Abstract
Prostate cancer (PCa) represents a significant clinical challenge because it is difficult to predict outcome and advanced disease is often fatal. We sequenced the whole genomes of 112 primary and metastatic PCa samples. From joint analysis of these cancers with those from previous studies, 930 cancers in total, we identified evidence for 22 novel putative coding driver genes, as well as evidence for NEAT1 and FOXA1 acting as drivers through non-coding mutations. Through the temporal dissection of aberrations, we identified driver mutations specifically associated with steps in the progression of PCa, for example establishing loss of CHD1 and BRCA2 as early events in cancer development of ETS fusion negative cancers. Computational chemogenomic (CanSAR) analysis of PCa mutations identified eleven targets of approved drugs, seven of investigational drugs and sixty-two compounds that may be active and should be considered candidates for future clinical trials.
Introduction
Prostate cancer is the most common solid cancer in men (diagnosed in 12%) and often fatal (9% of male cancer deaths). It is difficult to manage clinically due to a poor current understanding of what dictates its highly variable natural history, and of what underlies the development of castration-resistant disease1. Extensive data on the structure of prostate cancer genomes have been published2–6, including work from our own consortium7–10. These studies have identified a number of genetically distinct subgroups, including cancers with ERG, ETV1, ETV4, FLI1, SPOP, FOXA1 and IDH1 alterations. Overlapping with these categories, cancers may have alterations in PI3K and DNA repair pathways, with the latter significantly over-represented in advanced disease4. However, we have relatively limited understanding of the ordering of genetic events with the exception that ETS gene alteration appears to represent an early event, whilst mutations of AR are later, sometimes convergent, events, occurring in advanced and metastatic disease. Indeed, we have very little understanding of the evolution of mutational processes, the various genetic paths that cancers traverse on their way to progression, the levels of heterogeneity at different stages of development or the effect of these factors on clinical outcome.
Gene status has been used in studies designed to improve the poor predictive value of conventional clinical markers (PSA, Gleason sum, stage) and to develop disease management strategies. For example, genetic alteration of BRCA1/211, PTEN deletion12, amplification of AURKA together with the MYCN gene13, and coordinated loss of MAP3K7 and CHD114 have been reported to have prognostic value. A number of commercial prognostic tests based on gene expression profiles are also available15,16,17 and a classification framework has been proposed18. Improvements in the treatment of castration-resistant disease have been made through better targeting of AR regulation using abiraterone19 and enzalutamide20, whilst PARP inhibitors are effective against cancers harbouring BRCA1/2 mutations and other defects in DNA repair pathways21. However, significant advances have been made recently through the re-tasking of approved drugs22.
In the present study, we use previously unpublished whole genome DNA sequencing data in combination with published data to provide new insights into the mechanism of progression of prostate cancer to lethal disease, and to design novel molecular-based strategies for drug targeting.
Results
We whole genome sequenced cancerous and matched normal samples from 87 primary prostate cancers from the UK and 5 from China, together with 10 hormone-naïve prostate metastases and 10 castration-resistant metastases from the USA. Analysis (see Online Methods) reveals insights into the nature and order of acquisition of driver alterations, genomic heterogeneity in primary and metastatic cancers, changes in mutational signatures during progression, and potential drug targets. In addition, we identify coding and non-coding drivers by combining single nucleotide variants (SNVs) and small insertions/deletions (indels) within our dataset with those from TCGA4 (425 samples), the COSMIC database23 (243 samples) and Stand Up to Cancer24 (SU2C-PCF, 150 samples) to give a combined dataset, hereafter referred to as the ‘joint dataset’, comprising 710 primary cancers and 220 metastases. Supplementary Table 1 summarises the genes affected in both our study and the joint dataset.
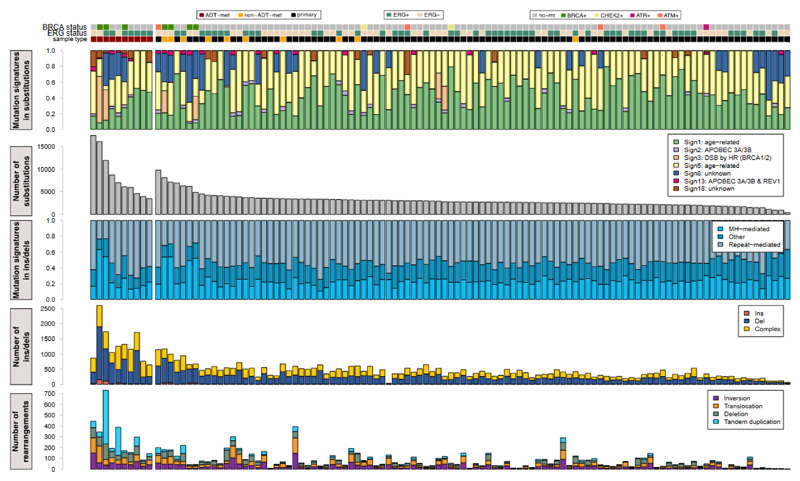
For the 112 cancer-normal pairs in our cohort, we identified 392,753 SNVs, 54,952 indels and 10,921 chromosomal rearrangements (Fig. 1). The mean genome-wide substitution rate was 1.23/Mb, with a significant difference in mutational burden between the primary (0.99) and metastatic (2.30) samples (P=4.4x10-15, Online Methods). Moreover, within the metastatic subset, mutation burden was higher in men treated with androgen deprivation therapy (ADT or CRPC) than treatment-naïve cases (2.98 vs 1.61, P=0.015). There were also significantly more rearrangements in metastatic than in primary samples (P=0.0059), whilst the proportion of breakpoints attributed to a chromoplexy-like event25 was indistinguishable between the two groups. Within the metastatic group, the ADT samples had more rearrangements than did the hormone-naïve (P=0.027), with no difference in the proportion of chromoplexy-like events (Fig. 1).
Figure 1. Mutational landscape of prostate cancers.
From top-to-bottom: mutation status of DNA repair genes, ETS fusion status and sample type; proportion of mutations assigned to each signature48; number of SNVs identified in each sample; proportion of small insertions/deletions associated with microhomology or repetitive regions; number of insertions, deletions and complex insertions/deletions in each sample; total number of structural variants in each sample, separated into inversions, translocations, deletions and tandem duplications. Sample ordering is reported in Supplementary Table 7.
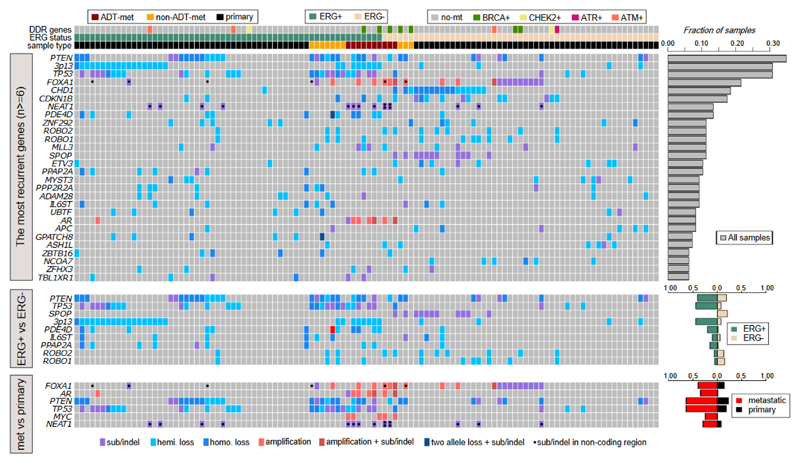
Genes of interest were identified through a comprehensive set of analyses to identify: excess non-synonymous mutations in coding regions; excess missense mutations within a gene, indicative of an oncogenic driver; excess mutations in non-coding regions; regions with an excess of structural variants in either ETS+ or ETS- cancers; regions with recurrent copy number aberrations in either ETS+ or ETS- cancers. Overall, we identified 73 genes with evidence for involvement in prostate cancer development (Fig. 2, Table 1, Supplementary Table 2). Based on a literature search, each gene was assigned a high, medium or low level of previous supporting evidence (Table 1, Supplementary Table 2). In addition to 22 genes with little or no previous evidence of involvement in prostate cancer (Table 1, ‘low’ previous evidence), we provide corroborating evidence for 8 further genes previously lacking strong evidence of driving prostate cancer (Table 1, ‘medium’ previous evidence).
Figure 2. Landscape of driver genes in prostate cancer.
Genes were identified using three different methods: upper panel shows genes that have undergone genetic aberration in at least 6 samples (n=112 biologically independent samples); middle panel shows genes with aberrations enriched in either ERG+ or ERG- cancers (Fisher exact test for PTEN, TP53, SPOP, 3p13, PDE4D, PPAP2A; ROBO1 and ROBO2 are in a region enriched for SVs in ETS- tumors; IL6ST is in a region enriched for SVs in ETS+ tumors; n=59 ETS+, n=53 ETS biologically independent samples); lower panel shows genes enriched in metastatic samples (Fisher exact test, n=20 metastatic, n=98 primary biologically independent samples). Right-hand bar graphs show the fraction of samples bearing each type of aberration. DDR = DNA damage response, ‘hemi.loss’ = loss of heterozygosity resulting from copy number change, ‘homo.loss’ = homozygous deletion resulting from copy number aberration, ‘two allele loss + sub/indel’ indicates genes in triploid regions bearing aberrations of all 3 gene copies. Sample ordering is reported in Supplementary Table 7.
Table 1. Putative driver genes.
