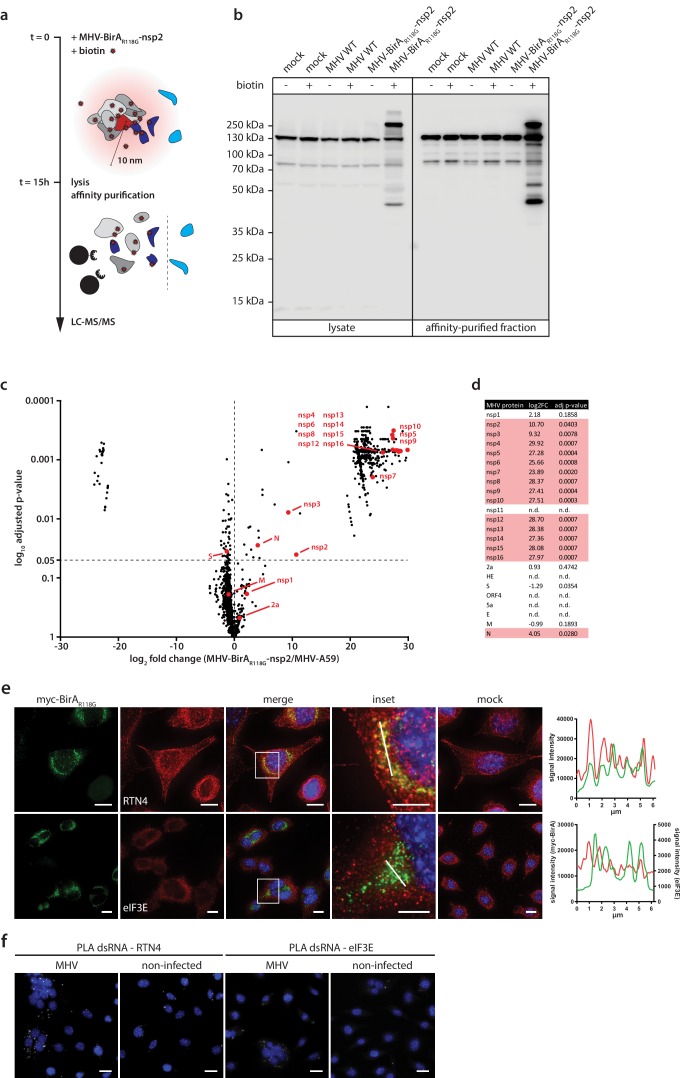
Figure 2. Determination of the coronavirus RTC-proximal proteome.
(a) Schematic overview of the BirAR118G-mediated proximity biotinylation assay using MHV-BirAR118G-nsp2. (b) Western blot analysis of MHV-BirAR118G-nsp2-infected L929 cells. L929 cells were infected with MHV-BirAR118G-nsp2, MHV-A59 or non-infected in medium with and without supplementation of 67 µM biotin. Cells were lysed 15 h.p.i. and biotinylated factors were subjected to affinity purification using streptavidin-coupled magnetic beads. Total cell lysates and affinity-purified fractions were separated by SDS-PAGE and analysed by western blot probed with horse radish peroxidase (HRP)-coupled Streptavidin. (c) Host and viral factors identified by LC-MS/MS. 4*107 L929 cells were infected with MHV-BirAR118G-nsp2 or MHV-A59 in medium supplemented with 67 µM biotin. 15 h.p.i., lysates were affinity purified and LC-MS/MS was performed from in-gel digested samples. MS identification of biotinylated proteins was performed in three independent biological replicates. Spectral interpretation was performed against a Mus musculus and MHV database and log2-transformed LFQ levels (x-axis) were used to determine significant differences in protein enrichment between sample groups (Student's T-test, y-axis). Identified cellular proteins are displayed as black dots, MHV proteins are highlighted in red (nsp: non-structural protein, N: nucleocapsid, S: spike, M: membrane, 2a: accessory protein 2a). (d) Summary of viral proteins identified by LC-MS/MS. nsp2-10, nsp12-16, and nucleocapsid were significantly enriched in fractions derived from MHV-BirAR118G-nsp2-infected cells whereas nsp1, nsp11, structural proteins spike (S), envelope (E) and membrane proteins (M) as well as all accessory proteins (NS2a, HE, ORF4, ORF5a) were either not significantly enriched or not detected. (e,f) Immunofluorescence analysis of RTC-proximal cellular factors. L929 cells were seeded on coverslips, infected with MHV-BirAR118G-nsp2 (e) or MHV-A59 (f), fixed at 9 h.p.i. and processed for immunofluorescence using anti-myc, anti-RTN4 and anti-eIF3E antibodies (e) or anti-dsRNA, anti-RTN4 and anti-eIF3E antibodies (f). Secondary fluorophore-coupled antibodies were used to detect the viral replicase and endogenous levels of RTN4 and eIF3E (e). Scale bars: 10 µm; insets 5 µm. Proximity ligations were performed using Duolink In Situ detection reagents (f). Nuclei are counterstained with DAPI. Z-projection of deconvolved z-stacks acquired with a DeltaVision Elite High-Resolution imaging system are shown. Intensity profiles highlighted in the magnified regions are shown. Scale bars: 20 µm (insets 5 µm).