Abstract
The genetic code is degenerate. With the exception of two amino acids (Met and Trp), all other amino acid residues are each encoded by multiple, so-called synonymous codons. Synonymous codons were initially presumed to have entirely equivalent functions, however, the finding that synonymous codons are not present at equal frequencies in genes/genomes suggested that codon choice might have functional implications beyond amino acid coding. The pattern of non-uniform codon use (known as codon usage bias) varies between organisms and represents a unique feature of an organism. Organism-specific codon choice is related to organism-specific differences in populations of cognate tRNAs. This implies that, in a given organism, frequently used codons will be translated more rapidly than infrequently used ones and vice versa. A theory of codon-tRNA co-evolution (necessary to balance accurate and efficient protein production) was put forward to explain the existence of codon usage bias. This model suggests that selection favours preferred (frequent) over un-preferred (rare) codons in order to sustain efficient protein production in cells and that a given un-preferred codon will have the same effect on an organism’s fitness regardless of its position within an mRNA’s open reading frame. However, many recent studies refute this prediction. Un-preferred codons have been found to have important functional roles and their effects appeared to be position-dependent. Synonymous codon usage affects the efficiency/stringency of mRNA decoding, mRNA biogenesis/stability, and protein secretion and folding. This review summarizes recent developments in the field that have identified novel functions of synonymous codons and their usage.
Introduction
In 1961, Crick, Barnett, Brenner and Watts-Tobin postulated key features of the genetic code (1). They suggested that “the genetic code is of the following general type: (a) A group of three bases (or, less likely, a multiple of three bases) codes one amino-acid. (b) The code is not of the overlapping type. (c) The sequence of the bases is read from a fixed starting point. This determines how the long sequences of bases are to be correctly read off as triplets. There are no special ‘commas' to show how to select the right triplets. If the starting point is displaced by one base, then the reading into triplets is displaced, and thus becomes incorrect. (d) The code is probably ‘degenerate'; that is, in general, one particular amino-acid can be coded by one of several triplets of bases” (1).
Further studies on the responses of aminoacyl (aa)-tRNAs to trinucleotide templates, allowed the general patterns of degeneracy to be established and confirmed that the genetic code is indeed redundant (2–8). Synonymous codons were originally presumed to be entirely equivalent and to mainly “help” alleviate the effects of mutations in the third position of codons (2–8) since in many instances such mutations would not change the identity of the encoded amino acid (Table 1). It was soon recognized, however, that synonymous codons, while encoding the same amino acid, might not necessarily be entirely equivalent (9). It was revealed that the frequencies of occurrence of synonymous codons would not be uniquely determined by the frequencies of corresponding amino acids in proteins and thus might substantially differ (9). In 1972, Goel and colleagues in their pioneering study noted that although “at the present time no specific functions are assignable to different synonymous codons”, synonymous codons might, nevertheless, be suspected of having different functions (9). They further suggested that knowledge of the frequencies of usage of different codons would be of great interest with respect to uncovering potential differences in codon functions and predicted that “codon frequency character” might also be useful “for purposes of numerical taxonomy” if different organisms showed differences in codon frequencies not solely “attributable to variations in DNA composition” (9).
Table 1.
Codon usage observed in Escherichia coli (EC), Saccharomyces cerevisiae (SC) and Homo sapiens (HS) ORFeomes (23). Representative examples showing substantial differences in usage of codons between the three ORFeomes are boxed. Frequency per thousand codons is shown
| EC | SC | HS | EC | SC | HS | EC | SC | HS | EC | SC | HS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UUU | Phe | 24.4 | 26.1 | 17.6 | UCU | Ser | 13.1 | 23.5 | 15.2 | UAU | Tyr | 21.6 | 18.8 | 12.2 | UGU | Cys | 5.9 | 8.1 | 10.6 |
| UUC | Phe | 13.9 | 18.4 | 20.3 | UCC | Ser | 9.7 | 14.2 | 17.7 | UAC | Tyr | 11.7 | 14.8 | 15.3 | UGC | Cys | 5.5 | 4.8 | 12.6 |
| UUA | Leu | 17.4 | 26.2 | 7.7 | UCA | Ser | 13.1 | 18.7 | 12.2 | UAA | Stop | 2.0 | 1.1 | 1.0 | UGA | Stop | 1.1 | 0.7 | 1.6 |
| UUG | Leu | 12.9 | 27.2 | 12.9 | UCG | Ser | 8.2 | 8.6 | 4.4 | UAG | Stop | 0.3 | 0.5 | 0.8 | UGG | Trp | 13.4 | 10.4 | 13.2 |
| CUU | Leu | 14.5 | 12.3 | 13.2 | CCU | Pro | 9.5 | 13.5 | 17.5 | CAU | His | 12.4 | 13.6 | 10.9 | CGU | Arg | 15.9 | 6.4 | 4.5 |
| CUC | Leu | 9.5 | 5.4 | 19.6 | CCC | Pro | 6.2 | 6.8 | 19.8 | CAC | His | 7.3 | 7.8 | 15.1 | CGC | Arg | 14.0 | 2.6 | 10.4 |
| CUA | Leu | 5.6 | 13.4 | 7.2 | CCA | Pro | 9.1 | 18.3 | 16.9 | CAA | Gln | 14.4 | 27.3 | 12.3 | CGA | Arg | 4.8 | 3.0 | 6.2 |
| CUG | Leu | 37.4 | 10.5 | 39.6 | CCG | Pro | 14.5 | 5.3 | 6.9 | CAG | Gln | 26.7 | 12.1 | 34.2 | CGG | Arg | 7.9 | 1.7 | 11.4 |
| AUU | Ile | 29.6 | 30.1 | 16.0 | ACU | Thr | 13.1 | 20.3 | 13.1 | AAU | Asn | 29.3 | 35.7 | 17.0 | AGU | Ser | 13.2 | 14.2 | 12.1 |
| AUC | Ile | 19.4 | 17.2 | 20.8 | ACC | Thr | 18.9 | 12.7 | 18.9 | AAC | Asn | 20.3 | 24.8 | 19.1 | AGC | Ser | 14.3 | 9.8 | 19.5 |
| AUA | Ile | 13.3 | 17.8 | 7.5 | ACA | Thr | 15.1 | 17.8 | 15.1 | AAA | Lys | 37.2 | 41.9 | 24.4 | AGA | Arg | 7.1 | 21.3 | 12.2 |
| AUG | Met | 23.7 | 20.9 | 22.0 | ACG | Thr | 13.6 | 8.0 | 6.1 | AAG | Lys | 15.3 | 30.8 | 31.9 | AGG | Arg | 4.0 | 9.2 | 12.0 |
| GUU | Val | 21.6 | 22.1 | 11.0 | GCU | Ala | 18.9 | 21.2 | 18.4 | GAU | Asp | 33.7 | 37.6 | 21.8 | GGU | Gly | 23.7 | 23.9 | 10.8 |
| GUC | Val | 13.1 | 11.8 | 14.5 | GCC | Ala | 21.6 | 12.6 | 27.7 | GAC | Asp | 17.9 | 20.2 | 25.1 | GGC | Gly | 20.6 | 9.8 | 22.2 |
| GUA | Val | 13.1 | 11.8 | 7.1 | GCA | Ala | 23.0 | 16.2 | 15.8 | GAA | Glu | 35.1 | 45.6 | 29.0 | GGA | Gly | 13.6 | 10.9 | 16.5 |
| GUG | Val | 19.9 | 10.8 | 28.1 | GCG | Ala | 21.1 | 6.2 | 7.4 | GAG | Glu | 19.4 | 19.2 | 39.6 | GGG | Gly | 12.3 | 6.0 | 16.5 |
Eight years later, Grantham and colleagues opened the Pandora’s box of codon usage studies by publishing the first catalogue of codon usage frequencies tabulated from “every published mRNA sequence of 50 or more codons” in length (10). Although their initial analysis was limited to just 90 open reading frames (ORFs) (mostly from genes of several Escherichia coli phages along with a few mouse, rat, rabbit and human genes among others), they concluded that different codons are indeed utilized with different frequencies (10). They also suggested that, because of the degeneracy of the genetic code, the information contained on the mRNA level may better reflect a protein’s evolution than the information provided by its amino acid sequence (10,11).
