Abstract
The autophagosome precursor membrane, termed the “isolation membrane” or “phagophore,” emerges adjacent to a PI3P-enriched transient subdomain of the ER called the “omegasome,” thereafter expanding to engulf cytoplasmic content. Uncovering the molecular events that occur in the vicinity of the omegasome during phagophore biogenesis is imperative for understanding the mechanisms involved in this critical step of the autophagy pathway. We recently characterized the ATG2A-WIPI4 complex, one of the factors that localize to the omegasome and play a critical role in mediating phagophore expansion. Our structural and biochemical studies revealed that ATG2A is a rod-shaped protein with membrane-interacting properties at each end, endowing ATG2A with membrane-tethering capability. Association of the PI3P-binding protein WIPI4 at one of the ATG2A tips enables the ATG2A-WIPI4 complex to specifically tether PI3P-containing membranes to non-PI3P-containing membranes. We proposed models for the ATG2A-WIPI4 complex-mediated membrane associations between the omegasome and surrounding membranes, including the phagophore edge, the ER, ATG9 vesicles, and COPII vesicles.
Keywords: ATG2, WIPI, ATG18, membrane tethering, autophagy, autophagosome
Main Text
Autophagy is the process by which cytoplasmic materials are sequestered within autophagosomes and delivered to lysosomes for degradation and recycling. This sequestration is achieved through the de novo formation of an autophagosome, beginning with the nucleation of a phagophore in close proximity to the ER (Sanchez-Wandelmer, Ktistakis, & Reggiori, 2015). The phagophore expands and adopts a cup-like shape as it engulfs cytoplasmic materials. During this process, the edge of the cup-shaped phagophore is associated with the ER, which has led to the hypothesis that the phagophore acquires lipid molecules from the ER to serve as “building blocks” as it grows. In mammalian cells, the ER-phagophore junction involves the elusive membrane domain omegasome, which emerges as a PI3P-enriched subdomain of the ER and later dynamically associates with the ER while scaffolding the growing phagophore (Axe et al., 2008). Recent EM images of resin-embedded cellular slices of this junction showed that omegasomes are connected to either the ER or the phagophore, and less frequently to both (Uemura et al., 2014), suggesting that omegasomes are independent membrane units linking the ER to the phagophore. It has been proposed that the protein molecules that localize to the omegasomes, including the poorly characterized ~200 kDa protein ATG2 (yeast Atg2p and mammalian ATG2A/B) and the PI3P-binding β-propeller proteins Atg18p/WIPIs (yeast Atg18p and mammalian WIPI1–4), play crucial roles in phagophore expansion. In order to uncover the molecular mechanisms underlying this central step of autophagosome formation, we investigated the structural and biochemical characteristics of one of these factors localized at the ER-phagophore junction: the human ATG2A-WIPI4 complex (Chowdhury et al., 2018).
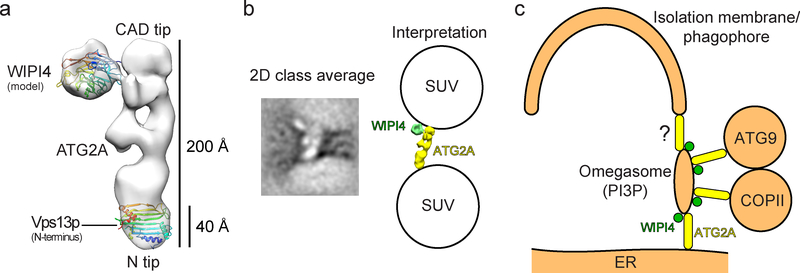
Single particle negative stain EM analyses of ATG2A and the ATG2A-WIPI4 complex revealed that ATG2A is a ~20 nm-long rod with a conserved N-terminal domain at one tip, and that the conserved CAD region (named after the characteristic Cys-Ala-Asp triad identified in the middle of ATG2A’s sequence) is located at the opposite tip. Notably, WIPI4 binds at CAD-containing tip of the rod (Fig. 1a). The positioning of conserved regions at the tips of the ATG2A rod suggests that the tips are functionally important. The C-terminus of the protein, which is also conserved and shown to be essential for cellular localization (Tamura et al., 2017), could not be unambiguously identified. A lack of density corresponding to this region was also observed for the ATG2B-WIPI4 complex (Zheng et al., 2017), suggesting that the C-terminus of ATG2A/B is flexible.
Figure 1.
Structural analyses of the ATG2A-WIPI4 complex. (a) The negative stain EM reconstruction (EMD-8899) of the ATG2A-WIPI4 complex with a docked WIPI4 homology model and the high-resolution structure of the N-terminal domain of Vps13p (left) (PDB: 6CBC). (b) A 2D class average of the ATG2A-WIPI4 complex bound to two SUVs (left) and the interpretation of the image (right). (c) A schematic of the proposed membrane-tethering events involving the omegasome (right).
