence of the two paralogues in vertebrates has been postulated to result from a duplication event more than 440 million years ago in the Palaeozoic era, during which the common ancestor of humans and zebrafish arose (Momand et al., 2011), or possibly even earlier if the duplication event happened around the period of vertebrate–invertebrate divergence ~550 million years ago (Smith and Keinath, 2015). The existence of two Mdm genes in both jawless and jawed vertebrates indicates that a duplication event possibly occurred in an ancestor prior to the invertebrate/vertebrate split or occurred separately in these two lines. Lamprey ‘Mdm4’ was labelled as such to differentiate it from lamprey Mdm2, but it is truncated compared to Mdm4 present in jawed vertebrates and thus could represent an intermediary protein (Mehta et al., 2013; Coffill et al., 2016).
In this review, we studied the homology of the Mdm2 family of 21 metazoans, comprising 16 vertebrate species and 5 invertebrate species (Supplementary Figure S1). The overall homology of Mdm2 and Mdm4 among mammals is high, with at least 80% identity compared to the human orthologues (Tables 1 and 2). Homology is reduced as the species further diverge from humans, with avian Mdm2 and Mdm4 sharing 55%−68% identity, amphibian Mdm2 and Mdm4 sharing 48% and 56%, and jawed fish Mdm2 and Mdm4 sharing 40%−48% with human Mdm2 and Mdm4. In the lamprey, one of the most distant vertebrates from humans, Mdm2 and Mdm4 share identities of 23% and 13%, respectively (Coffill et al., 2016). Among invertebrates, their single Mdm protein shares 18%−27% identity with human Mdm2 or Mdm4, including Mdm found in the ancient placozoa, arguably the simplest of all known animals and the most evolutionary distant species of invertebrates. Overall, neither Mdm2 nor Mdm4 within the same species is more divergent than the other—the evolutionary rates for both proteins are very similar. Despite the low overall homologies, most functions of Mdm2 and Mdm4 appear to be conserved, as evident in the comparison of homology among the different domains of the Mdm2 family (Tables 1 and 2) and through experimental analyses of proteins produced from ancient organisms (Siau et al., 2016).
Table 1.
Percentage identity of the domains and interdomain regions of Mdm2 in various species compared to Homo sapiens Mdm2.
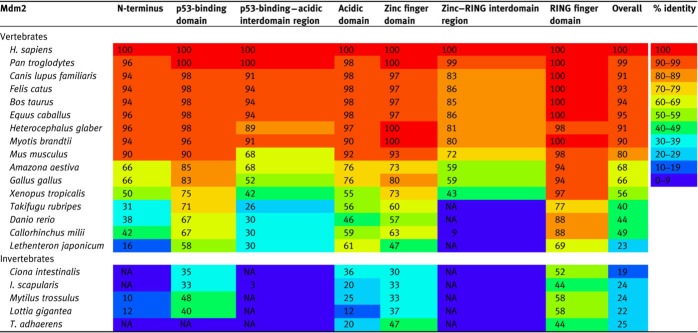 |
For invertebrate species, only one Mdm protein is present, and the numbers indicate the percentage identity between the single Mdm protein compared to H. sapiens Mdm2. ‘NA’ indicates that no significant alignment could be obtained. The species in the table are arranged in increasing taxonomic distance from H. sapiens (updated from Momand et al., 2011). The heat map represents the level of conservation across species for each region of Mdm2.
Table 2.
Percentage identity of the domains and interdomain regions of Mdm4 in various species compared to H. sapiens Mdm4.
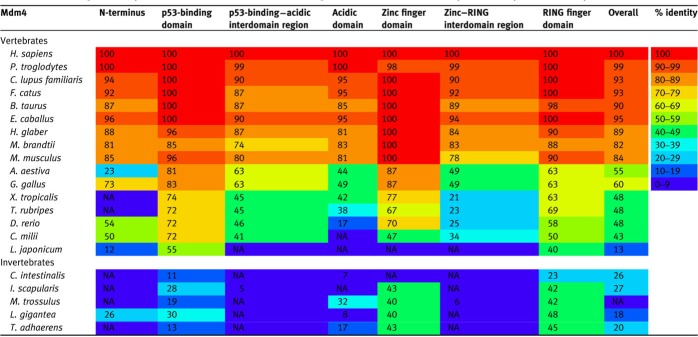 |
For invertebrate species, only one Mdm protein is present, and the numbers indicate the percentage identity between the single Mdm protein compared to H. sapiens Mdm4. ‘NA’ indicates that no significant alignment could be obtained. The species in the table are arranged in increasing taxonomic distance from H. sapiens (updated from Momand et al., 2011). The heat map represents the level of conservation across species for each region of Mdm4.
Two of the most important genetic models used in animal biology, Drosophila melanogaster and Caenorhabditis elegans, have lost the Mdm gene, and have evolved other strategies in regulating their respective p53 proteins. This will be discussed in a later section.
The Mdm2 family of genes is characterized by four main domains: the N-terminal p53-binding domain, the central acidic and zinc finger domains, and the C-terminal RING finger domain, summarized in Supplementary Figure S2. Among the species used in this review, it is clear that the defined domains of the Mdm2 family are highly conserved, whereas the interdomain regions exhibit more variability (Tables 1 and 2). Interestingly, many post-translational modification sites exist within these interdomain regions, and it is possible that the decreased homology reflects the complexity in the regulation of Mdm2 and Mdm4 across species.
The RING finger domain
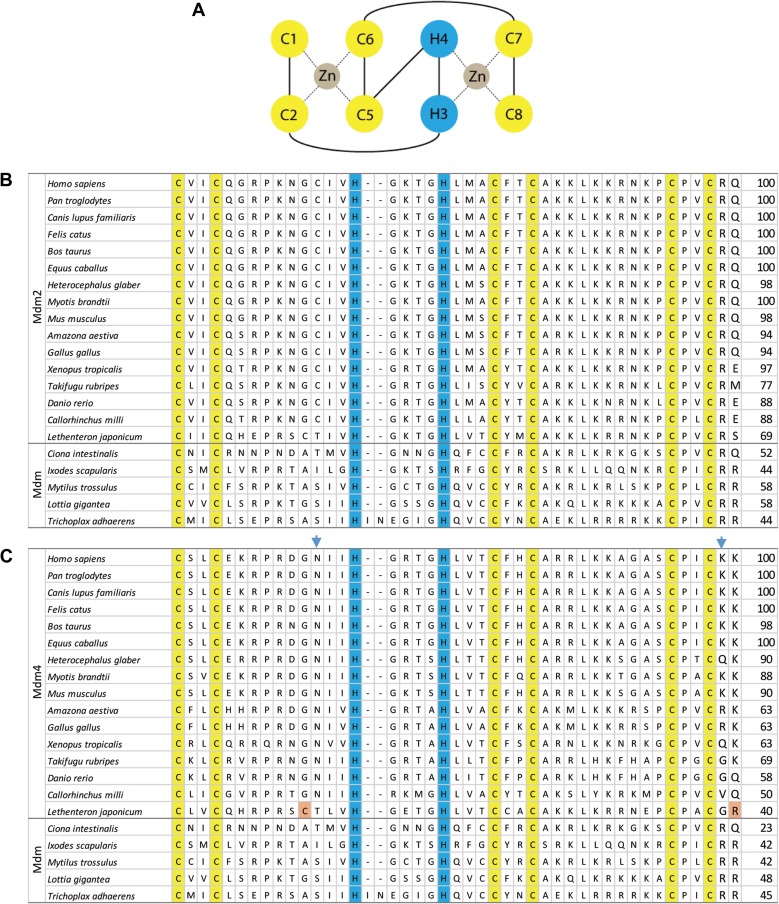
The RING finger domain consists of an octet of cysteines and histidines that form a cross-brace to facilitate zinc coordination. Unlike the typical C3HC4 or C4C4 zinc coordination, the cross-brace motifs in Mdm2 and Mdm4 have a unique C2H2C4 structure, as depicted in Figure 1A (Kostic et al., 2006; Linke et al., 2008). This atypical RING structure is conserved throughout metazoans with known Mdm proteins (Figure 1B and C). In vertebrates, the Mdm2 RING finger domain is responsible for the ubiquitination of p53 via the recruitment of ubiquitin-loaded E2 (UbcH5B/C and others) for the mediation of ubiquitin transfer to p53 (Saville et al., 2004; Hock and Vousden, 2014), and this function of Mdm2 is well conserved in human, mouse, zebrafish (Momand et al., 2011), and lamprey (Coffill et al., 2016).
