Abstract
Personalized drug screening (PDS) of approved drug libraries enables rapid development of specific small-molecule therapies for individual patients. With a multidisciplinary team including clinicians, researchers, ethicists, informaticians and regulatory professionals, patient treatment can be optimized with greater efficacy and fewer adverse effects by using PDS as an approach to find remedies. In addition, PDS has the potential to rapidly identify therapeutics for a patient suffering from a disease without an existing therapy. From cancer to bacterial infections, we review specific maladies addressed with PDS campaigns. We predict that PDS combined with personal genomic analyses will contribute to the development of future precision medicine endeavors.
Keywords: Drug repurposing, personalized-medicine, phenotypic-screen, personalized drug screening, personalized drug screening, iPS cells, glioblastoma
Introduction
Currently, medical treatment is centered on a specific disease normally categorized by its pathological changes and clinical symptoms. In the past two decades, it has been realized that many drugs are only effective for a subset of patients suffering from disease. Additionally, many medications cause varied adverse effects among these patients. Different patient genomic environments, lifestyle, treatment adherence and the drug itself contribute to variable drug efficacy and adverse effects [1]. Genetic differences include deleterious mutations and varying expression levels in drug targets and drug-metabolizing proteins. In the past decade, translational research has explored the development of personalized medicine tailored to an individual’s disease. Advances in genomics reveal how a patient’s genetic background can impact their risk, prognosis and response to a therapeutic intervention. One of the potential approaches in the development of personalized treatments is to combine the detailed personal health information with personalized drug screening (PDS); the goal of PDS is to use primary patient samples to rapidly identify effective therapeutic compounds with higher efficacy and lower toxicity.
We advocate that the drug libraries for PDS consist of drugs with FDA approval. Identification of an approved drug for a new indication has the advantage of immediately being used to treat patients or directly advanced to late-stage clinical trials, avoiding the long drug development process required for new drugs to move into clinical trials. For the purposes of this review, we will use PDS to describe a personalized screen carried out with an FDA-approved drug library. However, PDS is not exclusively for use with FDA-approved drug libraries.
To achieve these results, a multidisciplinary team of clinicians, researchers, informaticians, ethicists, informaticians and regulatory specialists within academia, government and industry need to work together to develop new methods and optimize the process of PDS. With a drug repurposing screen of a patient’s disease samples, whether it be cancer or a bacterial infection, more-effective drugs can quickly be identified in a new cyclical approach called ‘bedside-to-bench and back to patient’ [2].
To move PDS forward, a personalized medicine infrastructure must be fully embraced by all stakeholders in the healthcare system. A detailed pharmacoeconomic analysis of PDS is also required to determine the costs and risks in the context of established healthcare operations [3,4]. For example, the fifth major leading cause of death in the USA with up to 140 000 deaths annually is adverse drug toxicity [5,6]. We speculate that reduction of drug-induced toxicities will provide a significant decrease in the cost of healthcare and improve the ability of clinicians to accurately prescribe the best therapy for each patient. A personalized drug treatment approach including PDS and individual genome analysis, once well developed, can save resources and, most importantly, improve patient treatment. This review provides a perspective on how basic researchers and clinicians have worked together on translational science in the new era of precision medicine. Several examples will be detailed and discussed in this paper, as well as other important aspects of personalized drug screens.
Drug repurposing screens
To date, there are ~1500 drugs approved by the FDA to treat diseases [7]. Recently, drug repurposing emerged as an alternative approach for rapid identification of new treatments for diseases that lack effective therapies. Drug repurposing screens identify active compounds from an FDA-approved drug library for new indications using HTS methods. Once an active compound is identified, it can be used to treat patients as discussed in the examples provided in the following sections. Alternately, the active compound can move quickly into clinical trials, which dramatically reduces the time and costs of traditional drug development.
A unique example comes from infectious disease where the antibiotic susceptibility test has been in clinical practice for >50 years. Antibiotics identified as effective against patient bacterial isolates can be used to treat the patient immediately [7]. In oncology, active compounds identified from screens using patient-derived primary tumor samples can also be utilized to treat patients quickly [8]. Although some drugs with a new indication can be used in patients after Institutional Review Board (IRB) approval without clinical trials, clinical trials are required for FDA approval in most cases. Additionally, organoid models can be built with patient-specific mutations and genomic background. This establishes conditions more-relevant to an individual patient, and enables researchers to evaluate drug efficacy in vitro before making treatment decisions as described in the report for cystic fibrosis (CF) discussed below [9]. Therefore, drug repurposing screens can find low-hanging fruit among FDA-approved drugs with known properties and safety profiles, permitting faster development for new indications. The following four recent cases are examples of personalized medicine development with drug repurposing screens.