Having determined the overall structure of the ATG2A-WIPI4 complex, we next aimed to uncover the significance of its rod-like shape. Using liposome flotation assays, we found that ATG2A has a particular affinity for small unilamellar vesicles (SUVs). Given that the diameter of these SUVs is ~30 nm, which is only slightly larger than the axial length of ATG2A, we hoped to observe the nature of these ATG2A-liposome interactions and if they introduced a reshaping of either the protein or membrane structures. Remarkably, we were able to visualize the rod-shaped structures attached to the surfaces of SUVs by negative stain EM. Many of the rod-like shapes were found between two SUVs with each tip of the rod contacting an SUV. 2D analyses of these rods provided sufficient detail to confirm that the rod-shaped objects were indeed ATG2A (Fig. 1b), leading to our hypothesis that ATG2A is a membrane tether. We subsequently demonstrated ATG2A-mediated membrane tethering using biochemical membrane-tethering assays. Notably, we found that the ATG2A-WIPI4 complex can tether a PI3P-containing vesicle to a non-PI3P-containing vesicle, recapitulating the functionally important tethering of the omegasome to a neighboring membrane during phagophore expansion. The membrane-tethering activity by the ATG2A-WIPI4 complex has not yet been demonstrated in vivo. However, two recent reports showing that the yeast Atg2p-Atg18p complex tethers the membrane of the pre-autophagosomal site (PAS), which is enriched with PI3P, to the ER in vivo (Gomez-Sanchez et al., 2018; Kotani, Kirisako, Koizumi, Ohsumi, & Nakatogawa, 2018) complement our biochemical data in supporting our hypothesis that the ATG2A-WIPI4 complex acts as a membrane tether.
The reported ER-omegasome and omegasome-phagophore connections in mammalian cells (Uemura et al., 2014) led us to consider that the ATG2A-WIPI4 complex may mediate both of these membrane associations (Fig. 1c). The CAD tip and WIPI4 could simultaneously associate with the PI3P-rich omegasome while the N-terminal tip (N-tip) of ATG2A associates with the ER and the phagophore edge. The multivalent interactions involving the three components, ATG2A (CAD tip), WIPI4, and a PI3P-containing membrane, would maintain the ATG2A-WIPI4 complex stably bound to the omegasome. In contrast, the N-tip, which binds to membranes in a monovalent manner, could dissociate more frequently from the ER and/or the phagophore, allowing the omegasome to interact with either the ER or the phagophore in an alternating fashion. This model is consistent with the EM observations of ER-omegasome and omegasome-phagophore connections (Uemura et al., 2014) as well as observed dynamic associations between the omegasome and the ER (Axe et al., 2008). Our localization of the N-tip at the ER in our model is supported by the work of Kotani et al. (Kotani et al., 2018), which was published coincidentally with ours. Kotani et al. created an autophagy-defective Atg2p mutant by truncating the N-terminal 20 residues and showed that anchoring this mutant to the ER by fusing the transmembrane domain of Sec71, an ER resident, to the N-terminus of the mutant restored autophagic activity. These data, together with the intimate association observed between the omegasome and the phagophore (Axe et al., 2008), suggests that there exists another mechanism for omegasome-phagophore association – one that may not involve ATG2A. However, autophagic activity was only partially restored in the experiment by Kotani et al. Thus, their data are not solely sufficient to exclude the possibility that omegasome-phagophore association can be mediated by ATG2-WIPI4.
Yeast cell studies have shown that the ER-phagophore junction is enriched with not only the Atg2p-Atg18p complex, but also Atg9p and the components of COPII vesicles (Graef, Friedman, Graham, Babu, & Nunnari, 2013; Suzuki, Akioka, Kondo-Kakuta, Yamamoto, & Ohsumi, 2013). Atg9p is an integral membrane protein present on fast-moving small vesicles, and upon autophagy initiation, several “Atg9 vesicles” are recruited to the PAS (Graef et al., 2013; Suzuki et al., 2013), where they tether with COPII vesicles (Tan et al., 2013). Both Atg9 and COPII vesicles remain at the ER-phagophore junction during phagophore expansion, indicative of their participation in this process. Notably, Atg2p has been reported to associate with both Atg9p (Gomez-Sanchez et al., 2018) and COPII components (Graef et al., 2013). Mammalian ATG2A/B has not been demonstrated to interact directly with ATG9 or COPII, but ATG9 and COPII are located in the vicinity of the emerging phagophore/omegasome (Karanasios et al., 2016; Koyama-Honda, Itakura, Fujiwara, & Mizushima, 2013; Orsi et al., 2012). These considerations led us to suggest that the ATG2 N-tip could recruit these dynamic ATG9 and COPII vesicles to the omegasome (Fig. 1c).