Figure 1.
The RING finger domains of Mdm2 and Mdm4. (A) The Mdm2/Mdm4 RING finger motif consists of an octet of two cysteines (C1 and C2), two histidines (H3 and H4) and four cysteines (C5–C8). Two zinc ions are bound in coordinate bonds (dotted lines) by half of the motif (C1+C2+C5+C6 and H3+H4+C7+C8). (B) This C2H2C4 cross-brace motif is conserved in vertebrate Mdm2 and invertebrate Mdm. The cysteine and histidine residues involved are numbered and highlighted in yellow and blue, respectively. (C) The blue arrowheads indicate the asparagines (N448 in humans) that can be substituted to cysteines, as well as lysines (K478 in humans) to arginines, to make the Mdm4 functional E3 ligases. The residues highlighted in orange present in L. japonicum Mdm4 potentially make them functional E3 ligases. The numbers on the right of the table indicate the percentage identity compared to H. sapiens Mdm2 or Mdm4.
The RING finger domain of Mdm2 retains a high level of homology among tetrapods, maintaining at least 94% identity compared to human Mdm2 (Figure 1B). The homology between human and jawed fish Mdm2 is moderately high as well, ranging from 77% to 88%. Even the lamprey Mdm2 exhibits a 69% identity with human Mdm2, suggesting a high level of conservation of E3 ligase function among vertebrates. Most strikingly, invertebrate Mdm shows moderate homology with human Mdm2, and biochemical assays have demonstrated that even placozoa Mdm retains its inhibitory function on placozoa p53 (Siau et al., 2016), despite a billion years of evolutionary distance between placozoa and humans (Lane and Verma, 2012). Placozoa p53 is degraded in an Mdm-dependent manner, and the degradation is abrogated by a mutation in the RING domain (Siau et al., 2016). Furthermore, in those assays, placozoa Mdm RING domain has sufficient homology to recruit mouse ubiquitin-conjugating E2 proteins (Saville et al., 2004; Hock and Vousden, 2014) to ubiquitinate placozoa p53 and lead to its degradation in a proteasome-dependent manner. This level of conservation of the RING finger domain highlights its importance in the negative regulation of p53 by ubiquitination and proteasomal degradation throughout the evolution of metazoans.
The RING finger domain of Mdm4 in most vertebrates, however, is unable to function as an E3 ligase, although heterodimerization with Mdm2 enhances the polyubiquitination of p53, Mdm2, or Mdm4 (Linares et al., 2003; Linke et al., 2008; Okamoto et al., 2009). Mutational studies have demonstrated that converting asparagine 448 of Mdm4 (human numbering) to cysteine can restore its E3 ubiquitin ligase activity in vitro, although further modifications of other domains are required to restore full E3 ligase activity in cellular assays (Iyappan et al., 2010). Interestingly, the lamprey Mdm4 has two of these residues (C237 and R269, Figure 1C), and indeed lamprey Mdm4 destabilized lamprey p53 in p53−/−Mdm2−/− mouse embryonic fibroblasts (Coffill et al., 2016). Conversely, the analogous cysteine residue in invertebrate Mdm is absent, yet placozoa Mdm still retains its E3 ligase function (Siau et al., 2016).
The p53-binding domain
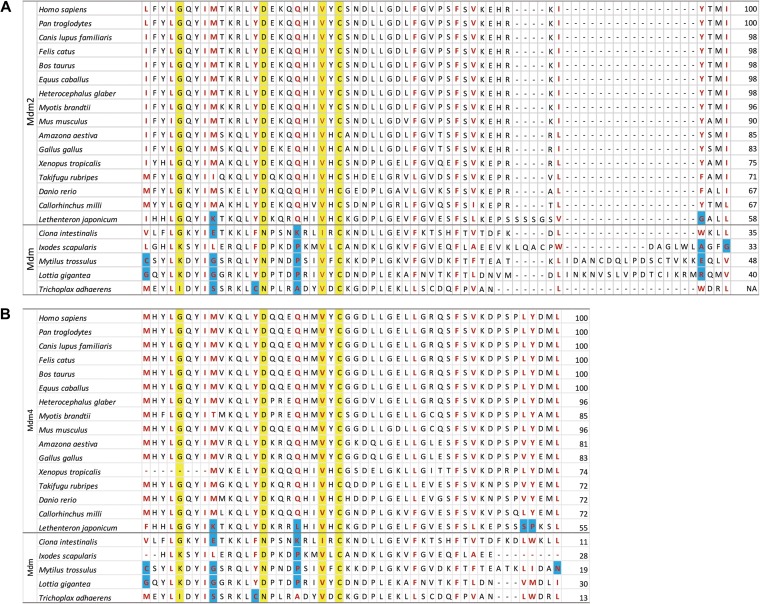
Following the RING finger domain, the p53-binding domains of Mdm2 and Mdm4 are the next most conserved among metazoans. This region interfaces with the transactivation domain of p53 (residues 15−29, human numbering), in particular F19, W23, and L26, which are the critical residues for Mdm2 interaction and have been exploited to develop antagonists against Mdm2 and/or Mdm4 (Kussie et al., 1996; Chène, 2004; Pazgier et al., 2009; Brown et al., 2013; Chang et al., 2013; Tan et al., 2015). This region in p53 is highly conserved, thus it is unsurprising that the corresponding regions in Mdm2 and Mdm4 are similarly highly conserved throughout evolution (Coffill et al., 2016; Siau et al., 2016). Thirteen residues on the corresponding interaction surfaces of Mdm2 and Mdm4 have also been identified to form the hydrophobic cleft in which p53 binds (Chène, 2004; Toledo and Wahl, 2007), and a number of these residues are highly conserved in evolution (Figure 2A and B).
Figure 2.