Subsequent studies by Ikemura, Sharp, Kurland and many others established that synonymous codons are distributed non-uniformly and non-randomly in different genes/genomes (12–23). This phenomenon is now referred to as codon usage bias (Table 1). It was also found that codon frequencies usually correlate with the concentration of cognate tRNAs (12,13,16) and thus it was suggested that codon bias affects translation elongation rates (17–22), as frequently used codons were, as a rule, found to be translated more rapidly than infrequently used ones due to the more ready availability (during decoding of the message) of corresponding frequent cognate tRNAs (20–22,24,25). Optimal/frequent codons differ among species (Table 1), coordinated with changes in the population of tRNA genes revealing that bias in codon usage is adaptive (16).
Numerous studies in recent years have been aimed at gaining a better understanding of the origin, evolution and function(s) of codon usage bias (26–30). The available data strongly indicate a role for synonymous codons beyond that of amino acid coding and support of the predefined protein expression levels for the sequence of codons in an mRNA. Codon usage has been found to contain many novel layers of information. Synonymous codon usage was found to affect mRNA stability (31–33), protein secretion (34–36) and protein folding (37–43) and to have an impact on many other aspects of mRNA/protein function and cellular physiology, such as e.g. mRNA splicing (44,45 and references therein), exonic transcription factor binding (46) and the origin of diseases (44). Obviously, many opposite yet complementary forces shape codon usage bias, however, how exactly natural selection shapes patterns of codon usage remain unresolved.
The purpose of this short review is to provide insight into the current extent of our knowledge regarding synonymous codon usage and its physiological significance and to present some of the open questions surrounding this topic.
Codon Usage Bias
The genetic code is generally conserved among different organisms, although alternative codes have been reported in both nuclear and mitochondrial genomes (47,48). The majority of codon reassignments in these alternative codes involve sense to nonsense codon changes (or vice versa) (47,48). Incidences of genetic code alterations are, however, relatively rare and will not be considered here. By the mid-1980s, it became clear that global codon usage patterns vary between organisms, leading to organism-specific biases in the frequencies of synonymous codons (Table 1) (13–19). It was also found that organism-specific codon choice is related to organism-specific populations of cognate tRNAs and that in both unicellular and multicellular organisms there exists a strong positive correlation between codon usage and cellular tRNA content (12,13,16). It should be noted, however, that tRNA content is not entirely constant in either unicellular or multicellular organisms. There also exist intercellular and tissue variations in tRNA abundance and tRNA content in a given cell/tissue has been shown to change upon cell growth, differentiation and/or development, thus responding to the changing pattern/repertoire of the expressed genes and the differences in their codon usage (49–56).
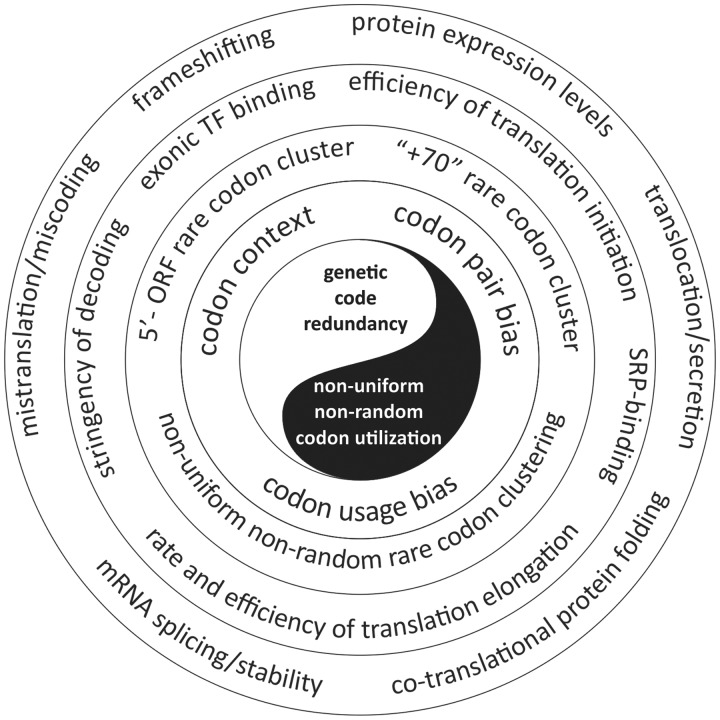
The exact molecular mechanisms linking tRNA abundance to biased codon usage remain largely undefined and represent one of the biggest enigmas in the field. A theory of codon-tRNA co-evolution (to balance accurate and efficient protein production) has been put forward to explain the existence of codon usage bias and the correlation between tRNA content and codon usage (57–61). Given that both mutational and selection pressures are involved in shaping the codon bias in a given species, the so-called mutation-selection-drift balance model became the core of this theory (57–61). This model suggests that selection favours preferred (frequent) over un-preferred (rare) codons (to sustain efficient protein production in cells) and that a particular un-preferred codon will have the same effect on fitness regardless of its position within an mRNA’s ORF (57–61). However, many recent studies refute these predictions. Un-preferred (rare) codons have been found to have important functional roles (Figure 1) and their effects appeared to be position-dependent (36,38,62,63). Therefore, many key questions still remain largely open. These questions include (i) What forces exactly determine the choice of codons in a given gene and across the genome? (ii) What accounts for differences in codon usage biases between different organisms and how shifts in codon usage between them occur? (iii) What is the mechanism linking tRNA abundance to codon usage? and (iv) How do cells adjust changes in tRNA content to the repertoire of expressed genes at any given moment in time depending on the cellular status and environmental conditions?