Personalized antitumor therapy identified by a drug repurposing screen using patient cells
Yuan et al. identified a personalized medicine to treat respiratory papillomatosis in a report from The New England Journal of Medicine [8]. A 24-year-old male patient was unsuccessfully treated for a chemoresistant and progressive viral-induced tumor in the neck and throat over a period of 20 years. Researchers at the Georgetown University Medical Center created a new cell line from his excised tumor. By sequencing the tumor DNA, they were able to detect human papilloma virus (HPV) gene expression responsible for tumor growth. The tumors contained viral genome mutations that differed from the prototype HPV-11 sequence along with a viral genome duplication event that occurred during tumor extension into the lung. Before this study, respiratory papillomatosis did not have an approved drug therapy; repeated surgery was the only option to slow down tumor progress. To identify a better therapeutic solution, researchers tested three anticancer drugs on the reprogrammed patient-derived cells: cidofovir, dihydroartemisinin and vorinostat. These drugs were selected because of their historical use in treating this disease and previous efficacy against HPV-positive cell lines. Only vorinostat demonstrated a threefold selectivity for cancer cells compared with healthy cells with a half-maximal inhibitory concentration (IC50) of 4.2 μM. Treatment of the patient with vorinostat, an old and inexpensive anticancer drug, produced positive results with tumor shrinkage, stabilized tumor size and no new tumors.
This case study shows how collaboration between clinicians and basic researchers led to a new treatment for a disease that lacked an effective therapy. In this example, only three compounds were screened; nevertheless, it provided a useful approach to identify effective therapeutics using an FDA-approved drug library for treatments of an otherwise incurable disease. It is important to note, however, that this drug might not be effective for other patients with similar respiratory papillomatosis because of different individual genetic backgrounds, tumor subtypes or other environmental factors. Individualized drug repurposing screens with cells derived from a patient tumor sample should be adopted to better identify effective anticancer drugs for individual patients.
Individualized drug repurposing screens with cells derived from glioblastoma tumor samples
Glioblastoma multiforme (GBM) is an aggressive brain cancer that arises from astrocytes. The current standard-of-care is surgical debulking followed by radiotherapy with concomitant adjuvant temozolimide (TMZ) administration. However, the 5-year survival rate stands at an abysmal 10% and the common survival length after the diagnosis is ~1 year [10]. In clinical trials, the addition of temozolimide treatment to radiotherapy only resulted in a 2.5 month increase in median survival time [11]. To find new chemotherapeutic agents to treat GBM, we screened the approved drug library using cells derived from patient tumor samples to identify compounds that selectively killed patient tumor cells over normal human astrocyte controls.
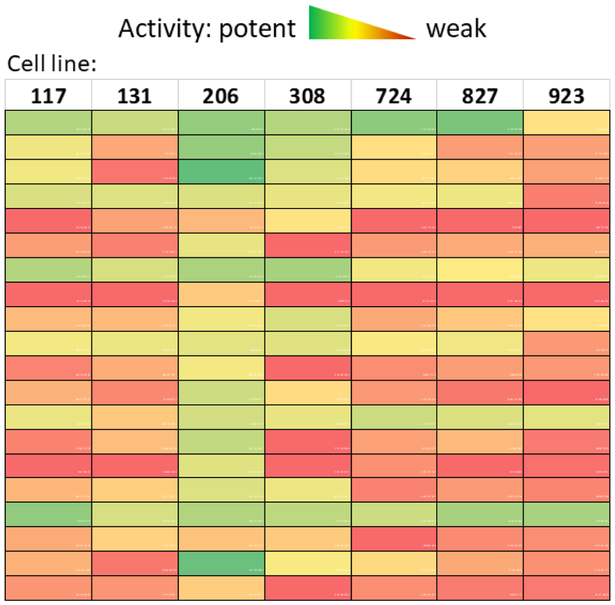
We conducted a drug repurposing screen using patient-derived glioblastoma cells that led to the identification of active compounds with different potencies against individual patient cells (unpublished data). Tumor stem cells were isolated from GBM patient biopsies and cultured as neurospheres [12]. Four of these patient-derived cells were used to screen the collection of 4000 approved drugs and bioactive compounds. We further tested 120 primary hit compounds in cells derived from seven patient samples and observed variable activities (potencies) among these hits. Some compounds potently suppressed GBM cells from one or two patients but lacked activity against the other patients. The results also revealed variability in that some patient GBM cells are generally more susceptible to chemotherapy. For example, patient cell line 206 responded to the greatest number of compounds, whereas patient cell line 923 responded to the least number of compounds (Figure 1). Altogether, the results demonstrate the variable drug responses from seven GBM cell lines derived from individual patient tumor samples. Therefore, personalized drug repurposing screens using patient-derived tumor cells represent a useful approach to identify effective drugs for the treatment of GBM.
Figure 1.