Alignments of the p53-binding domains of Mdm2 and Mdm4. (A) The residues in red form the hydrophobic cleft in which an α helix from the p53 transactivation domain binds to Mdm2, while the residues highlighted in yellow represent the genetically determined amino acids that are crucial in p53 interaction. The blue highlight indicates amino acids that differ in physical properties from the consensus, for instance a charged lysine in place of a hydrophobic methionine. (B) The homologous residues on Mdm4 are similarly highlighted. The numbers on the right of the table indicate the percentage identity compared to H. sapiens Mdm2 or Mdm4.
Among vertebrate Mdm2, the 13 amino acids that form the hydrophobic cleft are largely conserved, and substitution of these residues throughout vertebrate evolution involves mostly hydrophobic amino acids. The exception is lamprey Mdm2, where a charged lysine at codon 53 and a small glycine at codon 95 are in place of the consensus methionine and tyrosine, respectively (Figure 2A, blue highlights). In vertebrate Mdm4, the homologous 13 residues are also conserved in terms of their physical properties, except lamprey Mdm4, where lysine 57 takes the place of the consensus methionine, leucine 67 replacing a polar glutamine, polar serine 94 replacing hydrophobic leucine, and the substitution of proline 95 for the consensus tyrosine (Figure 2B, blue highlights). Remarkably, the interaction between p53 and Mdm2 is sufficiently conserved that human p53 has been shown to interact with lamprey Mdm2. Furthermore, the interactions between lamprey p53 and lamprey Mdm2 and between human p53 and lamprey Mdm2 can be disrupted by Staplin (PM2), a stapled peptide antagonist based on the human p53 transactivation domain (Brown et al., 2013; Tan et al., 2015; Coffill et al., 2016).
For invertebrate Mdm, 2–4 of the 13 hydrophobic residues, compared to either vertebrate Mdm2 or Mdm4, have been replaced with polar, charged, or small amino acids. Nonetheless, placozoa Mdm can interact with placozoa p53 and human p53, albeit weakly, while human Mdm2 binds strongly with placozoa p53 (Siau et al., 2016). In those assays, Staplin-2 (MO11), a modified version of Staplin with a chlorinated tryptophan (Brown et al., 2013; Tan et al., 2015), can disrupt the interactions of placozoa p53 with either placozoa Mdm or human Mdm2, while Staplin can only disrupt the interaction between placozoa p53 and human Mdm2 but not between placozoa p53 and placozoa Mdm. These experiments demonstrate a high level of conservation within the p53-binding domains among vertebrate and invertebrate Mdm2 family members, even as p53, Mdm2, and Mdm4 co-evolved.
The acidic and zinc finger domains
The central region of Mdm2 and Mdm4 contains the acidic domain and the zinc finger domain. As the name suggests, the acidic domain is a partially unstructured region rich in acidic residues, resulting in isoelectric points (pI) ranging from 2.8 to 3.6 for this section of the protein. Adjacent to the acidic domain is a C4 zinc finger domain. Unlike the p53-binding and RING finger domains, the acidic and zinc finger domains are not as highly conserved (Tables 1 and 2). Together, the acidic and zinc finger domains form binding sites for a number of regulatory proteins, including p300 (Grossman et al., 1998; Sabbatini and McCormick, 2002), YY1 (Grönroos et al., 2004; Sui et al., 2004), ARF (Kamijo et al., 1998; Jackson et al., 2001), and PML (Bernardi et al., 2004). Deletions within the acidic domain of Mdm2 result in the deregulation of p53 degradation (Dolezelova et al., 2012; Leslie et al., 2015). Unfortunately, many of these studies involve human and murine Mdm2 and Mdm4, thus the lack of comparative functional data across multiple species make it difficult to discern the evolution of the acidic and zinc finger domains.
The acidic and zinc finger domains are implicated in interactions with ribosomal proteins (RPs), a highly conserved function of Mdm2 and Mdm4 that has been investigated across species (Zheng et al., 2015). Over the past decade, numerous higher eukaryote RPs have been implicated in the regulation of tumourigenesis, immune signalling, development, and disease (Zhou et al., 2015). At least 17 mammalian RPs have been implicated in tumour suppression through activation of p53. The majority of these have been shown to directly bind to and inhibit Mdm2 function, with a subset also binding to Mdm4 (Zhou et al., 2015; He et al., 2016). Numerous lines of evidence including mouse models highlight the key roles of RPL11/uL11 and RPL5/uL5 as transducers coupling impaired ribosome biogenesis/nucleolar stress to p53 activation (Macias et al., 2010; Bursać et al., 2012; Liu et al., 2016; Nicolas et al., 2016). Together with 5S rRNA, these form a ribosomal sub-complex (5S RNP) capable of sequestering Mdm2 (Horn and Vousden, 2008; Donati et al., 2013; Sloan et al., 2013). The complex ribosome structure and its primary protein translation function are conserved throughout evolution (Fox, 2010; Ramakrishnan, 2014). It is, therefore, highly likely that extra-ribosomal functionality can be attributed to RPs from divergent species. Human RPL11 has been shown to interact with lamprey Mdm2 both in vitro and in a heterologous cell line (Coffill et al., 2016). Similar results were obtained for the placozoan Mdm (Siau et al., 2016). Given the high sequence homology between human RPL11 and lamprey/placozoan RPL11 (>94%; Mehta et al., 2013), it is highly likely that this interaction prevails in these organisms. Furthermore, amino acids in the Mdm2 acid and zinc finger domains that form critical contacts with RPL11 (Zheng et al., 2015) are highly conserved in placozoan Mdm and lamprey Mdm2. Orthologues of the RPL11 inhibitor oncoprotein PICT1/GLTSCR2 (Sasaki et al., 2011) are also present in these extant organisms, again suggesting evolutionary conservation of the RPL11–Mdm2 interaction.
Whilst RPL11 interacts with Mdm2 and placozoan Mdm, it does not bind to Mdm4 (Gilkes et al., 2006). These functional similarities between Mdm2 and placozoan Mdm (which also displays p53-specific E3 ubiquitin ligase activity) suggest that the ancestral Mdm gene that underwent duplication 550–440 million years ago (Momand et al., 2011; Smith and Keinath, 2015; Coffill et al., 2016) was more Mdm2-like, and the duplicated Mdm4 evolved to lose its E3 ligase function. This may have occurred to prevent over-inhibition of p53 and enable stricter regulation co-dependent on Mdm2 and Mdm4. Functional characterization of singleton Mdm genes identified in other invertebrates is necessary to confirm this.
Human RPL5 did not interact with either placozoan Mdm or lamprey Mdm2, although this may be due to its lower sequence homology with the RPL5 proteins found in these organisms (60% and 80%, respectively). Further experiments with the endogenous proteins will shed light on the roles of RPL5 and ribosomal sub-complexes regulating Mdm2 function.
Mdm4 auto-inhibitory sequence
It was reported that the N-terminal domain of Mdm4 alone has a binding affinity to p53 transactivation domain ~100 times stronger than full-length Mdm4, and this has been attributed to the presence of an auto-inhibitory sequence element within full-length Mdm4 (Bista et al., 2013). This so-called WWW element is within amino acids 190–210 in human Mdm4, centred around tryptophans 200 and 201, and is surprisingly highly conserved (Supplementary Figure S3) among jawed vertebrates. No corresponding region is present in Mdm2, nor is it found in lamprey Mdm4, suggesting that this is a divergence in Mdm2 and Mdm4 function following duplication of the Mdm gene. Investigations into lamprey Mdm4 would yield greater insights into the roles of this region in the dampening of Mdm4-mediated inhibition of p53.