Figure 1.
The yin and yang of codon usage. The genetic code is degenerate. Synonymous codons (encoding the same amino acids) were initially presumed to have equivalent functions, however, they are not present at equal frequencies in genes/genomes and codon choice is found to have functional implications beyond amino acid coding. Genetic code redundancy and non-uniform/non-random codon utilization are tightly interconnected and interdependent and shape organism-specific codon usage bias. Non-random codon utilization is reflected in specific codon context and non-random codon pair bias. Codon context analyses also reveal hierarchical clustering of codons along mRNAs. Codon usage is believed to shape the tRNA pool of organisms, resulting in concentrations of cognate tRNAs correlating with codon frequencies. This correlation implies that in a given organism, frequently used codons are translated more rapidly than infrequently used ones. Codon usage bias impacts the efficiency and speed of mRNA decoding and is widely believed to determine efficiency of the protein production. Natural selection shapes patterns of codon usage to maintain robust protein expression, however maximizing the speed and output of translation may put conflicting demands on the protein synthesis machinery resulting in e.g. improper protein folding. Therefore, codon choice has to be carefully controlled to balance these and other processes in cells, such as e.g. mRNA stability. Thus, differential codon usage provides multiple layers of information beyond amino acid coding and support of protein expression levels predefined by the sequence of codons in mRNA. Optimal/frequent codons differ among species, coordinated with changes in the population of tRNA genes revealing that bias in codon usage is adaptive. Natural selection simultaneously exploits numerous possibilities and directions to optimize expression of the genetic information. Many opposite yet complimentary forces shape codon usage bias.
Genome vs Gene Level: Codon Bias and Codon Context
Earlier studies concluded that natural selection distinguishing between synonymous codons shapes the organism-specific codon usage bias and that this constraint further reflects differences in translational efficiency of different codons and, thus, different genes (16,17,57–61). While a causal link between codon usage and level of gene expression was not always straightforward, it was nevertheless concluded that highly expressed genes tend to harbour more preferred codons compared to lowly expressed genes, which were found to be enriched in synonymous un-preferred codons (17,64). It appeared, however, that the patterns of codon preference within each group (e.g., genes with high vs low expression) are also not random (62). Close examination of mRNA sequences revealed biases in the distribution of nucleotides/codons within ORFs, a phenomenon known as codon context (65–69). It was suggested that the translation of each individual codon responds to its sequence environment though the mechanism of this context (65–69). Nonsense and missense suppression, elongation rate, precision of tRNA selection and polypeptide chain termination all appeared to be affected by codon context (70–74). The contexts of neighbouring nucleotides and neighbouring codons are revealed in the non-random juxtaposition of codons in mRNA ORFs, referred to as “codon pair bias” (68,69,73,75). There have been many recent studies analysing codon pair biases in various species (68,69,73–79). It was found that the most frequently preferred codon pairs in all analyzed species contain the patterns nnGCnn, nnCAnn or nnUnCn, while most ORFeomes tend to avoid codon pairs containing the patterns nnUAnn, nnGGnn, nnGnnC, nnCGCn, GUCCnn, CUCCnn, nnCnnA or UUCGnn (68). The most conserved preferred codon pair, GGGCUU, was over-represented in 76% of the organisms studied and the least preferred pair, UUCGCA, was under-represented in 86% of the studied genomes (68). Interestingly, independently of the overall differences in codon biases between organisms, the most avoided and the most preferred codon pairs appeared to be uniformly distributed across all three domains of life (bacteria, archaea and eukaryotes), suggesting strong evolutionary conservation (68). These findings suggested that codon context could be even more important than variations in single codon usage for translational efficiency/gene expression (68). Nevertheless, the functional significance of many of the observed context effects remains far from being completely understood.
Codon Clustering
In addition to biases in synonymous codon usage relative to neighbouring codons in an mRNA (codon context/codon pair bias as described above), it has also been established that codon choice (particularly for rare codons) is biased according to a codon’s specific conserved location in an mRNA (36,38,62,63,80–83). Thus, regardless of an organism’s identity and the kingdom to which it belongs, rare codons have been shown to occur in clusters, enriched at a number of specific locations in gene ORFs rather than being randomly scattered across genes (36,38, 62,63,80–83). These include clusters of rare codons located at 5’ and 3’ ORF termini (62,63,80–83), a so-called “+70” rare codon cluster (located ∼35–40 codons downstream of the signal sequences (or transmembrane segments) in secreted proteins (36,80), and many other internal clusters located within gene ORFs (sometimes these internal clusters were found to encode linkers of protein structural domains) (38,62,63,84,85). Such placement of specific synonymous codons (especially rare codons) in gene ORFs suggests a functional role conserved in evolution beyond what would be expected by random chance. Below, evidence linking the clustering of rare codons in particular positions in mRNAs to translation initiation, protein secretion and protein folding is presented. This evidence makes it clear that codon choice has functional implications beyond amino acid coding and support of the predefined translation levels for the sequence of codons in an mRNA.
Codon clustering and initiation of protein translation
Clustering of rare codons at the 5' termini (typically at codon positions 1 to ∼20) of genes from E. coli and many other prokaryotes, as well as genes from some eukaryotes such as the yeast Saccharomyces cerevisiae (63), has been linked to the efficiency of translation initiation (a major rate-limiting step in protein synthesis (86–88)) and to the overall control of protein expression (63,81–83). Rare codons in many bacteria are mostly AT-rich (23); thus, mRNA regions harbouring these codons tend to possess reduced secondary structure. mRNA secondary structure at the 5' termini of ORFs can negatively affect protein expression by limiting access of ribosomes to the ribosome binding site (RBS) on the mRNA (63,81–83). Therefore, reduction of secondary structure in these 5’ regions due to clustering of AT-rich rare codons results in enhanced protein expression (81,82). Despite differences in translation apparatus and the mechanism of protein synthesis between prokaryotes and eukaryotes, many eukaryotic ORFeomes also display reduced 5’-terminal mRNA secondary structure near the start codon (89). In eukaryotes, this feature may potentially facilitate start-codon recognition by the scanning ribosome (89). In addition to enhancing translation initiation, preferential use of rare synonymous codons at the 5’ termini of genes/ORFs has also been proposed in certain cases to promote slow co-translational formation of the N-terminal folding nucleus, thus facilitating overall correct protein folding in the cell (38,63) (see below). Interestingly, preferential clustering of rare codons at 5’ ORF termini at yet another conserved location ∼70 codons downstream of the initiation codon (specific for secretory proteins) has been linked to protein secretion/membrane integration (36,80) (see below).