Genes were identified in our study using several methods, detailed in the last column: dN/dS; enrichment for SVs or CNAs in ETS+ or ETS- cancers; enrichment for truncating mutations or homozygous deletions, clinical correlation. From a PubMed literature search, prior evidence for each gene being a driver of prostate cancer was classified as ‘low’ if the gene has not been previously reported as playing a role in prostate cancer tumorigenesis or progression. Isolated alterations may have been observed or biological evidence for importance may have been presented as indicated in the prior evidence column. Prior evidence was classified as ‘medium’ for genes reported previously as playing a role in prostate carcinogenesis or progression but currently lacking statistical support based on genetic alterations. Evidence considered included presence of multiple genetic alterations, SNP associations, and known cancer genes in other tissues. The high confidence genes are those that are widely accepted to represent cancer genes and to be altered in prostate cancer: this would include genes such as HRAS, SPOP, IDH1 etc. In each case there are two or more of the following: statistical verification of higher incidence, biological experiments, clinical correlations, confirmation in multiple studies, recognition as cancer genes in other cancer types. dN/dS = non-synonymous: synonymous ratio, calculated for all SNVs and indels; dN/dS (missense) = non-synonymous: synonymous ratio, calculated for missense SNVs only; SV = structural variant; CNA = copy number aberration; SNV = single nucleotide variant; indel = small insertion/deletion; ETS = E26 transformation-specific.
| gene | Mutation type(s) | Previous evidence | Prior evidence | Evidence in our study |
|---|---|---|---|---|
| ADAM28 | SV, CNA | low | 59biological evidence | SVs and CNA in ETS+ |
| ANTXR2 | SV, SNV/indel | low | none | clinical correlation |
| ASH1L | SV, SNV/indel | low | 25 | truncating mutations, SVs in ETS- |
| CDH12 | SV | low | none | clinical correlation |
| FOXOl | CNA | low | 60biological evidence | CNA in ETS- |
| IL6ST | SV | low | 61biological evidence | dN/dS, SVs and CNA in ETS+, clinical correlation |
| LCE2B | SNV/indel | low | none | dN/dS (missense) |
| MAP3K1 | SV, CNA | low | none | SVs, CNA in ETS+ |
| MYST3 | SV | low | 25 | SVs in ETS-, RNA expression |
| NCOA7 | SV | low | none | SVs in ETS- |
| NDST4 | SNV/indel | low | none | dN/dS (missense) |
| NEAT1 | non-coding | low | 31biological evidence | non-coding |
| PDE4D | SV | low | 62SNP data | SVs and CNA in ETS+ |
| PPAP2A | SV | low | 62SNP data | SVs and CNA in ETS+ |
| PPP2R2A | SV | low | 63biological evidence | SVs and CNAs in ETS+ |
| ROBO1 | SV | low | 64biological evidence | SVs in ETS+ |
| R0B02 | SV | low | 25 | SVs in ETS+ |
| RPL11 | SNV/indel | low | 25 | dN/dS (missense) |
| SENP6 | SV | low | 42biological evidence | enriched SVs, RNA expression |
| TBL1XR1 | SNV/indel, SV | low | 65known AR co-regulator biological evidence | dN/dS |
| USP28 | SV, CNA, SNV/indel | low | none | SVs, CNA, SNV/indel |
| ZNF292 | SV, CNA SNV/indel, | low | 25 | enriched SVs, homozygous deletions, truncating mutations |
| ARID1A | SNV/indel | medium | 55 | dN/dS |
| CASZ1 | SNV/indel | medium | COSMIC, TCGA and SU2C | dN/dS |
| CNOT3 | SNV/indel | medium | 67Mut. in leukemia | dN/dS (missense) |
| LRP1B | SV, CNA | medium | 62SNP data | SVs and CNA in ETS- |
| PIK3R1 | SNV/indel | medium | 24 | dN/dS |
| RGMB | CNA | medium | 38deletions | CNA in ETS- |
| TBX3 | SNV/indel | medium | known breast cancer gene | dN/dS |
| ZMYM3 | SNV/indel | medium | COSMIC SU2C | dN/dS |
Coding drivers
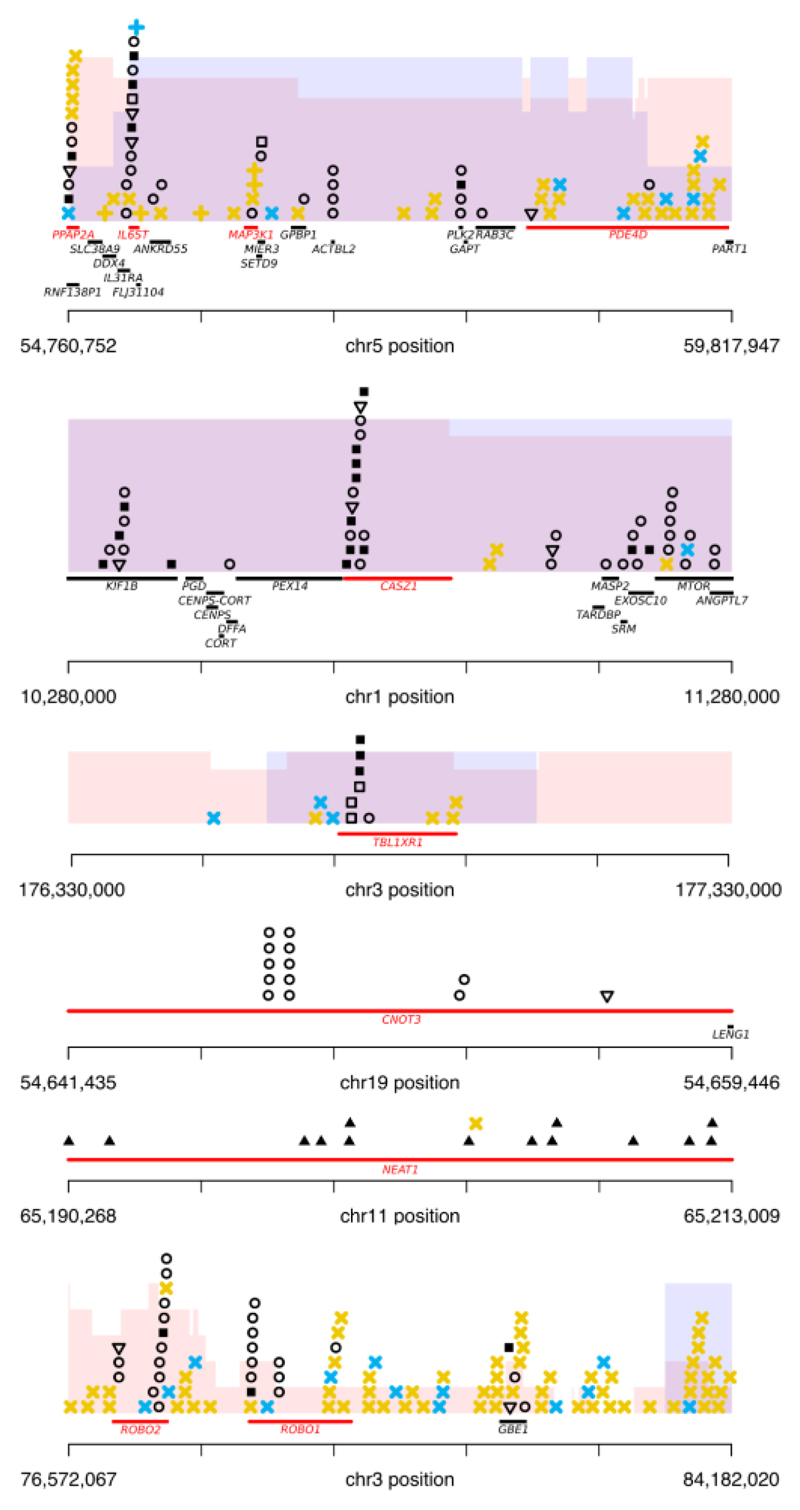
We identified 28 genes with an excess of non-synonymous coding mutations, five of which are previously unknown drivers in prostate cancer (Supplementary Table 2). TBL1XR1 was enriched in truncating SNVs and indels and is also located in a genomic region enriched for rearrangements in ETS+ cancers (chr3: 172-179Mb) (Fig. 3). These rearrangements result in loss of heterozygosity (LOH) or, in one case, homozygous deletion, suggesting a cancer suppressor role for this gene. Another significantly mutated gene primarily affected by truncating mutations was ZMYM3, which encodes a component of CoREST, a transcriptional repressor complex including REST (RE-1 silencing transcription factor) and involved in suppression of neuronal differentiation-related genes in non-nervous tissues26. In addition, two further CRPC samples from the SU2C-PCF study24 had nonsense mutations and one sample within our study had a 70kb exonic deletion in REST.
Figure 3. Putative novel driver genes.
Putative drivers are shown in red and genomic aberrations are displayed as: missense SNVs – circles; nonsense SNVs – open triangles; essential splice site mutations – open squares; indels – closed squares; non-coding mutations – closed triangles; simple SV - yellow cross; chromoplexy event – blue cross; region enriched for loss of heterozygosity, with height proportional to the number samples containing LOH - pink shading; region enriched for homozygous deletions, with height proportional to the number of samples containing homozygous deletion – blue shading.