The role of Mdm2 and Mdm4 through evolution—insights from in vivo models
Research involving genetic models has provided significant insights into the evolution of Mdm2 and Mdm4 function, and more recent experiments with knock-in models have allowed more precise dissection of the roles Mdm2 and Mdm4 play in the regulation of p53 pathway and the impact of tissue specificity on tumour spectrum. The range and depth of experiments with knock-in and knock-out mouse models are too numerous and extensive to be compiled within this review, and only a selection is described to illustrate the similarities and differences between the different model organisms studied.
The related p53 family members p63 and p73 play important roles in development (Moll and Slade, 2004) and can interact with Mdm2 and/or Mdm4 (Wang et al., 2001; Zdzalik et al., 2010; Tashakori et al., 2016). p63 and p73 degradation by Mdm2 are respectively co-mediated by the Fbw7 and Itch E3 ubiquitin ligases (Galli et al., 2010; Wu and Leng, 2015). Putative orthologues of these can be found in the Trichoplax genome, suggesting a distant evolutionary conservation of function that requires experimental validation. Furthermore, Mdm2 and Mdm4 have been reported to have p53-independent roles (Melo and Eischen, 2012; Li and Lozano, 2013; Carrillo et al., 2015). Here, we shall focus on the regulation of p53 by the Mdm2 family.
Insights from mouse models
The most incontrovertible finding obtained from the mouse models is that Mdm2 and Mdm4 are the key regulators of p53. Mdm2 knock-out in mice is embryonic lethal and embryos die at 3.5 days post-coitum (dpc) prior to implantation due to activation of p53-dependent apoptosis, and this embryonic lethality can be rescued by the concomitant deletion of the Trp53 gene (Jones et al., 1995; Montes de Oca Luna et al., 1995). Mdm4 knock-out is similarly lethal for embryos, but embryos fail to progress in development due to a failure of cells to proliferate 7.5–8.5 dpc (Parant et al., 2001). Similarly, the loss of Trp53 rescues Mdm4-null embryos from lethality. While Mdm2 and Mdm4 are close paralogues, Mdm2 does not complement the loss of Mdm4 and vice versa, suggesting that both Mdm2 and Mdm4 require the other for optimal p53 degradation and regulation, in addition to less well-understood or studied independent roles.
Experiments in mice have also revealed that the levels of p53 and Mdm2 are critically controlled. While hemizygous Mdm2+/− mice are normal under homoeostatic conditions, they are more sensitive to ionizing radiation (IR) compared to wild-type animals. Earlier lethality brought upon hemizygous Mdm4+/− mice revealed that they were even more radiosensitive than Mdm2 hemizygotes (Terzian et al., 2007). Expectedly, these instances of IR-induced lethality are dependent on p53 activity.
Double heterozygotes (Mdm2+/−Mdm4+/−) are not viable, as they are anaemic and smaller than wild-type embryos. In addition, the embryos that survive to 20 dpc suffer from neural and other developmental defects, in addition to a general depletion of all blood cell types (Terzian et al., 2007). Interestingly, the loss of a single copy of Trp53 can rescue the defects of the Mdm2+/−Mdm4+/− double heterozygotes, highlighting the importance of stoichiometry and gene dosage in the p53 pathway.
The proto-oncogene human Mdm2 is highly amplified in sarcomas and is overexpressed in a diverse range of cancers. Likewise, overexpression of Mdm2 leads to increased chromosomal instability and enhanced tumourigenesis in mice (Jones et al., 1998; Lushnikova et al., 2011). Mice with an extra copy of Mdm2 under its native promoter show increased incidences of sarcoma in the presence or absence of functional p53, pointing to a p53-independent role for Mdm2 in tumourigenesis. The development of additional sophisticated models that allowed more precise manipulation of Mdm2 levels in vivo has deepened our understanding into how subtle increases in Mdm2 expression can have profound impact in the rates of tumourigenesis. For instance, analysis of human tumours that have elevated levels of Mdm2 revealed that a single nucleotide polymorphism, SNP309, found in the HDM2 promoter increases the transcription and translation of the protein. This leads to attenuation of the p53 pathway and increased incidences of tumourigenesis (Bond et al., 2004). These findings were subsequently verified by experiments with knock-in models comparing murine Mdm2 (SNP309G) with Mdm2 (SNP309T) (Post et al., 2010). The Mdm2 (SNP309G/G) allele had increased the risk of tumourigenesis and altered tumour spectrum in mice with a p53 hotspot mutation. More recent work that varied murine Mdm2 levels in a Trp53–/– background allowed the deconvolution of tissue specificity and activity to reveal abnormalities in skin pigmentation and in reproductive tissue architecture that resulted in lower fertility (Pant et al., 2016).
Insights from zebrafish models
Mdm2 in zebrafish is expressed in the developing embryo in brain, somites, and the migrating posterior lateral line (Thisse et al., 2000). Just like the other vertebrates, no other homologue other than Mdm2 and Mdm4 has been found in the assembled zebrafish genome (GRCz10).
Our laboratory exploited the development of genome editing tools to generate mutations within the zebrafish mdm2 and mdm4 genes (Chua et al., 2015). We created mutants that contain small insertions or deletions within the coding region of mdm2 and mdm4 that result in frame-shifts and the introduction of premature stop codons to produce severely truncated forms of Mdm2 and Mdm4 proteins (Chua et al., 2015).
The p53-regulatory functions of Mdm2 is conserved in mouse and zebrafish; the loss of mdm2 (in Δ5, +2, and +4 mutants) resulted in embryonic lethality arising from the unchecked accumulation of p53 protein, which triggered apoptosis in the developing embryos. These embryos did not survive beyond 3 days post-fertilization. The deficiency in Mdm2 can be rescued by a single missense mutation that inactivates p53 transcriptional activity (Guo et al., 2013; Chua et al., 2015).
In sharp contrast, the loss of mdm4 in zebrafish did not result in embryonic lethality as observed in mice (Chua et al., 2015). It also did not lead to the accumulation of p53 or growth arrest (our unpublished data). Furthermore, homozygous mdm4 mutants were physically indistinguishable from wild-type embryos. However, we observed that their fecundity reduced with age more rapidly than wild-type, although this needs to be more thoroughly investigated.