Codon clustering and protein secretion/membrane integration
A common and conserved location of rare codon clustering ∼56 to 75 codons downstream of the initiation codon in genes encoding secreted/transmembrane proteins has been originally identified in the late-1990s based on the analysis of 66 genes encoding bacterial or yeast membrane proteins (80). With signal sequences typically being 20 to 30 amino acids in length (90), this corresponds to a location ∼35–40 codons downstream of the signal sequences and/or transmembrane segments of these proteins (80) A functional role for this type of rare codon clustering was suggested by the so-called “+70 pause” hypothesis (80). This hypothesis proposed that, due to the generally lower cellular content of tRNAs corresponding to rare codons, “+70” rare codon clustering “might have evolved to give time for recognition of targeting signals by the proper machinery, and to temporally separate the insertion and folding of hydrophobic domains in a membrane from other protein assembly events” (80). Indeed, it has been experimentally shown that presence of rare codons at the “+70” position promotes nascent-chain recognition by the signal recognition particle (SRP), which assists in protein translocation across membranes (36). Similarly, strategically placed Shine-Dalgarno–like elements were identified in ORFeomes of E. coli secretory proteins (91,92). The Shine-Dalgarno (SD) sequence is generally located around 7-8 bases upstream of the start codon in prokaryotic mRNAs. It plays an important role in the formation of the translation initiation complex by base-pairing with the anti-SD sequence located at the 3′ end of 16S rRNA. SD sequences are generally avoided in the coding region of E. coli ORFeome, however the low-affinity SD–like sequences have variable rates of occurrence inside ORFeome and were found to transiently slow down the translation of the respective genes (93). The presence of these elements at the 5’ ORF termini was hypothesized to slow down translation elongation (similarly to “+70” rare codon cluster) in order to allow efficient integration of the transmembrane helix of membrane proteins (91,92).
Codon clustering and protein folding
Analyses of ORFeomes from prokaryotic and eukaryotic organisms revealed that rare codon clustering is not limited to a particular set of genes (high vs low expression), is not related to the GC content of the organism, and is significantly more prevalent than would be expected based on random selection (62,91). The locations of rare codon-rich regions along mRNAs seem to have been highly conserved throughout evolution as evidenced by their similarity across homologous protein families from different organisms (38,62,84,85,91,94–98). This led to the proposal that the placement of these rare codon clusters may be linked to protein structure (38,62,84,85,91,94–98). It has been suggested that synonymous codon usage may actively guide protein folding in the cell by modulating sequential folding events that take place during co-translational folding of proteins (37–43). It is believed that changes in the rate of translation determined by synonymous codon usage in an mRNA (rapid translation of regions with primarily preferred codons vs slow translation of regions with rare codon clusters) may facilitate co-translational folding (38). Strategically placed rare codon clusters (such as those encoding domain linkers) were suggested to allow temporal separation of folding events (such as domain and/or sub-domain folding) on the ribosome (84,85,91,94–98). In this regard, the observed enrichment of rare codons at 3' termini of ORFs of E. coli and 11 other prokaryotes (albeit less pronounced than clustering at 5’ termini of ORFs) was proposed to play a role in slowing down protein folding before release from the ribosome (38). The higher than expected frequency of rare codons observed at the 3' terminus of ORFs (e.g., in E. coli) may also be required for more robust termination of translation in some cases (63).
While the precise functional roles of many conserved codon clusters has yet to be determined, the overall significance of synonymous codon usage for protein folding and biogenesis has been highlighted by findings showing that synonymous codon changes can affect proteins’ activity (99–101), interactions with drugs and inhibitors (100), phosphorylation profiles (102), sensitivity to limited proteolysis (43,100,102,103), spectroscopic properties (104), aggregation propensity (43,104–106) and, ultimately, structure (43). Thus, differential synonymous coding use appears to provide a secondary code that guides in vivo protein folding and, through this mechanism, produces a multitude of effects on protein production and function.
Synonymous Codons and mRNA Stability
In addition to the effects of codon usage on translation and the encoded proteins described above, the synonymous codon choice can also impact the mRNA template itself. A link between codon usage and mRNA turnover rates has long been recognized in both prokaryotes and eukaryotes (107–109). mRNA turnover is a critical determinant of gene expression, and mRNAs with longer half-lives typically produce more protein. It was generally believed that mRNAs enriched in GC-rich codons would be more resistant to degradation (longer lived) than those with lower GC content due to their greater thermodynamic stability. However, recent work has shown that the efficiency of mRNA translation guided by codon usage (31–33) has a more broad and powerful influence on in vivo mRNA degradation rates than mRNA thermodynamic stability. This phenomenon has been demonstrated for unicellular (prokaryotic and eukaryotic) organisms such as E. coli (32) and the yeast S. cerevisiae (31), as well as a multicellular metazoan organism – zebrafish (33). It was found that stable/long-lived mRNAs harbour mainly preferred/optimal codons within their ORFs while many unstable/short-lived mRNAs have a higher frequency of un-preferred/non-optimal codons (31–33). Substitution of preferred codons with synonymous, un-preferred codons resulted in dramatic destabilization of the mRNA and vice versa. Given the established correlation between codon preference and translation rate, these findings suggested that transcript-specific translation elongation rate is an important determinant of mRNA stability and, thus, rapidly translated mRNAs would generally be expected to be more stable than slowly translated mRNAs (31–33). While the mechanism(s) underlying the connection between codon usage and mRNA stability have yet to be fully defined, it was shown in a zebrafish model that rare codons promote (in a translation- and 3’-UTR dependent manner) rapid deadenylation and subsequent degradation of mRNAs enriched in such codons by the deadenylase (CCR4-NOT) complex (33).