Two other genes with recurrent truncating mutations were IL6ST and CASZ1 (Fig. 3). The latter is a putative cancer suppressor in neuroblastoma27 while the former encodes glycoprotein 130, the signal-transducing subunit of the interleukin 6 (IL6) receptor. The pattern of mutations we observe in the joint dataset for IL6ST is dominated by truncating events. Moreover, this gene is located in a genomic region recurrently rearranged in ETS+ cancers, resulting in either LOH or homozygous deletion (four cases of each), suggesting a cancer suppressive role. TBX3, previously reported to harbor mutations in breast cancer28, exhibited a mixed pattern of mutations with mostly missense mutations and two cancers harbouring truncating events.
Analysis of missense mutations identified recurrent mutations in seven further genes, of which two are newly reported (Supplementary Table 2). CNOT3 exhibited mutation hotspots in two amino acid positions, p.E20K (4/932 samples) and p.E70K (5/932 samples), as well as a nonsense mutation in a single sample (Fig. 3). CNOT3 has a known cancer suppressive function in T-cell acute lymphoblastic leukaemia29. Enrichment for missense mutations was identified in RPL11 a ribosomal protein and putative cancer suppressor upstream of the MDM2/TP53 pathway30. In contrast to previous studies, the enrichment for missense mutations in both CNOT3 and RPL11 suggests oncogenic, rather than tumor suppressor, roles in prostate cancer.
A comparison between coding mutations in metastatic and primary samples within the joint dataset identified enrichment in metastases for mutations in TP53, AR, KMT2C, KMT2D, RB1, APC, BRCA2, CDK12, ZFHX3, CTNNB1, PIK3CB (Supplementary Table 2), confirming previous studies3,24.
Non-coding drivers
Analysis of non-coding components of the genome identified two regions significantly enriched for mutations. NEAT1, a lncRNA recently reported to be associated with PCa progression31, was mutated in 13/112 ICGC cases with significant over-representation in patients with metastatic disease (6/20 metastases vs. 7/91 primaries, Fisher exact test, P=0.012, Fig. 3). Interestingly, out of the metastatic cases NEAT1 mutations were found only in patients that had undergone ADT, consistent with the link between high NEAT1 expression and resistance to AR-targeting therapies31. Notably, two of these six cases had two separate NEAT1 mutations. The FOXA1 promoter also had significant evidence of selection. This gene modulates AR-regulated transcriptional signalling32 and has previously been found to harbor recurrent coding mutations5. In our series, we identified 14 samples with coding and 6 samples with non-coding mutations, with two samples (PD14721a and PD12813a) bearing both a coding and a non-coding mutation. Interestingly, we also identified mutations in the FOXP1 promoter, a gene with known cancer-suppressive effect in prostate tumorigenesis33, in three samples, but this was insufficient to reach statistical significance.
Structural variant enrichment in ETS+ and ETS- cancers
The density of rearrangements varies across the genome as a result of various factors including chromatin state, GC content, gene density, replication timing and repetitive sequence. In order to remove the effect of these factors, we segmented inter-breakpoint distance across the genome separately in ETS+ and ETS- cancers and identified regions with differential enrichment for rearrangements between the two subtypes. The functional importance of many of these regions was supported by an excess of truncating mutations or CNAs.
In addition to regions previously identified as enriched for rearrangements in ETS+ cancers (FOXP1, RYBP, SHQ1, PTEN, and TP53)34–37, two unreported regions were identified. The region chr5:55-59Mb covers the genes PPAP2A, PDE4D, MAP3K1 and IL6ST (Fig. 3). In IL6ST we also detected significant enrichment for coding mutations, suggesting this is the main target of the aberrations. In chr3:171:178Mb, TBL1XR1 is similarly enriched for both rearrangements and truncating mutations.
In ETS- cancers, we confirmed a previously reported enrichment for rearrangements containing CHD1(38,39). A target of enriched rearrangements in the region chr1:149-158Mb is likely ETV3. In 5/9 cancers, ETV3 was exclusively affected by these events (4 LOH by deletion and 1 by translocation). Additionally, one cancer had a truncating mutation (p.R413fs*3) and two had missense mutations (p.A73V and p.L37Q). In total, 12 patients had localised alteration, 10 of whom had ETS- cancers. Moreover, within the joint dataset, there are four cancer samples with truncating mutations in this gene. In contrast to ETV4, the nature of variants in ETV3 is indicative of a tumor suppressive role in PCa. Manual inspection of the recurrently rearranged region chr3:76-84Mb identified ROBO1 and ROBO2 as possible targets (Fig. 3). In total 16/112 samples had an event affecting one or other of these genes, and in four samples both were affected. Previously implicated in pancreatic ductal adenocarcinoma40, these two genes have not been previously reported in the context of PCa.
Events enriched at chr6:80-114Mb indicate that ZNF292 is a possible target. 11/112 patients (5 ETS+ and 6 ETS-) had loss of at least one chromosome copy and in two patients there was a homozygous loss specifically targeting ZNF292. Moreover, the joint dataset contained 5/932 samples with a truncating mutation, further suggesting a cancer suppressive function for this gene in PCa. Another gene affected by recurrent rearrangements on 6q was SENP6, a small ubiquitin-like modifier (SUMO)-specific protease that removes SUMO polypeptides from conjugated proteins41, and possibly plays a role in AR function42. Of note, 4/5 rearrangements in this region affected SENP6 only, leading to a significant reduction in expression (Supplementary Fig. 1). Finally, located at chr6:126Mb, the nuclear receptor co-activator NCOA7 was altered in six samples, one sample having homozygous loss.
Further regions enriched in ETS- cancers were chr2:133-144Mb (LRP1B), chr8:112-114 (CSMD3) and chr8:40-41Mb (MYST3). The first two genes are very large and fall within reported fragile sites43. Nevertheless, preferential enrichment of breakpoints in ETS- cancers may suggest either that underlying structure, such as AR binding sites or nucleosome structure, or epistatic interactions between ETS fusion and other rearrangements affect the occurrence of rearrangements at these loci. Samples containing structural variants affecting MYST3 were found to have significantly reduced RNA expression (Supplementary Fig. 1).
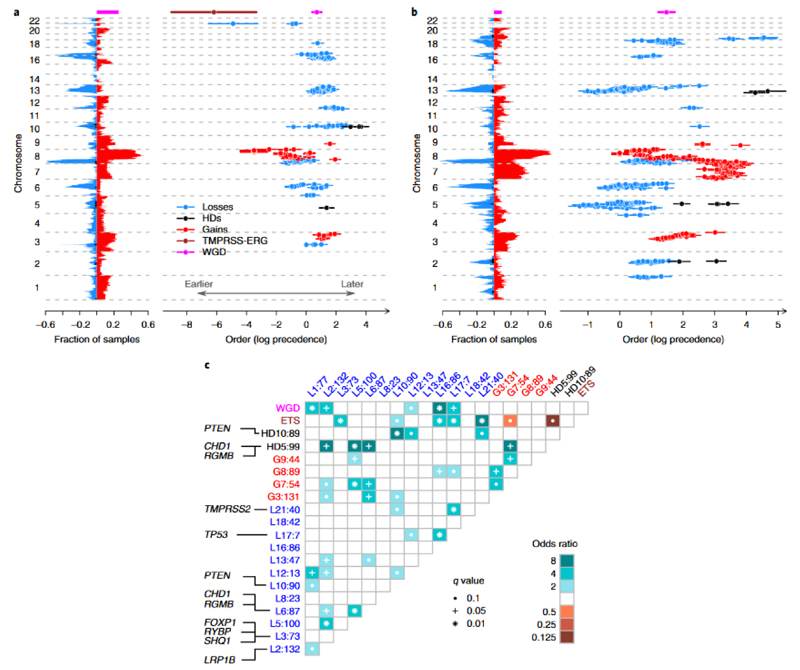
Timing of copy number aberrations
In order to identify routes to progression in PCa, we developed a novel approach to order the occurrence of copy number aberrations by combining information on: the clonality of copy number aberrations; timing relative to whole genome duplication; timing of homozygous deletions relative to neighboring hemizygous losses. Information from all tumors was combined using a Bradley-Terry model, to give the most likely ordering of events. By applying a set of logical rules (see Online Methods), we deciphered the temporal ordering of subclonal CNAs within each cancer. In general, homozygous deletions appear late in oncogenesis, corroborating previous findings that homozygous deletions are associated with advanced disease44–46. Clear differences emerge in the evolution of ETS+ and ETS- PCa’s. Where present, the deletion between the TMPRSS2 and ERG genes in ETS+ cancers was an early (generally clonal) event, as was gain of chr8q within the locus 112 – 137Mb (Fig. 4a). The earliest homozygous deletions in ETS+ cancers include chr5: 55Mb-59Mb, corroborating the rearrangements targeting PPAP2A, PDE4D, MAP3K1 and IL6ST, and chr10:89Mb-90Mb, which covers PTEN (Figs. 3 and 4a).
Figure 4. Temporal evolution of copy number aberrations in ETS+ and ETS- prostate cancer.
For (a) ETS+ cancers (n=45 biologically independent primary cancer samples), and b) ETS- cancers (n=47 biologically independent primary cancer samples): Left: The landscape of copy number aberrations with genomic loci plotted against fraction of cancers. Loss-of-heterozygosity is depicted in blue, homozygous deletions in black, gains in red, TMPRSS2-ERG deletion in brown and whole genome duplication in green. Right: The temporal evolution of significantly recurrent (p < 0.05, permutation test with Benjamini-Hochberg procedure) copy number aberrations by genomic loci over time (mean with 95% confidence intervals, log precedence relative to arbitrary reference). Lower values indicate earlier events (c) Pairwise associations among copy number aberrations. Recurrently aberrant regions with a false discovery rate < 0.1 are shown. Associations are indicated by odds ratio (OR) with brown colors depicting mutually exclusive events and blue-green colors depicting correlated events. Genomic loci annotated by: type of aberration (G=gain, L=loss, HD=homozygous deletion); chromosome; median position in Mb. For focal events the putative target genes are annotated.