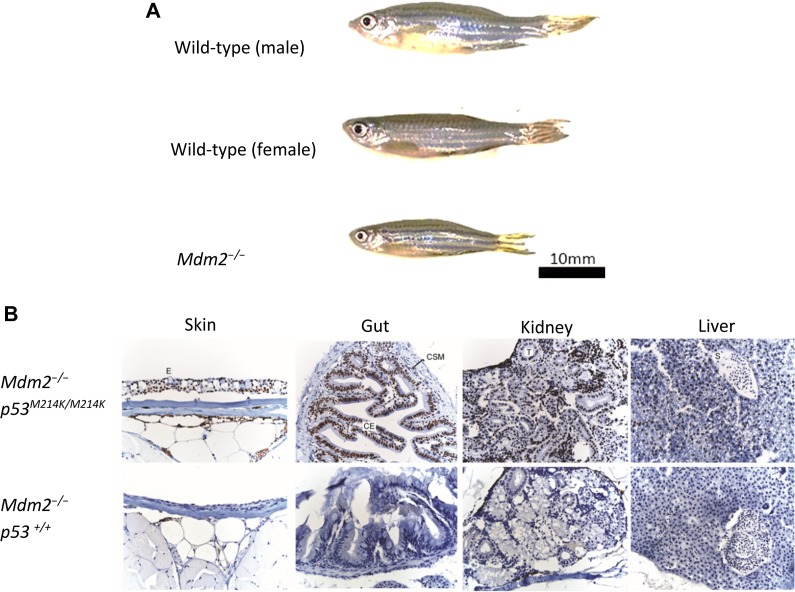
Gene-specific morpholino oligonucleotides (MO) are commonly injected into zebrafish embryos to transiently suppress gene expression (Nasevicius and Ekker, 2000). The transient knockdown of tp53 by MO in mdm2−/− embryos rescued embryonic lethality (Chua et al., 2015) and allowed their survival beyond their expected lifespan of 3 days (Supplementary Figure S4). We were further surprised that these mdm2−/− fish, which still possess wild-type p53, could grow into adulthood (beyond 3 months) (Figure 3A and unpublished data). These morphants are smaller than wild-type fish and lack testes or ovaries, suggesting that these fish are sterile; there have not been any successful matings between these morphants. Immunohistochemical analyses of these adult mdm2−/− fish demonstrated that they did not spontaneously accumulate p53 in any of the tissues examined (Figure 3B). These fascinating observations suggest that Mdm2 plays a critical role in controlling p53 activity during embryogenesis, but beyond a certain point during development, the Mdm2-mediated control on p53 can be alleviated without life-threatening consequences to the animal. It would be interesting to temporally silence p53 during/after embryogenesis in Mdm2−/− mice to test if this phenomenon is reproducible.
Figure 3.
m dm2 knockout in p53-morphant zebrafish is not embryonic lethal if tp53 is temporarily repressed during embryogenesis. (A) mdm2−/− fish grow to adulthood, albeit physically smaller, and lack developed reproductive organs. (B) Immunohistochemical staining reveals that wild-type p53 does not accumulate in mdm2−/− adult fish while, in contrast, mutant p53 protein is stabilized in mdm2−/−p53M214K/M214K fish.
Mdm2 absent in fruit flies and nematodes but present in some other invertebrates
The absence of Mdm2 homologues in fruit fly D. melanogaster and nematode C. elegans prompted the postulation that invertebrates do not possess Mdm2. This was shown otherwise with the discovery of Mdm genes in organisms such as the placozoan Trichoplax adhaerens and the deer tick Ixodes scapularis (Lane et al., 2010a, b). Although orthologues for the Mdm2 family are absent in fruit flies and nematodes, functional homologues can still provide similar roles in regulating p53. For instance, while Drosophila may not possess an Mdm orthologue, Dm-p53 protein levels are negatively regulated by the gene product of companion of reaper (Corp) (Chakraborty et al., 2015). Corp does not appear to have a readily identifiable orthologue within vertebrates and it may represent a protein that acquired a p53-modulating function if the Mdm gene was lost from the genome of an ancestor of fruit flies. BLAST analysis of reference proteomes for Corp orthologues revealed an uncharacterized protein with 24% identity in the parasitic nematode Thelazia callipaeda, and this may be a possible orthologue of human WBP11, a protein involved in DNA damage-responsive mRNA splicing. Advancements in the application of CRISPR technology in gene editing would allow the investigation of the roles of Mdm orthologues in more primitive organisms like Ixodes or Trichoplax, to determine how the functions of the Mdm2 family evolved, and to explore how proteins such as Corp and WBP11 may play a role in organisms that possess p53 but lack Mdm.
Insights from viruses
The p53-binding domains of Mdm2 and Mdm4 are also found within proteins forming the ‘B’ complex of SWI/SNF (SWIB), and these domains can be found in many diverse organisms (Bennett-Lovsey et al., 2002). The SWI/SNF family of complexes was first identified in yeast, functioning to remodel chromatin and aid transcription in an ATP-dependent manner (Winston and Carlson, 1992). The mammalian BAF60 (BRG1-associated factor) family members represent a similar chromatin-remodelling complex including SWIB (Wang et al., 1996; Nomoto et al., 1997; Bennett-Lovsey et al., 2002) and have been shown to interact with p53, p63, and p73 (Lunardi et al., 2010). SWIB domain proteins have been found in phycodnaviruses, while SWI2/SNF2 ATPases and other chromatin-remodelling enzymes have orthologues that are present across a number of lineages of dsDNA viruses (de Souza et al., 2010).
Viral integration into host genomes and resulting mobile genomic elements are assisted by chromosomal remodelling, and the restriction of this through p53 may represent an ancient function (Leonova et al., 2013; Levine et al., 2016; Wylie et al., 2016). Retroviruses rely on integrating their DNA into the host genome and thus benefit from an environment in which DNA repair is hampered and p53 function is moderated. Indeed, the binding of viral proteins to p53 led to the discovery of this important tumour suppressor (Kress et al., 1979; Lane and Crawford, 1979; Linzer and Levine, 1979). Thus, it seems plausible that domains of Mdm2 and Mdm4 important for decreased p53 function may have been present in viruses and ancient organisms.
Although adenovirus E1B-55 K does not have a readily identifiable SWIB domain, it does bind to the same or very similar binding site within the N-terminus of p53 (Lin et al., 1994). E1B-55 K and its viral partner E4orf6 function together as an E3 ligase with host proteins Cullin-2/5, RING-box1, and elongins to lead to the degradation of p53 and the MRE11–Rad50–NBS1 (MRN) complex in adenovirus-infected cells (Querido et al., 2001; Harada et al., 2002), although there appear to be evolutionary differences among the adenoviral serotypes (Cheng et al., 2011). Mdm2 and/or Mdm4 heterodimers also bind to the MRN complex and when Mdm2 is overexpressed, there is a delay in DNA repair leading to chromosome abnormalities and genome instability (Melo and Eischen, 2012). Modification of chromatin through DNA-interacting enzymes of the MRN complex has been observed with orthologous proteins across all cellular life forms, suggesting that many of these proteins and motifs were already present in a universal common ancestor (de Souza et al., 2010). Ancient roles are similarly implied for the proteins that modulate the activity of MRN, such as Mdm2, Mdm4, and E1B-55 K.
While the binding site for p53 within E1B-55 K is still not clearly defined (Blackford and Grand, 2009), the region was initially proposed to be between amino acids 224 and 354 of E1B-55 K (Kao et al., 1990), and the interaction of these two proteins could be decreased in the presence of peptides containing either amino acids 205–221 or 219–233 from E1B-55 K (Grand et al., 1999). Mutations that decrease p53 binding and/or degradation include amino acids from K104 to H215, H474, and F484, further demonstrating that the interaction is complex and may involve more than one site (Blackford and Grand, 2009). Alignment and homology modelling (Supplementary Figure S5) suggest that the region around 220–280 of E1B-55 K could potentially mimic two of the key α helices and hydrophobic cleft of Mdm2–p53 interaction and thus may represent a major binding site.