Synonymous codons and mistranslation
The numerous effects of synonymous codon usage on mRNA/protein biogenesis described above assume that the identity of the amino acid specified by a codon is not altered by use of one synonymous codon or another (hence the term “synonymous”). However, it is not always the case that “synonymous” changes in codons are silent; such changes can lead to incorporation of an amino acid that is different from the one encoded by the supposedly synonymous codons (70,71,110–113). This phenomenon is referred to as mistranslation or miscoding. Different codons demonstrate different levels of miscoding/mistranslation, varying by almost 20-fold (110 and references therein). For example, the frequency of miscoding of the AAU (Asn) codon in E. coli leading to incorporation of Lys (encoded by AAG and AAA) instead of Asn is about 4-fold higher than that for the AAC (Asn) codon (110). While mistranslation errors during protein synthesis are generally rare (occurring with frequencies ranging from 10−3 to 10−4 per codon; 110–113 and references therein), codon identity/sequence (as mentioned above), specific codon context, mispairing at the wobble position, and scarcity of cognate tRNAs can increase mistranslation levels by an order of magnitude (107–110). While these errors do occur and are affected by synonymous codon choice, computational and evolutionary analyses indicate that natural synonymous codon usage is biased toward specific patterns that reduce mistranslation as well as frameshifting (114).
Exonic Transcription Factor Binding and Codon Choice
Genomes contain many regulatory sequences specifying transcription factor (TF) binding. The regulatory code that specifies TF binding and the genetic code have long been assumed to operate independently of one another, as they were largely presumed to be segregated into the distinct (coding and non-coding) genomic regions. Nevertheless, the potential for some coding regions to accommodate transcriptional enhancers or splicing signals has been recognized (115–119). Recent analysis of positional occupancy patterns of specific TFs within the human exome in 81 diverse cell types revealed that preferred/frequent codons are also preferentially occupied by TFs (46). This reveals yet additional layer of information embedded in the genetic code and suggests that natural selection simultaneously exploits numerous possibilities and directions to optimize expression of the genetic information.
Concluding Remarks
Redundancy of the genetic code provided by multiple synonymous codons specifying a given amino acid was initially presumed to serve simply as a mechanism to minimize errors during translation of mRNAs into proteins, with synonymous codons having entirely identical functions. However, the finding that synonymous codons are not used with equal frequencies in different organisms, in different genes in a given organism, or even in different locations within a given gene suggested that they might have functional significance beyond amino acid coding. Codon use correlates with cognate tRNA levels and translation speed, and recent work has revealed that the relationship between codon use and translation rate impacts many important processes in the cell ranging from mRNA stability to protein folding. The mechanisms involved in all of the recently identified levels of information contained in the specific non-random utilization of synonymous codons in mRNAs remain to be fully defined and it is likely that additional effects of codon choice will be uncovered as well. The rapid progress in evolutionary, computational and synthetic biology should facilitate these efforts aimed at understanding this additional complex aspect of genetic code.
Acknowledgements
I apologize to those whose work or original publications could not be cited in this short article because of space limitations.
Conflict of Interest statement. None declared.
Funding
This work was supported by grants of the Human Frontier Science Program to A.A.K.: grant #RGP0024/2010, 13GRNT17070025 (AHA) and HL121779-01A1 (NIH).
References
- 1. Crick F.H., Barnett L., Brenner S., Watts-Tobin R.J. (1961) General nature of the genetic code for proteins. Nature, 192, 1227–1232. [DOI] [PubMed] [Google Scholar]
- 2. Nirenberg M., Leder P. (1964) RNA codewords and protein synthesis. The effect of trinucleotides upon the binding of sRNA to ribosomes. Science, 145, 1399–1407. [DOI] [PubMed] [Google Scholar]
- 3. Leder P., Nirenberg M. (1964) RNA codewords and protein synthesis. II. nucleotide sequence of a valine RNA codeword. Proc Natl Acad Sci U S A., 52, 420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernfield M.R., Nirenberg M.W. (1965) RNA codewords and protein synthesis. The nucleotide sequences of multiple codewords for phenylalanine, serine, leucine, and proline. Science, 147, 479–484. [DOI] [PubMed] [Google Scholar]
- 5. Trupin J.S., Rottman F.M., Brimacombe R.L., Leder P., Bernfield M.R., Nirenberg M.W. (1965) RNA codewords and protein synthesis, VI. On the nucleotide sequences of degenerate codeword sets for isoleucine, tyrosine, asparagine, and lysine. Proc. Natl. Acad. Sci. U. S. A., 53, 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nirenberg M., Leder P., Bernfield M., Brimacombe R., Trupin J., Rottman F., O'Neal C. (1965) RNA codewords and protein synthesis, VII. On the general nature of the RNA code. Proc. Natl. Acad. Sci. U. S. A., 53, 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brimacombe R., Trupin J., Nirenberg M., Leder P., Bernfield M., Jaouni T. (1965) RNA codewords and protein synthesis, 8. Nucleotide sequences of synonym codons for arginine, valine, cysteine, and alanine. Proc. Natl. Acad. Sci. U. S. A., 54, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Söll D., Ohtsuka E., Jones D.S., Lohrmann R., Hayatsu H., Nishimura S., Khorana H.G. (1965) Studies on polynucleotides, XLIX. Stimulation of the binding of aminoacyl-sRNA's to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids. Proc. Natl. Acad. Sci. U. S. A., 54, 1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goel N.S., Rao G.S., Ycas M., Bremermann H.J., King L. (1972) A method for calculating codon frequencies in DNA. J. Theor. Biol., 35, 399–457. [DOI] [PubMed] [Google Scholar]
- 10. Grantham R., Gautier C., Gouy M., Mercier R., Pavé A. (1980) Codon catalog usage and the genome hypothesis. Nucleic Acids Res., 8, r49–r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grantham R., Gautier C., Gouy M. (1980) Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res., 8, 1893–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ikemura T. (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol., 146, 1–21. [DOI] [PubMed] [Google Scholar]
- 13. Ikemura T. (1982) Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J. Mol. Biol., 158, 573–597. [DOI] [PubMed] [Google Scholar]
- 14. Bennetzen J.L., Hall B.D. (1982) Codon selection in yeast. J. Biol. Chem., 257, 3026–3031. [PubMed] [Google Scholar]