In ETS- cancers, losses at chr5:60–100Mb (CHD1 and RGMB), chr13:32-91Mb (which includes BRCA2, RB1 and FOXO1), and chr6:73-120Mb are followed by losses at chr2:124-142Mb, then by gains at chr3:100-187Mb, and then whole chromosome gain of chr7 (Fig. 4b). Loss of CHD1 has been previously implicated in the initiation of ETS- prostate cancers, preventing ERG re-arrangement in the prostate38 and our data confirm the exclusivity between ETS positivity and homozygous loss of CHD1 (Fig. 4c).
In both ETS+ and ETS- cancers, whole genome duplication (WGD) correlated with loss of chromosomal segments at: chr1:94Mb, chr2:140Mb, chr12:12Mb, chr16:85Mb and chr17:7Mb (Fig. 4c). From timing analysis, these losses appear to occur co-synchronously with WGD in most cases. Gains at chr8:101Mb occurred prior to WGD, chr3:131Mb occurred synchronously, and gains at chr7:88Mb tended to follow WGD.
Timing point mutations and indels
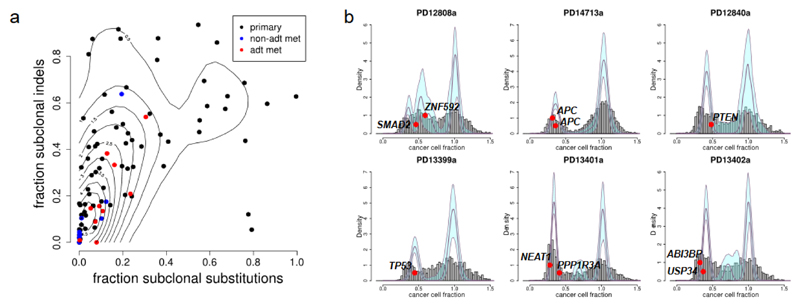
SNVs and indels were clustered according to their cancer cell fraction (CCF) using a Bayesian Dirichlet process47. The proportion of SNVs identified as subclonal showed considerable variation across cancers, but was significantly higher in primary than metastatic samples (Fig. 5a, P=0.022, Wilcoxon rank sum test), as was the proportion of subclonal indels (P=0.00033) and the fraction of the genome with subclonal copy number aberrations (P=0.0037, Supplementary Fig. 2). This is apparent evidence for a bottleneck in acquiring metastatic potential rather than a response to treatment, since levels of heterogeneity in untreated metastases are no lower than in androgen-deprived metastases (Fig. 5a).
Figure 5. Heterogeneity and subclonal mutations.
(a) Metastatic tumors have less heterogeneity than primary tumors, whether assessed from SNVs or indels. Each dot represents a different sample, colored by sample type. x-axis = fraction of SNVs that are subclonal, y-axis = fraction of indels that are subclonal, contour lines calculated using R package kde2d. n= 93 biologically independent samples (10 ADT metastases, 9 hormone naïve metastases, 74 primary tumors) (b) Samples with multiple subclonal mutations in driver genes. Fraction of cancer cells carrying mutation is shown as grey histogram for all mutations and as red ovals for mutations in known driver genes. Mutations are clustered using a Dirichlet process as previously described47, with thick plum-colored lines indicating fitted distribution and pale blue regions indicating 95% posterior confidence intervals. Peaks with a subclonal fraction close to 1 are clonal, whereas peaks at lower subclonal fractions indicate subclonal mutations.
The levels of heterogeneity observed in SNVs and indels were correlated (Fig. 5a, Pearson r = 0.57, P=2.3x10-9). Higher levels of heterogeneity were observed amongst indels than SNVs (P=2.4x10-8). However, it cannot be ruled out that variant calling of indels may have greater sensitivity for low allele frequency variants than calling of SNVs.
Driver SNVs were identified as clonal or subclonal in each sample according to the cluster to which they were assigned, with 84 classified as clonal and 22 (21%) as subclonal. Our power to detect subclonal mutations is limited by sequencing depth and the real number of subclonal driver mutations is likely much higher. The driver mutations identified as subclonal include two mutations in APC in the same sample, PD14713a. Interestingly, this cancer has undergone clonal loss of one copy of chr5q, followed by mutations in APC in 2 different subclones (Fig. 5b and Supplementary Fig. 3), suggesting convergent evolution. Five other samples each have two subclonal drivers: PD12808a has a missense mutation in ZNF292 and an essential splice site mutation in SMAD2; PD13401a has a nonsense mutation in PPP1R3A and a mutation in the promoter of NEAT1; PD13402a has a nonsense mutation in USP34 and an essential splice site mutation in ABI3BP (Fig. 5b); PD12820a has a missense mutation in USP48 and an essential splice site mutation in ASXL2; PD13389a has a frameshift mutation in PHF12 and an essential splice site mutation in TBX3 (not shown).
Subclonal mutations are also seen in several common drivers including one in TP53 (PD13339a) and one in PTEN (PD12840a). On the other hand, SPOP was mutated in 10 samples, always clonally and always in ETS- tumors (Fig. 2).
Mutational signatures
Analysis of the mutational signatures by non-negative matrix factorisation (NMF) revealed that, in addition to the ubiquitous ‘clock-like’ signatures 1 and 5, there was presence of the previously described signatures 2, 3, 8, 13 and 1848. Signature-3-positive samples were enriched for germline/somatic mutations in BRCA1/2 genes (4/6 samples) as reported previously48 (Fig. 1). However, the presence of high levels of microhomology (MH)-mediated deletions was even more strongly correlated with the presence of BRCA mutations (6/6 samples). Separating the mutations into early clonal, late clonal and subclonal epochs, as described in Online Methods, revealed that the proportion of signature 1 mutations decreases over time, suggesting an increase of cancer-associated mutagenic processes relative to innate processes (P=2.2x10-16, test for trend in proportions).
Signature 13, previously associated with the activity of the AID/APOBEC family of cytidine deaminases, was over-represented in advanced disease, 45% (9/20) in metastases vs. 14% (14/92) in primaries (Fisher exact test, P=5.6x10-3). Similarly, signature 18, which has been previously associated with failure of base excision repair and to the accumulation of mutations from 8-Oxoguanine damage49, was enriched in advanced disease, 40% (8/20) in metastases vs. 11% (10/92) in primaries (Fisher exact test, P=3.8x10-3). In a recent report of 560 breast cancer whole-genomes, signature 8 correlated with DNA damage repair deficiency50. Androgen signalling is known to positively regulate multiple genes involved in DNA repair51,52, while androgen deprivation impairs DNA double-strand break repair53. In support of these previous reports, the proportion of mutations assigned to signature 8 is consistently higher amongst later appearing (subclonal) populations of cells (55% ± 24%) than earlier (clonal) populations (28% ± 12%) (t-test, P=1.3×10-4, Supplementary Table 3). The proportion of metastases with evidence for the action of signature 8 was higher than that for primary tumors, although not reaching statistical significance (8/20 metastases, 25/92 primaries, Fisher exact test P=0.28). Increased prevalence of DNA-damage related genes in metastatic prostate cancer as well as the observations made in this study warrant an extensive study of mutational signatures in therapy-naïve disease and CRPC in a larger dataset to explore the relevance of check-point inhibition as an alternative therapy for advanced prostate cancer.
Clinical correlates
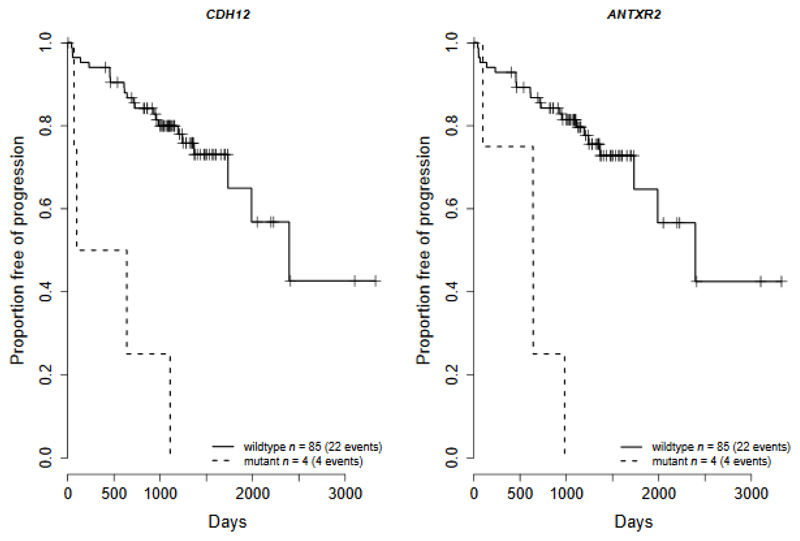
CDH12 and ANTXR2 alterations were significantly associated with time to biochemical recurrence (Benjamin-Hochberg adjusted P = 0.0060 (CDH1) & 0.012 (ANTXR2), HR = 9.3 & 7.7, Cox regression model, Fig. 6), and were significant predictors of biochemical recurrence independent of cofactors Gleason, PSA at prostatectomy, and pathological T-stage (P = 0.00061 (CDH1) & 0.0015 (ANTXR2), HR = 7.3 & 6.5, Cox regression model, Supplementary Table 4). A Cox regression model containing a combination of CDH12, ANTXR2, SPOP, IL6ST, DLC1 & MTUS1 mutations was determined to be an optimal predictor of time to biochemical recurrence and was a significant improvement over a baseline model of Gleason, PSA at prostatectomy, and pathological T-stage (model χ2 test, P = 0.00053). The number of mutational signatures identified in a cancer was negatively correlated with time to biochemical recurrence in prostatectomy patients (P = 0.014, HR = 3.0; Cox proportional hazards model on number of processes greater than 3, Supplementary Fig. 4) and is an independent predictor (P = 0.0061, HR = 3.6; Cox proportional hazards model). The number of SNVs detected was also an independent prognostic biomarker (P=0.031, HR=1.005; Cox proportional hazards model). The numbers of both samples and events within this study are modest and further analysis of larger cohorts is required to establish firmly these findings.