Conclusion
Evolutionary studies are limited by the availability of high-quality genome sequences with good DNA sequence coverage, but this is steadily being overcome with high throughput sequencing. With the current set of genomes of extant organisms, we are able to discern that despite high homology between Mdm2 and Mdm4 in vertebrates, the two proteins function differently based on the small differences in their protein sequences in their conserved domains. There is a decreasing disparity in function with greater divergence from our species, most obvious in that both Mdm2 and Mdm4 are necessary and non-redundant for development in mice, but the loss of Mdm4 alone is tolerated in zebrafish development. This is also evident in the case when lamprey Mdm4 can destabilize lamprey p53, but the Mdm4 of higher vertebrates cannot degrade their p53. Such evidence supports the hypothesis that following the duplication of the Mdm gene, the two new genes evolved to take up different roles in the regulation of p53.
Even viruses have evolved means to disrupt the regulation of p53/Mdm2/Mdm4. Inactivation of p53 is often a critical step in viral infection involving inhibition of transactivation, PML nuclear body sequestering, and RING and HECT domain-mediated ubiquitination and targeting to the proteasome (de Souza et al., 2010; Gustin et al., 2011; Chao, 2015; Miciak and Bunz, 2016; Wimmer et al., 2016). These activities often result in decreased DNA repair or delayed apoptosis and potentially lead to oncogenesis in human hosts. Further investigation will be required to determine whether common ancestors acquired protein motifs that eventually formed Mdm following viral infection or, alternatively, whether host protein functions are mimicked by viral motifs (Chemes et al., 2015). Additional studies of Mdm2 and Mdm4 of other organisms, especially species of ancient lineages, would yield greater insight in the relationship among Mdm2, Mdm4, and p53. Tractable model organisms would also need to be established to biochemically verify the functions of Mdm2 and Mdm4 in their native contexts.
The interaction between Mdm2 and/or Mdm4 and p53 has become an important model system for the design and development of drug-like molecules that disrupt intracellular protein–protein interactions, leading to substantial development in drug discovery. These include stapled peptides (Brown et al., 2013; Chang et al., 2013), cyclotides (Ji et al., 2013), and small molecule inhibitors (Vassilev et al., 2004; Ding et al., 2006; Koblish et al., 2006; Shangary et al., 2008; Lakoma et al., 2015). Evolution can offer clues in discerning new functions of the Mdm2 family and provide insights in drug design and discovery. The growing understanding of novel roles for Mdm2 and Mdm4 suggests that the new molecules may have wider therapeutic use beyond oncology (Li et al., 2016), and these are critical test systems of novel drugs that exploit the exquisite specificity of protein degradation in eukaryotic cells.
Supplementary Material
Acknowledgements
We apologize to those colleagues whose work has not been cited owing space limitations. Lamprey RPL11 sequence was kindly provided by Byrappa Venkatesh and his team (Institute of Molecular and Cell Biology, A*STAR). We thank Chandra Verma (Bioinformatics Institute, A*STAR) for helpful discussions with the E1B-55 K homology model and Weiyun Villien Zhang (p53 Laboratory, A*STAR) for assistance in editing the manuscript.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by the Biomedical Research Council of Agency for Science, Technology and Research (A*STAR), Singapore.
Conflict of interest: none declared.
References
- Barak Y., Juven T., Haffner R., et al. (1993). mdm2 expression is induced by wild type p53 activity. EMBO J. 12, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey R., Hart S.E., Shirai H., et al. (2002). The SWIB and the MDM2 domains are homologous and share a common fold. Bioinformatics 18, 626–630. [DOI] [PubMed] [Google Scholar]
- Bernardi R., Scaglioni P.P., Bergmann S., et al. (2004). PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6, 665–672. [DOI] [PubMed] [Google Scholar]
- Bista M., Petrovich M., and Fersht A.R (2013). MDMX contains an autoinhibitory sequence element. Proc. Natl Acad. Sci. USA 110, 17814–17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford A.N., and Grand R.J.A (2009). Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation. J. Virol. 83, 4000–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond G.L., Hu W., Bond E.E., et al. (2004). A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119, 591–602. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Quah S.T., Jong J., et al. (2013). Stapled peptides with improved potency and specificity that activate p53. ACS Chem. Biol. 8, 506–512.23214419 [Google Scholar]
- Bursać S., Brdovčak M.C., Pfannkuchen M., et al. (2012). Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc. Natl Acad. Sci. USA 109, 20467–20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahilly-Snyder L., Yang-Feng T., Francke U., et al. (1987). Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat. Cell Mol. Genet. 13, 235–244. [DOI] [PubMed] [Google Scholar]
- Carrillo A.M., Bouska A., Arrate M.P., et al. (2015). Mdmx promotes genomic instability independent of p53 and Mdm2. Oncogene 34, 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R., Li Y., Zhou L., et al. (2015). Corp regulates p53 in Drosophila melanogaster via a negative feedback loop. PLoS Genet. 11, e1005400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.S., Graves B., Guerlavais V., et al. (2013). Stapled α-helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl Acad. Sci. USA 110, E3445–E3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C.C. (2015). Mechanisms of p53 degradation. Clin. Chim. Acta 438, 139–147. [DOI] [PubMed] [Google Scholar]
- Chemes L.B., de Prat-Gay G., and Sánchez I.E (2015). Convergent evolution and mimicry of protein linear motifs in host-pathogen interactions. Curr. Opin. Struct. Biol. 32, 91–101. [DOI] [PubMed] [Google Scholar]
- Chène P. (2004). Inhibition of the p53-MDM2 interaction: targeting a protein-protein interface. Mol. Cancer Res. 2, 20–28. [PubMed] [Google Scholar]
- Cheng C.Y., Gilson T., Dallaire F., et al. (2011). The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity. J. Virol. 85, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua J.S., Liew H.P., Guo L., et al. (2015). Tumor-specific signaling to p53 is mimicked by Mdm2 inactivation in zebrafish: insights from mdm2 and mdm4 mutant zebrafish. Oncogene 34, 5933–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffill C.R., Lee A.P., Siau J.W., et al. (2016). The p53–Mdm2 interaction and the E3 ligase activity of Mdm2/Mdm4 are conserved from lampreys to humans. Genes Dev. 30, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding K., Lu Y., Nikolovska-Coleska Z., et al. (2006). Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem. 49, 3432–3435. [DOI] [PubMed] [Google Scholar]
- Dolezelova P., Cetkovska K., Vousden K.H., et al. (2012). Mutational analysis reveals a dual role of Mdm2 acidic domain in the regulation of p53 stability. FEBS Lett. 586, 2225–2231. [DOI] [PubMed] [Google Scholar]
- Donati G., Peddigari S., Mercer C.A., et al. (2013). 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 4, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakharzadeh S.S., Trusko S.P., and George D.L (1991). Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 10, 1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei P., and El-Deiry W.S (2003). P53 and radiation responses. Oncogene 22, 5774–5783. [DOI] [PubMed] [Google Scholar]
- Fox G.E. (2010). Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2, a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli F., Rossi M., D'Alessandra Y., et al. (2010). MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J. Cell Sci. 123, 2423–2433. [DOI] [PubMed] [Google Scholar]
- Gembarska A., Luciani F., Fedele C., et al. (2012). MDM4 is a key therapeutic target in cutaneous melanoma. Nat. Med. 18, 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes D.M., Chen L., and Chen J (2006). MDMX regulation of p53 response to ribosomal stress. EMBO J. 25, 5614–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand R.J., Parkhill J., Szestak T., et al. (1999). Definition of a major p53 binding site on Ad2E1B58K protein and a possible nuclear localization signal on the Ad12E1B54K protein. Oncogene 18, 955–965. [DOI] [PubMed] [Google Scholar]