- 15. Hastings K.E.M., Emerson C.P., Jr. (1983) Codon usage in muscle genes and liver genes. J. Mol. Evol., 19, 214–218. [DOI] [PubMed] [Google Scholar]
- 16. Ikemura T. (1985) Codon usage and tRNA content in unicellular and multicellularorganisms. Mol. Biol. Evol., 2, 13–34. [DOI] [PubMed] [Google Scholar]
- 17. Sharp P.M., Tuohy T.M., Mosurski K.R. (1986) Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res, 14, 5125–5143., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharp P.M., Li W.H. (1987) The Codon Adaptation Index - a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharp P.M., Cowe E., Higgins D.G., Shields D.C., Wolfe K.H., Wright F. (1988) Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res., 16, 8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sørensen M.A., Kurland C.G., Pedersen S. (1989) Codon usage determines translation rate in Escherichia coli. J. Mol. Biol., 207, 365–377. [DOI] [PubMed] [Google Scholar]
- 21. Andersson S.G., Kurland C.G. (1990) Codon preferences in free-living microorganisms. Microbiol Rev., 54, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurland C.G. (1991) Codon bias and gene expression. FEBS Lett., 285, 165–169. [DOI] [PubMed] [Google Scholar]
- 23. Nakamura Y., Gojobori T., Ikemura T. (2000) Codon usage tabulated from the international DNA sequence databases: status for the year 2000. (2000). Nucleic Acids Res., 28, 292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bonekamp F., Andersen H.D., Christensen T., Jensen K.F. (1985) Codon-defined ribosomal pausing in Escherichia coli detected by using the pyrE attenuator to probe the coupling between transcription and translation. Nucleic Acids Res., 13, 4113–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Curran J.F., Yarus M. (1989) Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J. Mol. Biol., 209, 65–77. [DOI] [PubMed] [Google Scholar]
- 26. Ermolaeva M.D. (2001) Synonymous codon usage in bacteria. Curr. Issues Mol. Biol., 3, 91–97. [PubMed] [Google Scholar]
- 27. Duret L. (2002) Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev., 12, 640–649. [DOI] [PubMed] [Google Scholar]
- 28. Hershberg R., Petrov D.A. (2008) Selection on codon bias. Annu. Rev. Genet., 42, 287–299., 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 29. Sharp P.M., Emery L.R., Zeng K. (2010) Forces that influence the evolution of codon bias. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 365, 1203–1212. 10.1098/rstb.2009.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Behura S.K., Severson D.W. (2013) Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes. Biol. Rev. Camb. Philos. Soc., 88, 49–61. 10.1111/j.1469-185X.2012.00242.x. Epub 2012 Aug 14. [DOI] [PubMed] [Google Scholar]
- 31. Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R., Coller J. (2015) Codon optimality is a major determinant of mRNA stability. Cell, 160, 1111–1124. 0.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boël G., Letso R., Neely H., Price W.N., Wong K.H., Su M., Luff J.D., Valecha M., Everett J.K., Acton T.B., et al. (2016) Codon influence on protein expression in E. coli correlates with mRNA levels. Nature, 529, 358–363. 10.1038/nature16509. Epub 2016 Jan 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mishima Y., Tomari Y. (2016) Codon Usage and 3' UTR Length Determine Maternal mRNA Stability in Zebrafish. Mol Cell, 61, 874–885. 10.1016/j.molcel.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 34. Zalucki Y.M., Beacham I.R., Jennings M.P. (2009) Biased codon usage in signal peptides: a role in protein export. Trends Microbiol., 17, 146–150. 10.1016/j.tim.2009.01.005. Epub 2009 Mar 21. [DOI] [PubMed] [Google Scholar]
- 35. Zalucki Y.M., Beacham I.R., Jennings M.P. (2011) Coupling between codon usage, translation and protein export in Escherichia coli. Biotechnol. J., 6, 660–667. 10.1002/biot.201000334. [DOI] [PubMed] [Google Scholar]
- 36. Pechmann S., Chartron J.W., Frydman J. (2014) Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol., 21, 1100–1105. 10.1038/nsmb.2919. Epub 2014 Nov 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsai C.J., Sauna Z.E., Kimchi-Sarfaty C., Ambudkar S.V., Gottesman M.M., Nussinov R. (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol., 383, 281–291. 10.1016/j.jmb.2008.08.012. Epub 2008 Aug 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Komar A.A. (2009) A pause for thought along the co-translational folding pathway. Trends Biochem. Sci., 34, 16–24. 10.1016/j.tibs.2008.10.002. Epub 2008 Nov 6. [DOI] [PubMed] [Google Scholar]
- 39. Zhang G., Ignatova Z. (2011) Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol., 21, 25–31. 10.1016/j.sbi.2010.10.008. Epub 2010 Nov 24. [DOI] [PubMed] [Google Scholar]
- 40. O'Brien E.P., Ciryam P., Vendruscolo M., Dobson C.M. (2014) Understanding the influence of codon translation rates on cotranslational protein folding. Acc. Chem. Res., 47, 1536–1544., 0.1021/ar5000117. Epub 2014 May 1. [DOI] [PubMed] [Google Scholar]
- 41. Gloge F., Becker A.H., Kramer G., Bukau B. (2014) Co-translational mechanisms of protein maturation. Curr. Opin. Struct. Biol., 24, 24–33. [DOI] [PubMed] [Google Scholar]
- 42. Chaney J.L., Clark P.L. (2015) Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys., 44, 143–166. 10.1146/annurev-biophys-060414-034333. Epub 2015 Feb 26. [DOI] [PubMed] [Google Scholar]
- 43. Buhr F., Jha S., Thommen M., Mittelstaet J., Kutz F., Schwalbe H., Rodnina M.V., Komar A.A. (2016) Synonymous codons direct cotranslational folding towards different protein conformations. Mol. Cell, 61, 341–351. 10.1016/j.molcel.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sauna Z.E., Kimchi-Sarfaty C. (2011) Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet., 12, 683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 45. Hunt R.C., Simhadri V.L., Iandoli M., Sauna Z.E., Kimchi-Sarfaty C. (2014) Exposing synonymous mutations. Trends Genet., 30, 308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 46. Stergachis A.B., Haugen E., Shafer A., Fu W., Vernot B., Reynolds A., Raubitschek A., Ziegler S., LeProust E.M., Akey J.M., et al. (2013) Exonic transcription factor binding directs codon choice and affects protein evolution. Science, 342, 1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Santos M., Santos M.A. (2012) Structural and molecular features of non-standard genetic codes In Cannarozzi G.M., Schneider A. (eds), Codon Evolution: Mechanisms and Models. Oxford University Press, New York, NY, USA, pp. 258–271. [Google Scholar]
- 48. Bezerra A.R., Guimarães A.R., Santos M.A. (2015) Non-Standard Genetic Codes Define New Concepts for Protein Engineering. Life (Basel), 5, 1610–1628. 0.3390/life5041610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith D.W., McNamara A.L. (1971) Specialization of rabbit reticulocyte transfer RNA content for hemoglobin synthesis. Science, 171, 577–579. [DOI] [PubMed] [Google Scholar]