The tethering events proposed above do not explain how lipids or membranes could actually be transferred into the phagophore, a prerequisite for its expansion. A recent report by Kumar et al. on VPS13, a large (>3000 A.A.) protein known to be involved in the biogenesis of organelles at contact sites between the ER and other organelles (Murley & Nunnari, 2016), has provided new insights into this mechanism (Kumar et al., 2018). VPS13 is sequentially similar to ATG2 at the N- and C-termini and their crystal structure revealed that the N-terminal ~300 residues of C. thermophilum Vps13p are folded into a mixed-αβ structure containing an extended hydrophobic cavity, a characteristic architecture of lipid binding/transferring proteins (Wong, Gatta, & Levine, 2018). Docking Vps13p N-terminal structure into the N-tip of our EM density shows that this fragment would occupy ~20–25% of the entire density (Fig. 1a). An extended central β-sheet that traversed the entirety of the full-length protein would accommodate a large number of lipids. Indeed, a larger Vps13p fragment containing the N-terminal ~1350 A.A., which is also rod-shaped, was found to sequester ~10 glycerolipids per protein molecule. Notably, it was shown that this fragment can transfer these lipids between membranes. Membrane tethering was shown to be necessary for efficient lipid transfer by Vps13p, and reliant on an ER-anchoring FFAT motif, which is absent in ATG2. While the structural mechanisms of membrane-tethering by ATG2 and VPS13 may not be identical, the sequential and architectural similarities between them raise the intriguing and probable possibility that ATG2 is also a lipid transfer protein.
If such transfer were to be demonstrated, ATG2A-mediated lipid transfer, together with the ATG2A-WIPI4 complex-mediated membrane tethering events proposed above, would serve as key processes in a long-sought mechanic model for phagophore expansion. The ATG2A-WIPI4 complex tethers the omegasome to the ER and the surrounding vesicles, such as ATG9 and COPII vesicles, allowing these tethered membranes to feed lipids to the omegasome. The lipids incorporated into the omegasomes could subsequently be transferred to the phagophore upon ATG2A-WIPI4 complex-mediated omegasome-phagophore association. Alternatively, the omegasome into which lipids are being incorporated may mature into the new phagophore edge by unknown means, resulting in an expansion of the phagophore. This model, based on the ATG2A-WIPI4 complex-mediated lipid transfer, explains why autophagosomes form adjacent to the ER and provides new insights into how PI3P/omegasome dynamics would control phagophore expansion and subsequent maturation into the autophagosome. Continued investigation of the membrane contact sites within the ER-phagophore junction promises to uncover the core molecular mechanisms of autophagosome biogenesis.
Acknowledgments
Funding
This research was supported by the grants from the National Institute of Health (GM092740 to T.O. and DP2EB020402 to G.C.L.). G.C.L. is supported as a Searle Scholar and a Pew Scholar. Computational analyses of EM data were performed using shared instrumentation at SR funded by NIH S10OD021634.
References
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, … Ktistakis NT (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol, 182(4), 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Otomo C, Leitner A, Ohashi K, Aebersold R, Lander GC, & Otomo T (2018). Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1811874115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Sanchez R, Rose J, Guimaraes R, Mari M, Papinski D, Rieter E, … Reggiori F (2018). Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J Cell Biol. doi: 10.1083/jcb.201710116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, & Nunnari J (2013). ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell, 24(18), 2918–2931. doi: 10.1091/mbc.E13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, … Ktistakis NT (2016). Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun, 7, 12420. doi: 10.1038/ncomms12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Kirisako H, Koizumi M, Ohsumi Y, & Nakatogawa H (2018). The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1806727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Honda I, Itakura E, Fujiwara TK, & Mizushima N (2013). Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy, 9(10), 1491–1499. doi: 10.4161/auto.25529 [DOI] [PubMed] [Google Scholar]
- Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li P, Lees JA, … De Camilli P (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol. doi: 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A, & Nunnari J (2016). The Emerging Network of Mitochondria-Organelle Contacts. Mol Cell, 61(5), 648–653. doi: 10.1016/j.molcel.2016.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, & Tooze SA (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell, 23(10), 1860–1873. doi: 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Wandelmer J, Ktistakis NT, & Reggiori F (2015). ERES: sites for autophagosome biogenesis and maturation? J Cell Sci, 128(2), 185–192. doi: 10.1242/jcs.158758 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, & Ohsumi Y (2013). Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci, 126(Pt 11), 2534–2544. doi: 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- Tamura N, Nishimura T, Sakamaki Y, Koyama-Honda I, Yamamoto H, & Mizushima N (2017). Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets. FEBS Lett, 591(23), 3819–3830. doi: 10.1002/1873-3468.12901 [DOI] [PubMed] [Google Scholar]
- Tan D, Cai Y, Wang J, Zhang J, Menon S, Chou HT, … Walz T (2013). The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci U S A, 110(48), 19432–19437. doi: 10.1073/pnas.1316356110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Yamamoto M, Kametaka A, Sou YS, Yabashi A, Yamada A, … Waguri S (2014). A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol Cell Biol, 34(9), 1695–1706. doi: 10.1128/MCB.01327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Gatta AT, & Levine TP (2018). Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat Rev Mol Cell Biol. doi: 10.1038/s41580-018-0071-5 [DOI] [PubMed] [Google Scholar]
- Zheng JX, Li Y, Ding YH, Liu JJ, Zhang MJ, Dong MQ, … Yu L (2017). Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy, 0. doi: 10.1080/15548627.2017.1359381 [DOI] [PMC free article] [PubMed] [Google Scholar]