- Grönroos E., Terentiev A.A., Punga T., et al. (2004). YY1 inhibits the activation of the p53 tumor suppressor in response to gienotoxic stress. Proc. Natl Acad. Sci. USA 101, 12165–12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman S.R., Perez M., Kung A.L., et al. (1998). p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell 2, 405–415. [DOI] [PubMed] [Google Scholar]
- Gu J., Kawai H., Nie L., et al. (2002). Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277, 19251–19254. [DOI] [PubMed] [Google Scholar]
- Guo L., Liew H.P., Camus S., et al. (2013). Ionizing radiation induces a dramatic persistence of p53 protein accumulation and DNA damage signaling in mutant p53 zebrafish. Oncogene 32, 4009–4016. [DOI] [PubMed] [Google Scholar]
- Gustin J.K., Moses A.V., Früh K., et al. (2011). Viral takeover of the host ubiquitin system. Front. Microbiol. 2, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Garcia-Manero G., McDonnell T.J., et al. (2007). HDM4 (HDMX) is widely expressed in adult pre-B acute lymphoblastic leukemia and is a potential therapeutic target. Mod. Pathol. 20, 54–62. [DOI] [PubMed] [Google Scholar]
- Harada J.N., Shevchenko A., Shevchenko A., et al. (2002). Analysis of the adenovirus E1B-55 K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76, 9194–9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., et al. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- He X., Li Y., Dai M.-S., et al. (2016). Ribosomal protein L4 is a novel regulator of the MDM2-p53 loop. Oncotarget 7, 16217–16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock A.K., and Vousden K.H (2014). The role of ubiquitin modification in the regulation of p53. Biochim. Biophys. Acta 1843, 137–149. [DOI] [PubMed] [Google Scholar]
- Honda R., Tanaka H., and Yasuda H (1997). Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27. [DOI] [PubMed] [Google Scholar]
- Horn H.F., and Vousden K.H (2008). Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene 27, 5774–5784. [DOI] [PubMed] [Google Scholar]
- Iyappan S., Wollscheid H.-P., Rojas-Fernandez A., et al. (2010). Turning the RING domain protein MdmX into an active ubiquitin-protein ligase. J. Biol. Chem. 285, 33065–33072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.W., Lindstrom M.S., and Berberich S.J (2001). MdmX binding to ARF affects Mdm2 protein stability and p53 transactivation. J. Biol. Chem. 276, 25336–25341. [DOI] [PubMed] [Google Scholar]
- Ji Y., Majumder S., Millard M., et al. (2013). In vivo activation of the p53 tumor suppressor pathway by an engineered cyclotide. J. Am. Chem. Soc. 135, 11623–11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Dai M.-S., Lu S.Z., et al. (2006). 14-3-3γ binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 25, 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.N., Hancock A.R., Vogel H., et al. (1998). Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl Acad. Sci. USA 95, 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.N., Roe A.E., Donehower L.A., et al. (1995). Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378, 206–208. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Weber J.D., Zambetti G., et al. (1998). Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl Acad. Sci. USA 95, 8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.C., Yew P.R., and Berk A.J (1990). Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55 K proteins. Virology 179, 806–814. [DOI] [PubMed] [Google Scholar]
- Koblish H.K., Zhao S., Franks C.F., et al. (2006). Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol. Cancer Ther. 5, 160–169. [DOI] [PubMed] [Google Scholar]
- Koster R., Timmer-Bosscha H., Bischoff R., et al. (2011). Disruption of the MDM2-p53 interaction strongly potentiates p53-dependent apoptosis in cisplatin-resistant human testicular carcinoma cells via the Fas/FasL pathway. Cell Death Dis. 2, e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic M., Matt T., Martinez-Yamout M.A., et al. (2006). Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J. Mol. Biol. 363, 433–450. [DOI] [PubMed] [Google Scholar]
- Kress M., May E., Cassingena R., et al. (1979). Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 31, 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat M.H.G., Jones S.N., and Vousden K.H (1997). Regulation of p53 stability by Mdm2. Nature 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Kussie P.H., Gorina S., Marechal V., et al. (1996). Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953. [DOI] [PubMed] [Google Scholar]
- Lakoma A., Barbieri E., Agarwal S., et al. (2015). The MDM2 small-molecule inhibitor RG7388 leads to potent tumor inhibition in p53 wild-type neuroblastoma. Cell Death Discov. 1, 15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.P., Cheok C.F., Brown C.J., et al. (2010. a). The Mdm2 and p53 genes are conserved in the Arachnids. Cell Cycle 9, 748–754. [DOI] [PubMed] [Google Scholar]
- Lane D.P., Cheok C.F., Brown C., et al. (2010. b). Mdm2 and p53 are highly conserved from placozoans to man. Cell Cycle 9, 540–547. [DOI] [PubMed] [Google Scholar]
- Lane D.P., and Crawford L.V (1979). T antigen is bound to a host protein in SV40-transformed cells. Nature 278, 261–263. [DOI] [PubMed] [Google Scholar]
- Lane D.P., and Verma C (2012). Mdm2 in evolution. Genes Cancer 3, 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie N.A., Donovan S.L., Shih C.S., et al. (2006). Inactivation of the p53 pathway in retinoblastoma. Nature 444, 61–66. [DOI] [PubMed] [Google Scholar]
- Leonova K.I., Brodsky L., Lipchick B., et al. (2013). p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc. Natl Acad. Sci. USA 110, E89–E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie P.L., Ke H., and Zhang Y (2015). The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2-MDMX protein heterodimerization. J. Biol. Chem. 290, 12941–12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J., Ting D.T., and Greenbaum B.D (2016). P53 and the defenses against genome instability caused by transposons and repetitive elements. Bioessays 38, 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., and Lozano G (2013). Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clin. Cancer Res. 19, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Stockton M.E., Bhuiyan I., et al. (2016). MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Sci. Transl. Med. 8, 336ra61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Chen J., Elenbaas B., et al. (1994). Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 8, 1235–1246. [DOI] [PubMed] [Google Scholar]
- Linares L.K., Hengstermann A., Ciechanover A., et al. (2003). HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl Acad. Sci. USA 100, 12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K., Mace P.D., Smith C.A., et al. (2008). Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 15, 841–848. [DOI] [PubMed] [Google Scholar]
- Linzer D.I., and Levine A.J (1979). Characterization of a 54 K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17, 43–52. [DOI] [PubMed] [Google Scholar]
- Liu D., and Xu Y (2011). p53, oxidative stress, and aging. Antioxid. Redox Signal. 15, 1669–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Tackmann N.R., Yang J., et al. (2016). Disruption of the RP-MDM2-p53 pathway accelerates APC loss-induced colorectal tumorigenesis. Oncogene, 10.1038/onc.2016.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardi A., Di Minin G., Provero P., et al. (2010). A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc. Natl Acad. Sci. USA 107, 6322–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushnikova T., Bouska A., Odvody J., et al. (2011). Aging mice have increased chromosome instability that is exacerbated by elevated Mdm2 expression. Oncogene 30, 4622–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E., Jin A., Deisenroth C., et al. (2010). An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell 18, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T.K., Ravi V., Yamasaki S., et al. (2013). Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc. Natl Acad. Sci. USA 110, 16044–16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo A.N., and Eischen C.M (2012). Protecting the genome from mdm2 and mdmx. Genes Cancer 3, 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miciak J., and Bunz F (2016). Long story short: p53 mediates innate immunity. Biochim. Biophys. Acta 1865, 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini D., Danovi D., Colombo E., et al. (2002). Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J. Biol. Chem. 277, 7318–7323. [DOI] [PubMed] [Google Scholar]