- 50. Smith D.W., Meltzer V.N., McNamara A.L. (1974) A comparison of rabbit liver and reticulocyte transfer RNA: evidence of unique species in reticulocytes. Biochim. Biophys. Acta, 349, 366–375. [DOI] [PubMed] [Google Scholar]
- 51. Weil S.C., Hirata R.K., McNamara A.L., Smith D.W. (1984) Changes in tRNA levels during the induction of hemoglobin synthesis in Friend leukemia cells. Biochem. Biophys. Res. Commun., 120, 707–713. [DOI] [PubMed] [Google Scholar]
- 52. Dong H., Nilsson L., Kurland C.G. (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol., 260, 649–663. [DOI] [PubMed] [Google Scholar]
- 53. Dittmar K.A., Sørensen M.A., Elf J., Ehrenberg M., Pan T. (2005) Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep., 6, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dittmar K.A., Goodenbour J.M., Pan T. (2006) Tissue-specific differences in human transfer RNA expression. PLoS Genet., 2, e221.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gingold H., Tehler D., Christoffersen N.R., Nielsen M.M., Asmar F., Kooistra S.M., Christophersen N.S., Christensen L.L., Borre M., Sørensen K.D., et al. (2014) A dual program for translation regulation in cellular proliferation and differentiation. Cell, 158, 1281–1292. 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 56. Topisirovic I., Sonenberg N. (2014) Distinctive tRNA repertoires in proliferating versus differentiating cells. Cell, 158, 1238–1239. 10.1016/j.cell.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 57. Bulmer M. (1987) Coevolution of codon usage and transfer RNA abundance. Nature, 325, 728–730. [DOI] [PubMed] [Google Scholar]
- 58. Duret L. (2000) tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet., 16, 287–289. [DOI] [PubMed] [Google Scholar]
- 59. Li W.H. (1987) Models of nearly neutral mutation with particular implications for nonrandom usage of synonymous codons. J. Mol. Evol., 24, 337–345. [DOI] [PubMed] [Google Scholar]
- 60. Shields D.C. (1990) Switches in species-specific codon preferences: the influence of mutation biases. J. Mol. Evol., 31, 71–80. [DOI] [PubMed] [Google Scholar]
- 61. Bulmer M. (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics, 129, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Clarke T.F., Clark P.L. (2008) Rare codons cluster. PLoS One, 3, e3412.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Clarke T.F., Clark P.L., 4th (2010) Increased incidence of rare codon clusters at 5' and 3' gene termini: implications for function. BMC Genomics, 11, 118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gouy M., Gautier C. (1982) Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res., 10, 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gutman G.A., Hatfield G.W. (1989) Nonrandom utilization of codon pairs in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A., 86, 3699–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Buckingham R.H. (1990) Codon context. Experientia, 46, 1126–1133. [DOI] [PubMed] [Google Scholar]
- 67. Buckingham R.H. (1994) Codon context and protein synthesis: enhancements of the genetic code. Biochimie, 76, 351–354. [DOI] [PubMed] [Google Scholar]
- 68. Tats A., Tenson T., Remm M. (2008) Preferred and avoided codon pairs in three domains of life. BMC Genomics, 9, 463. 10.1186/1471-2164-9-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moura G.R., Pinheiro M., Freitas A., Oliveira J.L., Frommlet J.C., Carreto L., Soares A.R., Bezerra A.R., Santos M.A. (2011) Species-specific codon context rules unveil non-neutrality effects of synonymous mutations. PLoS One., 6, e26817. 10.1371/journal.pone.0026817. Epub 2011 Oct 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Precup J., Parker J. (1987) Missense misreading of asparagine codons as a function of codon identity and context. J. Biol. Chem., 262, 11351–11355. [PubMed] [Google Scholar]
- 71. Parker J. (1989) Errors and alternatives in reading the universal genetic code. Microbiol. Rev., 53, 273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Irwin B., Heck J.D., Hatfield G.W. (1995) Codon pair utilization biases influence translational elongation step times. J. Biol. Chem., 270, 22801–22806. [DOI] [PubMed] [Google Scholar]
- 73. Coleman J.R., Papamichail D., Skiena S., Futcher B., Wimmer E., Mueller S. (2008) Virus attenuation by genome-scale changes in codon pair bias. Science, 320, 1784–1787. 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pacho F., Zambruno G., Calabresi V., Kiritsi D., Schneider H. (2011) Efficiency of translation termination in humans is highly dependent upon nucleotides in the neighbourhood of a (premature) termination codon. J. Med. Genet., 48, 640–644. 10.1136/jmg.2011.089615. Epub 2011 Jun 21. [DOI] [PubMed] [Google Scholar]
- 75. Kunec D., Osterrieder N. (2016) Codon pair bias is a direct consequence of dinucleotide bias. Cell Rep., 14, 55–67. 10.1016/j.celrep.2015.12.011. Epub 2015 Dec 24. [DOI] [PubMed] [Google Scholar]
- 76. Fedorov A., Saxonov S., Gilbert W. (2002) Regularities of context dependent codon bias in eukaryotic genes. Nucleic Acids Res., 30, 1192–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Boycheva S., Chkodrov G., Ivanov I. (2003) Codon pairs in the genome of Escherichia coli. Bioinformatics, 19, 987–998. [DOI] [PubMed] [Google Scholar]
- 78. Moura G., Pinheiro M., Silva R., Miranda I., Afreixo V., Dias G., Freitas A., Oliveira J.L., Santos M.A. (2005) Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol., 6, R28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Buchan J.R., Aucott L.S., Stansfield I. (2006) tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res., 34, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Képès F. (1996) The “+70 pause”: hypothesis of a translational control of membrane protein assembly. J. Mol. Biol., 262, 77–86. [DOI] [PubMed] [Google Scholar]
- 81. Kudla G., Murray A.W., Tollervey D., Plotkin J.B. (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science, 324, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goodman D.B., Church G.M., Kosuri S. (2013) Causes and effects of N-terminal codon bias in bacterial genes. Science, 342, 475–479. [DOI] [PubMed] [Google Scholar]
- 83. Bentele K., Saffert P., Rauscher R., Ignatova Z., Bluthgen N. (2013) Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol., 9, 675.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Krasheninnikov I.A., Komar A.A., Adzhubeĭ I.A. (1989) Frequency of using codons in mRNA and coding of the domain structure of proteins. Dokl. Akad. Nauk. SSSR, 305, 1006–1012. [PubMed] [Google Scholar]