Figure 6. Clinical outcome. Kaplan-Meier plots for biochemical recurrence.
Kaplan-Meier plots of recurrent mutated genes where there is a significant correlation with time to biochemical recurrence after prostatectomy, CDH12 (left, p=0.006) and ANTXR2 (right, p=0.012) (Cox regression model; Benjamini-Hochberg multiple testing correction). Clinical information was available for 89 prostatectomy samples with WGS data, with a median follow up of 1108 days in which biochemical recurrence occurred in 26 patients. The mutations in both genes consisted of a frameshift deletion in one sample and structural variants in the remaining samples.
Druggable targets in the prostate cancer disease network
A key opportunity arising from systematic analyses of cancer genomics is the early identification of therapeutic intervention strategies. To this end, we applied established chemogenomic technologies using the canSAR knowledgebase54 to map and pharmacologically annotate the cellular network of the prostate disease genes identified in this study. We derived the network using curated protein-protein and transcriptional interaction data. We included the protein products of the genes identified in this study and other key proteins that directly interact with these proteins or affect their function (see Online Methods and Supplementary Fig. 5 for details). This resulted in a focussed prostate network of 156 proteins. Each protein was annotated based on multiple assessments of ‘druggability’, i.e. the likelihood of the protein being amenable to small molecule drug intervention (Table 2 and Supplementary Table 5). We find that PCa driver genes are embedded in a highly druggable cellular network that contains eleven targets of approved therapies and seven targets of investigational drugs. As well as the Androgen Receptor (AR) and the Glucocorticoid Receptor (GR), the network contains targets of drugs approved for other indications, several of which (e.g. BRAF, ESR1, RARA, RXRA, HDAC3) are under clinical investigation for PCa.
Table 2. Drug targets identified from CanSAR analysis.
Proteins in bold typeface are derived from genes identified as prostate drivers in this study or proteins that have a significant known interaction with these proteins.
| Target of approved drug |
| AR, BRAF, ESR1, HDAC3, KCNH2, MAP2K1, NR3C1, RARA, RARB, RARG, RXRA |
| Target of investigational drug |
| AKT1, ATM, MDM2, PDE4D, PIK3CA, PIK3CB, TP53 |
| Target being investigated chemically |
| AHR, BRCA1, CTNNB1, HRAS, IDH1, JUN, MAP3K1, MEN1, NCOR1, NCOR2, NR4A1, PIK3R1, PPP2R2A |
| Predicted target by structure-based method |
| ANTXR2, APC, ARNT, ASH1L, BRCA2, CBFA2T2, CDH12, CDK12, CHD1, CREBBP, DLC1, DOCK10, ERG, ETV3, FOXA1, FOXG1, FOXO1, FOXO4, FOXP1, GATA1, GATA2, HDGF, HNF4A, IL6ST, KAT6A, KDM4A, KDM6A, KMT2C, KMT2D, NKX3-1, PIAS1, PIAS2, PTEN, RB1, RGMB, RNF43, SKI, SMAD2, SMAD3, SMAD4, SMARCA4, SPDEF, SPOP, TBL1X, TBL1XR1, TBX3, TP73, ZBTB16, ZHX2 |
Seven proteins within the prostate network are targets of drugs currently in clinical trials. In particular, the ataxia-telangiectasia mutated (ATM) inhibitor AZD-0156, currently in Phase 1 for safety assessment, is a likely candidate for exploration in PCa due to the recently described role of DNA damage repair, particularly in advanced PCas21,55. The network highlights targets of PI3 Kinase pathway inhibitors (PI3K, AKT1) that are undergoing clinical investigation in PCa, as well as IDH1 and MDM2 drug targets.
To give an indication of the potential of these drugs, we analysed the most recent drug sensitivity data (GDSC56, see URL below). Eighteen drugs acting on our network were tested in GDSC on PCa cell lines. Of these, 5 showed significant effect on growth inhibition and the remaining13 drugs showed weak activity in at least one cell line (Supplementary Table 6). However, to validate fully the potential of these drugs, extensive drug sensitivity testing needs to be performed in disease-relevant cancer models that correctly reflect the patient population.
Potential future opportunities for PCa therapy are also highlighted by 13 proteins that are under active chemical biology or drug discovery investigation (Table 2). These include Menin (MEN1), a component of the MLL/SET1 histone methyltransferase complex. Mice with MEN1 mutations develop PCa57 and recent data have shown that menin expression is involved in CRPC58. A further 49 proteins are predicted to be druggable and therefore potentially amenable to drug discovery. These include the known PCa protein SPOP, the transcription activator BRG1 (SMARCA4), CDK12, and the CREB binding protein CREBBP.
In summary, we find that 80 of the 156 proteins central to the prostate disease network are either targets of existing drugs or have the potential to be targeted in the future. To maintain an up-to-date-view of this analysis, we provide a link to a live-page in canSAR (see link below).
Discussion
The analysis of whole genome sequence data from 112 prostate cancers has revealed many of the genetic factors underlying the processes of carcinogenesis, progression, metastasis and the acquisition of drug resistance. Supporting evidence has been provided for thirty candidate driver genes with limited or no previous support, including the non-coding drivers NEAT1 and FOXA1.
Through the timing of genomic aberrations, we have a picture of the possible routes to progression in PCa. Most driver mutations may occur either clonally or subclonally, but mutations in SPOP and ETS-fusions occur early in cancer development and are exclusively clonal. Whereas the gain of 8q and ETS fusion appear to be sufficient to drive a dominant clonal expansion, ETS- cancers typically need a combination of large-scale losses, acquired over an extended period of time. Known cancer drivers are frequently observed subclonally and two competing drivers are seen in several cancers. Metastases have less genomic heterogeneity, likely resulting from a bottleneck in achieving metastatic potential.
We observe changes in the mutational processes operative upon cancers during progression. Signature 8 was enriched in subclonal expansions, and signatures 13 and 18 were enriched in metastatic cancers. Cancers with germline or somatic BRCA1/BRCA2 mutations were enriched for signature 3, demonstrating the effect of double-strand repair defects throughout cancer evolution.
Losses of CDH12 and ANTXR2 result in poorer recurrence-free survival. We identify 69 PCa associated proteins that are either targets for currently available drugs or new potential targets for therapeutic development.
Analysis of the whole-genome sequences of over a hundred prostate cancers has started to reveal the complex evolutionary pathways of these cancers. The early acquisition of driver aberrations including ETS-fusions and whole genome duplications strongly affects the acquisition of subsequent aberrations. Acquisition of individual mutations affects both the subsequent likelihood of metastasis and response to treatment. Network analyses identified, in addition to previously known drivers, targets that could be exploited for clinical investigation with existing drugs as well as targets for new drug discovery, giving potential for the results of genome analysis to be translated rapidly into therapeutic innovation and patient benefit.
Online Methods
Patient Cohorts, Samples and Ethics
We have complied with all relevant ethical regulations. 92 cancer samples from prostatectomy patients treated at The Royal Marsden NHS Foundation Trust, London, at the Addenbrooke’s Hospital, Cambridge, at Oxford University Hospitals NHS Trust, and at Changhai Hospital, Shanghai, China were collected as described previously68,69. Clinical details for the patients are shown in Supplementary Table 7. Ethical approval was obtained from the respective local ethics committees and from The Trent Multicentre Research Ethics Committee. All patients were consented to ICGC standards. (see link below). 20 men from PELICAN (Project to ELIminate lethal CANcer)70, an integrated clinical-molecular autopsy study of metastatic prostate cancer, were the subjects of the current study. Subjects consented to participate in the Johns Hopkins Medicine IRB-approved study between 1995 and 2005. (Supplementary Table 7). A17 had a germline BRCA1 mutation, as previously reported71.
DNA preparation and DNA sequencing
DNA from whole blood samples and frozen tissue was extracted and quantified using a ds-DNA assay (UK-Quant-iT™ PicoGreen® dsDNA Assay Kit for DNA) following the manufacturer’s instructions with a Fluorescence Microplate Reader (Biotek SynergyHT, Biotek). Acceptable DNA had a concentration of at least 50ng/μl in TE (10mM Tris/1mM EDTA), was between 1.8-2.0 with an OD 260/280. WGS was performed at Illumina, Inc. (Illumina Sequencing Facility, San Diego, CA USA) or the BGI (Beijing Genome Institute, Hong Kong), as described previously, to a target depth of 50X for the cancer samples and 30X for matched controls68.
The Burrows-Wheeler Aligner (BWA) was used to align the sequencing data to the GRCh37 reference human genome72. Sequencing data have been deposited at the European Genome-phenome Archive (EGAS00001000262).