- Moll U.M., and Slade N (2004). p63 and p73: roles in development and tumor formation. Mol. Cancer Res. 2, 371–386. [PubMed] [Google Scholar]
- Momand J., Jung D., Wilczynski S., et al. (1998). The MDM2 gene amplification database. Nucleic Acids Res. 26, 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J., Villegas A., and Belyi V.A (2011). The evolution of MDM2 family genes. Gene 486, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J., Zambetti G.P., Olson D.C., et al. (1992). The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R., Wagner D.S., and Lozano G (1995). Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206. [DOI] [PubMed] [Google Scholar]
- Nasevicius A., and Ekker S.C (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216–220. [DOI] [PubMed] [Google Scholar]
- Nicolas E., Parisot P., Pinto-Monteiro C., et al. (2016). Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 7, 11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto K., Nakazato S., Kazahari K., et al. (1997). Gene structure of rat BAF60b, a component of mammalian SW1/SNF complexes, and its physical linkage to the growth hormone gene and transcription factor SUG/proteasome p45 gene. Gene 202, 157–165. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Taya Y., and Nakagama H (2009). Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 583, 2710–2714. [DOI] [PubMed] [Google Scholar]
- Pant V., Xiong S., Chau G., et al. (2016). Distinct downstream targets manifest p53-dependent pathologies in mice. Oncogene 35, 5713–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J., Chavez-Reyes A., Little N.A., et al. (2001). Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29, 92–95. [DOI] [PubMed] [Google Scholar]
- Pazgier M., Liu M., Zou G., et al. (2009). Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc. Natl Acad. Sci. USA 106, 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y., Shkedy D., de Graaf P., et al. (2005). Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc. Natl Acad. Sci. USA 102, 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post S.M., Quintás-Cardama A., Pant V., et al. (2010). A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell 18, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E., Blanchette P., Yan Q., et al. (2001). Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev 15, 3104–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. (2014). The ribosome emerges from a black box. Cell 159, 979–984. [DOI] [PubMed] [Google Scholar]
- Ranaweera R.S., and Yang X (2013). Auto-ubiquitination of Mdm2 enhances its substrate ubiquitin ligase activity. J. Biol. Chem. 288, 18939–18946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayburn E., Zhang R., He J., et al. (2005). MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr. Cancer Drug Targets 5, 27–41. [DOI] [PubMed] [Google Scholar]
- Sabbatini P., and McCormick F (2002). MDMX inhibits the p300/CBP-mediated acetylation of p53. DNA Cell Biol. 21, 519–525. [DOI] [PubMed] [Google Scholar]
- Sasaki M., Kawahara K., Nishio M., et al. (2011). Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat. Med. 17, 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville M.K., Sparks A., Xirodimas D.P., et al. (2004). Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J. Biol. Chem. 279, 42169–42181. [DOI] [PubMed] [Google Scholar]
- Sermeus A., and Michiels C (2011). Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangary S., Qin D., McEachern D., et al. (2008). Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc. Natl Acad. Sci. USA 105, 3933–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts A., Steegenga W.T., Riteco N., et al. (1996). MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15, 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Siau J.W., Coffill C.R., Zhang W.V., et al. (2016). Functional characterization of p53 pathway components in the ancient metazoan Trichoplax adhaerens. Sci. Rep. 6, 33972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan K.E., Bohnsack M.T., and Watkins N.J (2013). The 5 S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 5, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., and Keinath M.C (2015). The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 25, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L.C., Trusko S.P., Freeman N., et al. (1988). A gene amplified in a transformed mouse cell line undergoes complex transcriptional processing and encodes a nuclear protein. J. Biol. Chem. 263, 17150–17158. [PubMed] [Google Scholar]
- de Souza R.F., Iyer L.M., and Aravind L (2010). Diversity and evolution of chromatin proteins encoded by DNA viruses. Biochim. Biophys. Acta 1799, 302–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel J.M., and Wahl G.M (2004). Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 23, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G., Affar el B., Shi Y., et al. (2004). Yin Yang 1 is a negative regulator of p53. Cell 117, 859–872. [DOI] [PubMed] [Google Scholar]
- Tan B.X., Brown C.J., Ferrer F.J., et al. (2015). Assessing the efficacy of Mdm2/Mdm4-inhibiting stapled peptides using cellular thermal shift assays. Sci. Rep. 5, 12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashakori M., Zhang Y., Xiong S., et al. (2016). p53 activity dominates that of p73 upon Mdm4 loss in development and tumorigenesis. Mol. Cancer Res. 14, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T., Wang Y., Van Pelt C.S., et al. (2007). Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol. Cell. Biol. 27, 5479–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Neel H., Thisse B., et al. (2000). The Mdm2 gene of zebrafish (Danio rerio): preferential expression during development of neural and muscular tissues, and absence of tumor formation after overexpression of its cDNA during early embryogenesis. Differentiation 66, 61–70. [DOI] [PubMed] [Google Scholar]
- Toledo F., and Wahl G.M (2007). MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int. J. Biochem. Cell Biol. 39, 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Mark D., Kong D., Loncarek J., et al. (2015). MDM1 is a microtubule-binding protein that negatively regulates centriole duplication. Mol. Biol. Cell 26, 3788–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B., et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848. [DOI] [PubMed] [Google Scholar]
- Wang W., Xue Y., Zhou S., et al. (1996). Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10, 2117–2130. [DOI] [PubMed] [Google Scholar]
- Wang X., Arooz T., Siu W.Y., et al. (2001). MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 490, 202–208. [DOI] [PubMed] [Google Scholar]
- Wimmer P., Berscheminski J., Blanchette P., et al. (2016). PML isoforms IV and V contribute to adenovirus-mediated oncogenic transformation by functionally inhibiting the tumor-suppressor p53. Oncogene 35, 69–82. [DOI] [PubMed] [Google Scholar]
- Winston F., and Carlson M (1992). Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8, 387–391. [DOI] [PubMed] [Google Scholar]
- Wu H., and Leng R.P (2015). MDM2 mediates p73 ubiquitination: a new molecular mechanism for suppression of p73 function. Oncotarget 6, 21479–21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie A., Jones A.E., D'Brot A., et al. (2016). p53 genes function to restrain mobile elements. Genes Dev. 30, 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzalik M., Pustelny K., Kedracka-Krok S., et al. (2010). Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle 9, 4584–4591. [DOI] [PubMed] [Google Scholar]
- Zheng J., Lang Y., Zhang Q., et al. (2015). Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation. Genes Dev. 29, 1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liao W.-J., Liao J.-M., et al. (2015). Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilfou J.T., and Lowe S.W (2009). Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 1, a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.