- 85. Thanaraj T.A., Argos P. (1996) Ribosome-mediated translational pause and protein domain organization. Protein. Sci., 5, 1594–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gold L. (1988) Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem., 57, 199–233., 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 87. Gualerzi C.O., Pon C.L. (2015) Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell. Mol. Life. Sci., 72, 4341–4367. 10.1007/s00018-015-2010-3. Epub 2015 Aug 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jackson R.J., Hellen C.U., Pestova T.V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol., 11, 113–127. 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gu W., Zhou T., Wilke C.O. (2010) A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol., 6, e1000664–e1000610. 1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Paetzel M., Karla A., Strynadka N.C., Dalbey R.E. (2002) Signal peptidases. Chem. Rev., 102, 4549–4580. [DOI] [PubMed] [Google Scholar]
- 91. Chartier M., Gaudreault F., Najmanovich R. (2012) Large-scale analysis of conserved rare codon clusters suggests an involvement in co-translational molecular recognition events. Bioinformatics, 28, 1438–1445. 10.1093/bioinformatics/bts149. Epub 2012 Mar 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fluman N., Navon S., Bibi E., Pilpel Y. (2014) mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. Elife, 3, eLife.03440. 10.7554/eLife.03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li G.W., Oh E., Weissman J.S. (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature, 484, 538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Purvis I.J., Bettany A.J., Santiago T.C., Coggins J.R., Duncan K., Eason R., Brown A.J. (1987) The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol., 193, 413–417. [DOI] [PubMed] [Google Scholar]
- 95. Krasheninnikov I.A., Komar A.A., Adzhubeĭ I.A. (1988) Role of the rare codon clusters in defining the boundaries of polypeptide chain regions with identical secondary structures in the process of co-translational folding of proteins. Dokl. Akad. Nauk. SSSR, 303, 995–999. [PubMed] [Google Scholar]
- 96. Krasheninnikov I.A., Komar A.A., Adzhubeĭ I.A. Role of the code redundancy in determining cotranslational protein folding. Biokhimiia, 54, 187–200. [PubMed] [Google Scholar]
- 97. Widmann M., Clairo M., Dippon J., Pleiss J. (2008) Analysis of the distribution of functionally relevant rare codons. BMC Genomics, 9, 207. 10.1186/1471-2164-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. McKown R.L., Raab R.W., Kachelries P., Caldwell S., Laurie G.W. (2013) Conserved regional 3' grouping of rare codons in the coding sequence of ocular prosecretory mitogen lacritin. Invest. Ophthalmol. Vis. Sci., 54, 1979–1987. 10.1167/iovs.12-10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Komar A.A., Lesnik T., Reiss C. (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett., 462, 387–391. [DOI] [PubMed] [Google Scholar]
- 100. Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science, 315, 525–528. Epub 2006 Dec 21. [DOI] [PubMed] [Google Scholar]
- 101. Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y. (2015) Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell, 59, 744–754. 10.1016/j.molcel.2015.07.018. Epub 2015 Aug 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhou M., Guo J., Cha J., Chae M., Chen S., Barral J.M., Sachs M.S., Liu Y. (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature, 495, 111–115. 10.1038/nature11833. Epub 2013 Feb 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang G., Hubalewska M., Ignatova Z. (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol., 16, 274–280. 10.1038/nsmb.1554. Epub 2009 Feb 8. [DOI] [PubMed] [Google Scholar]
- 104. Sander I.M., Chaney J.L., Clark P.L. (2014) Expanding Anfinsen's principle: contributions of synonymous codon selection to rational protein design. J. Am. Chem., 136, 858–861., 10.1021/ja411302m. Epub 2014 Jan 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hu S., Wang M., Cai G., He M. (2013) Genetic code-guided protein synthesis and folding in Escherichia coli. J. Biol. Chem., 288, 30855–30861. 10.1074/jbc.M113.467977. Epub 2013 Sep 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kim S.J., Yoon J.S., Shishido H., Yang Z., Rooney L.A., Barral J.M., Skach W.R. (2015) Protein folding. Translational tuning optimizes nascent protein folding in cells. Science, 348, 444–448. 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 107. Hoekema A., Kastelein R.A., Vasser M., de Boer H.A. (1987) Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol. Cell. Biol., 7, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Caponigro G., Muhlrad D., Parker R. (1993) A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol., 13, 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Deana A., Ehrlich R., Reiss C. (1996) Synonymous codon selection controls in vivo turnover and amount of mRNA in Escherichia coli bla and ompA genes. J. Bacteriol., 178, 2718–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kramer E.B., Farabaugh P.J. (2007) The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA, 13, 87–96. Epub 2006 Nov 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zaher H.S., Green R. (2009) Fidelity at the molecular level: lessons from protein synthesis. Cell, 136, 746–762. 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kramer E.B., Vallabhaneni H., Mayer L.M., Farabaugh P.J. (2010) A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA, 16, 1797–1808. 10.1261/rna.2201210. Epub 2010 Jul 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ribas de Pouplana L., Santos M.A., Zhu J.H., Farabaugh P.J., Javid B. (2014) Protein mistranslation: friend or foe? Trends Biochem Sci., 39, 355–362. 10.1016/j.tibs.2014.06.002. Epub 2014 Jul 8. [DOI] [PubMed] [Google Scholar]
- 114. Huang Y., Koonin E.V., Lipman D.J., Przytycka T.M. (2009) Selection for minimization of translational frameshifting errors as a factor in the evolution of codon usage. Nucleic Acids Res., 37, 6799–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hyder S.M., Nawaz Z., Chiappetta C., Yokoyama K., Stancel G.M. (1995) The protooncogene c-jun contains an unusual estrogen-inducible enhancer within the coding sequence. J. Biol. Chem, 270, 8506–8513. [DOI] [PubMed] [Google Scholar]
- 116. Lang G., Gombert W.M., Gould H.J. (2005) A transcriptional regulatory element in the coding sequence of the human Bcl-2 gene. Immunology, 114, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ritter D.I., Dong Z., Guo S., Chuang J.H. (2012) Transcriptional enhancers in protein-coding exons of vertebrate developmental genes. PLoS One, 7, e35202. doi: 10.1371/journal.pone.0035202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Khan A.H., Lin A., Smith D.J. (2012) Discovery and characterization of human exonic transcriptional regulatory elements. PLoS One, 7, e46098. doi: 10.1371/journal.pone.0046098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Birnbaum R.Y., Clowney E.J., Agamy O., Kim M.J., Zhao J., Yamanaka T., Pappalardo Z., Clarke S.L., Wenger A.M., Nguyen L., et al. (2012) Coding exons function as tissue-specific enhancers of nearby genes. Genome Res., 22, 1059–1068. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]