Variant Calling Pipeline
SNVs, insertions and deletions were detected using the Cancer Genome Project Wellcome Trust Sanger Institute pipeline as described previously68. In brief, SNVs were detected using CaVEMan with a cut-off ‘somatic’ probability of 95%. Post-processing filters were applied. Insertions and deletions were called using a modified version of Pindel73. Variant allele frequencies of all indels were corrected by local realignment of unmapped reads against the mutant sequence. Structural variants were detected using Brass68. A positive ETS status was assigned if a breakpoint between ERG, ETV1 or ETV4 and previously reported partner DNA sequences was detected.
Data availability
Sequencing data that support the findings of this study have been deposited in the European Genome-phenome Archive with the accession code EGAS00001000262.(see link below). See Supplementary Table 7 for sample specific EGA accession codes.
Code availability
Alignment and variant calling was performed using analysis pipelines in the Cancer Genome Project (CGP) at the Wellcome Trust Sanger Institute. Software versions applied to each sample are listed in Supplementary Table 9. The CGP pipelines may be downloaded (see link below).
Chromoplexy was called using Chainfinder version 1.0.1. Chainfinder may be downloaded (see link below)
The Battenberg algorithm was used to call clonal and subclonal copy number aberrations in all samples. The Battenberg pipeline may be downloaded (see link below).
Putative drug targets were identified using CanSAR version 3.0.
Data analysis was carried out using R, version 3.0.0.
Mutation burdens
Mutation burdens were compared between primary and metastatic samples and between ADT and hormone-naïve samples using a negative binomial generalised linear model (GLM), implemented with the R package MASS. Sample type was found to be an independent predictor of number of SNVs, as was age at time of sampling.
Timing of copy number events
We developed a novel approach to order the occurrence of copy number aberrations by combining three sources of information:
Clonality of copy number aberrations
Timing relative to whole genome duplication
Timing of homozygous deletions relative to neighboring hemizygous losses.
Information from all tumors was combined using a Bradley-Terry model, to give the most likely ordering of events during progression of PCa.
The Battenberg algorithm was used to detect clonal and subclonal somatic copy-number alterations (CNAs) and to estimate ploidy and cancer content from the next-generation sequencing data as previously described74. Briefly, germline heterozygous SNPs were phased using Impute2, and a- and b- alleles were assigned. Data were segmented using piecewise constant fitting75 and subclonal copy-number segments were identified via a t-test as those with b-allele frequencies that differed significantly from the values expected of a clonal copy number state. Ploidy and cancer purity were estimated with the same method used by ASCAT76.
In this cohort, we defined WGD samples as those that had an average ploidy greater than 3. For tumors that had not undergone WGD, gains were defined as those regions that had at least one allele with copy number greater than 1, while losses were defined as those segments that undergone LOH. For tumors that had undergone WGD, losses were called in those segments with at least one allele with copy number of less than 2, whereas gains were called for those with an allelic copy number greater than 2. An extension of this logic was used for subclonal copy number segments – the evolving cellular fraction was always defined as that which deviated away from overall ploidy (defined as 2 for non-WGD samples and 4 for WGD samples). For example, if 75% of cells within a non-WGD tumor have a copy number of 3 + 1 at a given genomic loci, with the remaining 25% of cells having a copy number of 2 + 1, then we assume there has been clonal gain to 2 + 1, and then a subclone containing 75% of cells has undergone a further gain.
Three independent approaches were used to extract evolutionary data from each cancer sample. The first involved ordering clustered sub-clonal cancer fractions, the second used implicit ordering of clonal HDs in relation to losses, and the third estimated the relative timing of whole genome duplication. The logical arguments used within each approach were considered in turn:
Battenberg algorithm-derived estimates for the cellular fraction and standard deviation of each subclonal aberration were input to a Markov Chain Monte Carlo hierarchical Bayesian Dirichlet process to group linked events together in an unsupervised manner. This defined clusters of different cell populations, each present at a calculated cancer cell fraction. The pigeonhole principle was then used to determine the hierarchical relationship between these clusters. Using this process, gains, losses and HDs were ordered with the following caveat to ensure that only independent events are ordered: if there was a clonal and subclonal gain (or loss) at the same locus, then only the clonal or initial gain (or loss) was ordered.
Homozygous deletions have implicitly occurred after loss of heterozygosity at the same locus.
The parsimony principle was used to define the allele counts that correspond to early and late changes in relation to WGD. For losses, if the minor allele copy number equals 0, then the loss occurred prior to WGD. Otherwise the loss occurred after WGD. Regarding gains, if the major allele copy number is twice or greater than ploidy, then the gain occurred prior to WGD. Otherwise, the gain occurred after WGD.
The above arguments allow us to gain insights into the order of copy number events within each individual tumor sample. To establish a consensus order across a cohort of tumor samples requires the ordering data to be integrated across all samples. As specific copy number events (location of breakpoints and the individual copy number states) tend to be unique to individual samples, we defined reference copy number segments that occurred recurrently. These were then used to build an overall contingency table.
The reference genomic segments were defined as regions that were recurrently aberrant. Regions of significant recurrence (false detection rate (FDR), P < 0.05) were determined by performing 100,000 simulations, placing the copy number aberrations detected from each sample in random locations within the genome. The process was repeated for gains, LOH and HDs and the randomly generated copy number landscape compared to that arising from this cohort provided significance levels. Each significantly aberrant region was initially segmented using all breakpoints from all the events that contributed to that region. For instance, the significantly enriched region for LOH: chr8: 0-44Mb contains over 300 breakpoints drawn from from all the samples which contain LOH at chromosome 8p. We computed significantly recurrent regions and reference segments for both ETS+ and ETS- sample subgroups.
Performing pair-wise comparisons between all segmented results using the Bradley-Terry method described below proved computationally expensive and therefore the total number of segments used in the pairwise comparison was rationalised by grouping reference segments to make combined segments of minimum length 1 MB.
We then considered each tumor sample in turn. If any copy number event overlapped the reference genomic segments and was ordered in relation to any other event (that also overlapped regions of significance), those overlapped reference segments were ordered in comparison to other overlapped reference segments. In addition to these reference segments, the TMRPSS2-ERG deletion was ordered more stringently by considering only those segments that could result in the gene fusion, and not merely overlap the locus. In this manner, a contingency table of contests was constructed, using reference genomic segments as the variables. We built contingency tables for both ETS+ and ETS- tumor samples to determine whether their evolutionary trajectory differed significantly.
An implementation of the Bradley-Terry model of pairwise comparison in R77 with bias reduced maximum likelihood estimated the ability or overall order of each individual reference segment.
Subclonal Analysis
The fraction of each cancer genome with subclonal copy number aberrations was calculated as the total amount of the genome with subclonal CNA, as identified by the Battenberg algorithm, divided by the total amount of the genome that had copy number aberrations. One sample (PD13397a, Supplementary Table 8) was identified as having very low cellularity, as it had a completely flat copy number profile and only 411 identified SNVs. Since CNAs could not called in this sample, it was not possible to adjust allele frequencies to CCFs and this sample was excluded from subclonality analysis. SNVs and indels were separately clustered using a Bayesian Dirichlet process, as previously described47. Clonal variants are expected to cluster at a CCF close to 1.0. However, in 18 tumors (Supplementary Table 8), there was no cluster in the range [0.95,1.05]. The likely cause of a shift in CCF is inaccuracy in copy number calling and these samples therefore failed quality control and were excluded from subclonality analysis. From Markov Chain Monte Carlo sampling carried out within the Dirichlet process model, the posterior probability of each variant having a CCF below 0.95 was estimated. Variants with a probability above 80% were designated as ‘subclonal’, those with probability below 20% were designated ‘clonal’ and those with intermediate probabilities were designated as ‘uncertain’. The fraction of subclonal variants used in Fig. 5 and Supplementary Fig. 2 was then calculated after excluding uncertain variants.
Mutational Spectra
The mutational spectra, defined by the triplets of nucleotides around each mutation of each sample were deconvoluted into mutational processes as previously described48,78. Clonal and subclonal variants were separated, as defined above. Further separation of clonal mutations was performed for mutations in genomic regions that had undergone copy number gains. These mutations were classified as ‘early’ or ‘late’ depending whether their observed allele frequencies were more likely to indicate their presence on 2 or 1 chromosome copies, respectively, as assessed by binomial probability. Assignment of mutations to mutational signatures was carried out on each subset of mutations (early, late, clonal, subclonal), as well as on all mutations from each sample (Supplementary Table 3).
Clinical survival analyses
A Cox regression model was fitted to 71 features: every gene with mutations (breakpoints, subs or indels) with a potential functional impact (missense, nonsense, start-lost, inframe, frameshift, or occurred in a non-coding transcript) or a CNA highlighted by the copy number aberration analysis that occurred in three of more prostatectomy patients. The endpoint was biochemical recurrence. P-values were adjusted for multiple testing using the Benjamini-Hochberg method. Multivariate analyses were performed on all genes found to be significant using discretised Gleason (6, 7, 8 or 9), pathological T-stage (T2, T3) and PSA at prostatectomy as cofactors. Gene selection for the optimal predictor of time to biochemical recurrence was determined using Lasso79, a shrinkage and selection method for linear regression, starting with all genes that had a significant association with time to biochemical recurrence. Standard algorithms were used for survival analyses and statistical associations.
Identifying novel oncogenes
The joint dataset was compiled from the aggregation of variants called within our samples with 3 other datasets, yielding a total of 930 samples, comprised of 710 primary and 220 metastatic samples:
TCGA4, 425 primary cancer samples, whole exome sequencing with SureSelect Exome v3 baits on Illumina HiSeq 2000, average coverage ~100X
COSMIC database22, 243 samples, curated set of mutations from several sources, http://cancer.sanger.ac.uk/cosmic
Stand Up to Cancer23 (SU2C-PCF), 150 metastatic castrate resistant samples, paired-end, whole exome sequencing with SureSelect Exome v4 baits on Illumina HiSeq2000, average coverage ~160X
To identify coding and non-coding drivers from SNVs and indels, we used two previously described methods50. Coding drivers on the joint dataset (930 cancers) were identified using dNdScv, a dN/dS method designed to quantify positive selection in cancer genomes. dNdScv models somatic mutations in a given gene as a Poisson process. Inferences on selection are carried out separately for missense substitutions, truncating substitutions (nonsense and essential splice site mutations) and indels, and then combined into a global P-value per gene. Non-coding recurrence was studied using NBR. Both dNdScv and NBR model the variation of the mutation rate across the genome using a negative binomial regression with covariates. First, Poisson regression is used to obtain maximum-likelihood estimates for the 192 rate parameters (rj) describing each of the possible trinucleotide substitutions in a strand-specific manner. rj = nj/Lj, where nj is the total number of mutations observed across samples of a given trinucleotide class (j) and Lj is the number of available sites for each trinucleotide. These rates are used to estimate the total number of mutations across samples expected under neutrality in each element considering the mutational signatures active in the cohort and the sequence of the elements (Eh = Σj rjLj,h). This estimate assumes no variation of the mutation rate across elements in the genome. Second, a negative binomial regression is used to refine this estimate of the background mutation rate of an element, using covariates and Eh as an offset. Both methods identify genes or non-coding regions with higher than expected mutation recurrence, correcting for gene length, sequence composition, mutation signatures acting across patients and for the variation of the mutation rate along the genome. A QQ-plot confirmed that P-values obtained from this method in this cohort were not subject to inflation and consequent over-calling of driver genes (Supplementary Fig. 6).
Chromoplexy, characterized by highly clustered genomic breakpoints that occur in chains and are sometimes joined by deletion bridges, has been shown to be prevalent in PCa25. To identify rearrangement drivers, we first used ChainFinder25 to account for any bias towards regions with chromoplexy and identified ‘unique’ rearranged regions per sample taking the mid-point between all the breakpoints ChainFinder assigns to the same chromoplexy event. Next, separately aggregating the ICGC samples with and without ERG fusions, we calculated inter-breakpoint distance and performed piecewise constant fitting (PCF)75 to identify genomic regions which were recurrently rearranged in multiple samples. Rearranged regions with potential functional impact were identified using two criteria: a minimum 3-fold difference in the number of SVs per MB of ERG+ and ERG- samples; region contains at least one gene with multiple samples with truncating events, i.e. homozygous deletion, stop codon, frameshift indel or essential splice site mutation. In addition, several identified regions were significantly enriched for LOH in either ETS+ or ETS- samples, from copy number analysis (see above). The variants identified in key regions are depicted in Fig. 3.
Chemogenomics annotation of the prostate cancer network
To construct the network, we used the 71 protein products of the 73 genes identified in this study (hereon referred to as Prostate Proteins) to seed a search for all possible interacting proteins in the canSAR interactome54. This interactome contains merged and curated data from the IMeX consortium80, Phosphosite, (see link below) and other databases. It includes:
-
1)
interactions where there were more than two publications reporting experiments demonstrating the binary interaction between the two proteins
-
2)
interactions where there is 3D protein structural evidence of a direct complex
-
3)
interactions where there are at least two publications reporting that one protein is a substrate of the other
-
4)
interactions where there are at least two papers reporting that one protein is the product of a gene under the direct regulatory control of the other
It excludes the following:
-
A)
interactions that were inferred from a large immunoprecipitation experiment without follow-up to demonstrate the specific binary interaction
-
B)
interactions inferred from text mining
-
C)
interactions inferred from co-occurrence in publications or from gene expression correlation.
The initial prostate cancer seeded network resulted in a large collection of 3290 proteins that have some experimental evidence of interacting with at least one Prostate Protein. When we added extra proteins into the network, we wanted to ensure that we only add proteins that are more likely to function primarily through interaction with the proteins in the network rather than just be generic major hubs. To this end, we carried out the following steps: Starting with the input (prostate protein) list, we obtained all possible first neighbours. We then computed, for each new protein, the proportion of its first neighbours that are in the original input list. To define the proteins that are most likely to function through our network, we calculated the chances of these proportions occurring in a random network. We did this by randomising our interactome 10,000 times and computing how often the observed proportions can be achieved by chance (empirical p-value). We corrected the p-values for multiple testing and retained only proteins that have corrected FDR p-values less than 0.05. (Supplementary Fig. 5). We performed network minimisation to maintain only proteins that are strongly connected to more than one Prostate Protein or whose only connection is to one of the Prostate Proteins. We identified a Prostate Cancer network of 156 proteins. Using canSAR’s Cancer Protein Annotation Tool (CPAT)81, we annotated the 156 proteins with pharmacological and druggability data. We labelled proteins that are: 1) targets of approved drugs; 2) targets of drugs under clinical investigation, 3) targets of preclinical or discovery stage compounds that are active at concentrations equal to or less than 100 nM against the protein of interest 4) proteins that we predict to be druggable using our structural druggability prediction protocols81–84 but that have few or no published active inhibitors – these are potential targets for future drug discovery.
Supplementary Material
Acknowledgements
We acknowledge support from Cancer Research UK C5047/A14835/A22530/A17528, C309/A11566, C368/A6743, A368/A7990, C14303/A17197 (ZKJ, SM, NC, SE, DL, TD, MA, EB, JB, GA, PW, BA, DSB, CSC, RAE), the Dallaglio Foundation, (CR-UK Prostate Cancer ICGC Project and Pan Prostate Cancer Group), and PC-UK/Movember (ZKJ). The NIHR support to The Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust; (ZKJ, ND, SM, NC, SE, DL, TD, ST, MA, EB, CF, NL, DN, VK, NA, PK, CO, DC, AT, EM, ER, TD, SH, JB,GA, PW, BA, DSB, CSC, RAE) Cancer Research UK funding to The Institute of Cancer Research and the Royal Marsden NHS Foundation Trust CRUK Centre; the National Cancer Research Institute (National Institute of Health Research (NIHR) Collaborative Study: “Prostate Cancer: Mechanisms of Progression and Treatment (PROMPT)” (grant G0500966/75466) (DN, VG); the Li Ka Shing foundation (DCW, DJW). The Academy of Finland and Cancer Society of Finland (GSB). We thank the National Institute for Health Research, Hutchison Whampoa Limited, University of Cambridge, the Human Research Tissue Bank (Addenbrooke’s Hospital) which is supported by the NIHR Cambridge Biomedical Research Centre, The Core Facilities at the Cancer Research UK Cambridge Institute, Orchid and Cancer Research UK, D Holland from the Infrastructure Management Team & P Clapham from the Informatics Systems Group at the Wellcome Trust Sanger Institute. DMB is supported by Orchid. CV’s academic time was supported by the NIHR Oxford Biomedical Research Centre (Molecular Diagnostics Theme/Multimodal Pathology sub-theme). We also acknowledge support from the Bob Champion Cancer Trust, The Masonic Charitable Foundation successor to The Grand Charity, The King Family and the Stephen Hargrave Trust (CSC, DB). P Workman is a Cancer Research Life Fellow. We acknowledge core facilities provided by CRUK funding to the CRUK ICR Centre, the CRUK Cancer Therapeutics Unit and support for canSAR C35696/A23187 (PW, GA). The authors would like to thank those men with prostate cancer and the subjects who have donated their time and their samples to the Cambridge, Oxford, The Institute of Cancer Research, John Hopkins, and University of Tampere BioMediTech Biorepositories for this study. We also would like to acknowledge support of the research staff in S4 who so carefully curated the samples and the follow-up data (J Burge, M Corcoran, A George, and S Stearn). We thank M. Stratton for the helpful discussions when setting up the CR-UK Prostate Cancer ICGC Project.
Footnotes
URLs
http://www.cancerrxgene.org (GDSC)
http://cansar.icr.ac.uk/cansar/publications/sequencing_prostate_cancers_identifies_new_canc er_genes_routes_progression_and_drug_targets/ (link via google chrome)
https://www.ebi.ac.uk/ega/studies/EGAS00001000262
https://github.com/cancerit/dockstore-cgpwgs
Author contribution
RAE, CSC, DN, DSB, CSF, SB, AGL, PW, BA, DCW, FCH and DFE designed the study.
RAE, CSC, DCW and DB wrote the paper, and all other authors contributed to revisions.
ZKJ, HW, CEM, DN, VG, AGL, RAE, FCH, SB, AYW, CSF, CV, DMB, ND, SM, SH, WH, Y-JL, AL, JK, KK, HL, LM, SE, LMatthews, AN, YY, HZ, ST, EB, CF, NL, SH, DNicol, PG, VK, NVA, PK, CO, DC, AT, EM, ER, TD, NCS, coordinated sample collection, pathology review and processing.
DCW, GG, TM, IM, DJW, DSB, MG, JZ, AB, LGB, SD, BK, NC, VB, DL, SM, TD, MA, STavare, CG, KR, DG, AM, LS, JT, AF, UM, supported, directed and performed the analyses.
CS and the TCGA, JdeB and GA provided data for the meta-analysis.
DFE, AGL, GSB, CSF, DSB, DN, CSC, RAE, joint PIs CR-UK Prostate Cancer ICGC Project.
Competing Financial Interests
There are no competing financial interests.
References
- 1.Attard G, et al. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 2.Weischenfeldt J, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23:159–70. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalonde E, et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5 year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 2014;15:1521–32. doi: 10.1016/S1470-2045(14)71021-6. [DOI] [PubMed] [Google Scholar]
- 8.Cooper CS, Eeles R, Wedge DC, Van Loo P. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–72. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutros PC, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–45. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 10.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–7. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castro E, et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur Urol. 2015;68:186–93. doi: 10.1016/j.eururo.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 12.Kluth M, et al. Concurrent deletion of 16q23 and PTEN is an independent prognostic feature in prostate cancer. Int J Cancer. 2015;137:2354–63. doi: 10.1002/ijc.29613. [DOI] [PubMed] [Google Scholar]
- 13.Mosquera JM, et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia. 2013;15:1–10. doi: 10.1593/neo.121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues LU, et al. Coordinate loss of MAP3K7 and CHD1 promotes aggressive prostate cancer. Cancer Res. 2015;75:1021–34. doi: 10.1158/0008-5472.CAN-14-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuzick J, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein EA, et al. Decipher Genomic Classifier Measured on Prostate Biopsy Predicts Metastasis Risk. Urology. 2016;90:148–52. doi: 10.1016/j.urology.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Bostrom PJ, et al. Genomic Predictors of Outcome in Prostate Cancer. Eur Urol. 2015;68:1033–44. doi: 10.1016/j.eururo.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Luca B-A, et al. DESNT: A Poor Prognosis Category of Human Prostate Cancer. European Urology Focus. doi: 10.1016/j.euf.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan CJ, et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015;16:152–60. doi: 10.1016/S1470-2045(14)71205-7. [DOI] [PubMed] [Google Scholar]
- 20.Loriot Y, et al. Effect of enzalutamide on health-related quality of life, pain, and skeletal-related events in asymptomatic and minimally symptomatic, chemotherapy-naive patients with metastatic castration-resistant prostate cancer (PREVAIL): results from a randomised, phase 3 trial. Lancet Oncol. 2015;16:509–21. doi: 10.1016/S1470-2045(15)70113-0. [DOI] [PubMed] [Google Scholar]
- 21.Mateo J, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James ND, et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387:1163–77. doi: 10.1016/S0140-6736(15)01037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svensson C, et al. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 2014;42:999–1015. doi: 10.1093/nar/gkt921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, et al. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ. 2011;18:1174–83. doi: 10.1038/cdd.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer K, Pflugfelder GO. Putative Breast Cancer Driver Mutations in TBX3 Cause Impaired Transcriptional Repression. Front Oncol. 2015;5:244. doi: 10.3389/fonc.2015.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–90. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki M, et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med. 2011;17:944–51. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakravarty D, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5 doi: 10.1038/ncomms6383. 5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang YA, Yu J. Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis. 2015;2:144–151. doi: 10.1016/j.gendis.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takayama K, et al. Integrative analysis of FOXP1 function reveals a tumor-suppressive effect in prostate cancer. Mol Endocrinol. 2014;28:2012–24. doi: 10.1210/me.2014-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krohn A, et al. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231:130–41. doi: 10.1002/path.4223. [DOI] [PubMed] [Google Scholar]
- 35.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King JC, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kluth M, et al. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer. 2014;135:1369–80. doi: 10.1002/ijc.28784. [DOI] [PubMed] [Google Scholar]
- 38.Burkhardt L, et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013;73:2795–805. doi: 10.1158/0008-5472.CAN-12-1342. [DOI] [PubMed] [Google Scholar]
- 39.Liu W, et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene. 2012;31:3939–48. doi: 10.1038/onc.2011.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heun P. SUMOrganization of the nucleus. Curr Opin Cell Biol. 2007;19:350–5. doi: 10.1016/j.ceb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Kaikkonen S, et al. SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol Endocrinol. 2009;23:292–307. doi: 10.1210/me.2008-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith DI, Zhu Y, McAvoy S, Kuhn R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 2006;232:48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 44.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams JL, Greer PA, Squire JA. Recurrent copy number alterations in prostate cancer: an in silico meta-analysis of publicly available genomic data. Cancer Genet. 2014;207:474–88. doi: 10.1016/j.cancergen.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolli N, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5 doi: 10.1038/ncomms3997. 2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pilati C, et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J Pathol. 2017 doi: 10.1002/path.4880. [DOI] [PubMed] [Google Scholar]
- 50.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polkinghorn WR, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–53. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodwin JF, et al. DNA-PKcs-Mediated Transcriptional Regulation Drives Prostate Cancer Progression and Metastasis. Cancer Cell. 2015;28:97–113. doi: 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarish FL, et al. Castration radiosensitizes prostate cancer tissue by impairing DNA double-strand break repair. Sci Transl Med. 2015;7:312re11. doi: 10.1126/scitranslmed.aac5671. [DOI] [PubMed] [Google Scholar]
- 54.Tym JE, et al. canSAR: an updated cancer research and drug discovery knowledgebase. Nucleic Acids Res. 2016;44:D938–43. doi: 10.1093/nar/gkv1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leongamornlert D, et al. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br J Cancer. 2014;110:1663–72. doi: 10.1038/bjc.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seigne C, et al. Characterisation of prostate cancer lesions in heterozygous Men1 mutant mice. BMC Cancer. 2010;10:395. doi: 10.1186/1471-2407-10-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malik R, et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med. 2015;21:344–52. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudnicka C, et al. Overexpression and knock-down studies highlight that a disintegrin and metalloproteinase 28 controls proliferation and migration in human prostate cancer. Medicine (Baltimore) 2016;95:e5085. doi: 10.1097/MD.0000000000005085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, et al. FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer Res. 2011;71:3257–67. doi: 10.1158/0008-5472.CAN-10-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malinowska K, et al. Interleukin-6 stimulation of growth of prostate cancer in vitro and in vivo through activation of the androgen receptor. Endocr Relat Cancer. 2009;16:155–69. doi: 10.1677/ERC-08-0174. [DOI] [PubMed] [Google Scholar]
- 62.FitzGerald LM, et al. Identification of a prostate cancer susceptibility gene on chromosome 5p13q12 associated with risk of both familial and sporadic disease. Eur J Hum Genet. 2009;17:368–77. doi: 10.1038/ejhg.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao W, Cao L, Zeng S, Qin H, Men T. Upregulation of miR-556-5p promoted prostate cancer cell proliferation by suppressing PPP2R2A expression. Biomed Pharmacother. 2015;75:142–7. doi: 10.1016/j.biopha.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 64.Parray A, et al. ROBO1, a tumor suppressor and critical molecular barrier for localized tumor cells to acquire invasive phenotype: study in African-American and Caucasian prostate cancer models. Int J Cancer. 2014;135:2493–506. doi: 10.1002/ijc.28919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daniels G, et al. TBLR1 as an androgen receptor (AR) coactivator selectively activates AR target genes to inhibit prostate cancer growth. Endocr Relat Cancer. 2014;21:127–42. doi: 10.1530/ERC-13-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones S, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–3. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collart MA, Kassem S, Villanyi Z. Mutations in the NOT Genes or in the Translation Machinery Similarly Display Increased Resistance to Histidine Starvation. Front Genet. 2017;8:61. doi: 10.3389/fgene.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooper CS, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015 doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mao X, et al. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70:5207–12. doi: 10.1158/0008-5472.CAN-09-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nickerson ML, et al. Somatic alterations contributing to metastasis of a castration-resistant prostate cancer. Hum Mutat. 2013;34:1231–41. doi: 10.1002/humu.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nilsen G, et al. Copynumber: Efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics. 2012;13:591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Firth D, Turner HL. Bradley-Terry models in R: the BradleyTerry2 package. Journal of Statistical Software. 2012;48 [Google Scholar]
- 78.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–59. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shin S, Fine J, Liu Y. Adaptive Estimation with Partially Overlapping Models. Stat Sin. 2016;26:235–253. doi: 10.5705/ss.2014.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orchard S, et al. Protein interaction data curation: the International Molecular Exchange IMEx consortium. Nat Methods. 2012;9:345–50. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patel MN, Halling-Brown MD, Tym JE, Workman P, Al-Lazikani B. Objective assessment of cancer genes for drug discovery. Nat Rev Drug Discov. 2013;12:35–50. doi: 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- 82.Bulusu KC, Tym JE, Coker EA, Schierz AC, Al-Lazikani B. canSAR: updated cancer research and drug discovery knowledgebase. Nucleic Acids Res. 2014;42:D1040–7. doi: 10.1093/nar/gkt1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitsopoulos C, Schierz AC, Workman P, Al-Lazikani B. Distinctive Behaviors of Druggable Proteins in Cellular Networks. PLoS Comput Biol. 2015;11:e1004597. doi: 10.1371/journal.pcbi.1004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Workman P, Al-Lazikani B. Drugging cancer genomes. Nat Rev Drug Discov. 2013;12:889–90. doi: 10.1038/nrd4184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data that support the findings of this study have been deposited in the European Genome-phenome Archive with the accession code EGAS00001000262.(see link below). See Supplementary Table 7 for sample specific EGA accession codes.