Heatmap of glioblastoma drug repurposing screen hits. Area under the curves (AUCs) of 20 confirmed hits were calculated from compound dose–response curves and color coded. Each AUC value is calculated as area under the dose–response curve fit from duplicate 11-point titration data from each cell line. Green indicates high activity whereas red indicates low activity.
Identifying effective drug combinations to combat infections with multidrug-resistant bacteria
Bacterial multidrug resistance puts a heavy burden on patients, providers and the healthcare system [13–15]. In some patients, treatments consisting of single antibiotics risk the growth of drug-resistant bacteria that can lead to severe complications and death [16]. Combination treatment is one solution to overcome antibiotic resistance but current antibiotic susceptibility tests are only suitable for approximately two-dozen compounds and are not able to identify effective drug combinations. In 2016, Sun et al. reported a rapid antibiotic susceptibility test for bacterial isolates with the capability of screening hundreds of approved drugs and drug combinations [17]. Several antibiotics not previously administered were found to be useful for combating the ten-most-common multidrug-resistant bacteria (MDRB) clinical strains, including Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Citrobacter freundii, Enterobacter cloacae and Escherichia coli. This new high-throughput method has greatly improved the current clinical antibiotic susceptibility test. First, hundreds of FDA-approved compounds can be screened against patient-derived cultures in a short period of time. Second, the targeted drug combination approach, which combines achievable drug plasma concentrations and the drug’s clinical breakpoint (the minimum inhibitory concentration that defines antimicrobial susceptibility) identifies two- or three-drug combinations. Some of these drug combinations showed synergistic effects. Combining drugs not only overcomes the antibiotic drug resistance of these MDRBs but also reduces a drug’s adverse effects. The researchers also applied this approach to a patient in the intensive care unit (ICU) with severe infection of MDRB and clinicians found that the new drug combinations effectively controlled the infection and treated the patient (unpublished data).
Utilizing organoid models with patient-specific mutations for drug discovery
Many diseases are caused by several distinct mutations in a single gene. Each class of genetic variants responds best to different approaches, complicating drug development and selection of medications for treatment. CF, for example, has >2000 mutations in the CF transmembrane conductance regulator (CFTR) protein that are separated into six classes of pathological effects on the CFTR protein including protein folding, synthesis and stability (http://www.genet.sickkids.on.ca/cftr/StatisticsPage.html) [18]. The most common mutation found in ~70% of patients is ΔF508 which prevents proper CFTR folding and trafficking to the cell surface membrane, thus preventing proper chloride channel function. The other mutations cause reduced chloride channel function, although the CFTR proteins are present in the plasma membrane. Two FDA-approved drugs indicated for CF are available: one CFTR channel activator and the other, a corrector, which improves the mutant CFTR protein folding and trafficking [19]. Different mutations of CFTR respond differently to these two drugs approved for CF treatment. Recently, organoid models using CFTR-mutant CRISPR knock-in iPS cells have been used to test the efficacy of these two approved CF drugs [9,20,21]. These organoids maintain the polarized spatial organization and demonstrate measurable parameters like the tissue swelling present in intestinal tissues.
In 2016, Dekkers et al. demonstrated how patient rectal-epithelia-derived intestinal organoid models can be used to characterize responses to the FDA-approved CFTR drugs lumacaftor and ivacaftor, or in a combination called Orkambi® (lumacaftor and ivacaftor) [19]. Ivacaftor activates channel gating, whereas lumacaftor acts as a corrector of protein folding and trafficking leading to proper localization of CFTR channels at the plasma membrane. Not all CFTR mutations responded equally to either of the two drugs or the combination, revealing the patient-to-patient variability in drug responses owing to the different CFTR mutations. The researchers discovered that rectal organoids with a ΔF508/G1249R mutation only responded positively to ivacaftor and not lumacaftor or the combination of both. Following this organoid study, the patient with this type of CFTR mutation was successfully treated with ivacaftor to relieve the CF symptoms with improved respiratory function and increased bodyweight [19]. This work demonstrates the usefulness of organoid models with different CFTR mutations to guide personalized treatment. Additionally, this information could potentially be used to treat other patients with CF suffering from this mutation.
A screening approach to personalized drug development
Personalized drug development can be initiated with a drug repurposing screen as overviewed in Figure 1. There are several major components in this process including selecting patients, collecting patient cells or samples, developing the screening assay, screening the compounds with the FDA-approved drug library (possibly testing the compounds in combination), identifying hits and verifying the ideal compound or group of compounds.
Selecting patients for PDS
Given that PDS uses the patient’s own cells in the screening assay, the first step is identifying which patients might be amenable to sampling and would benefit the most from PDS. Prioritizing selection within the boundaries of healthcare ethics requires understanding which patients undergoing traditional treatment are not fully benefiting from their current therapy or those for which the established therapy is detrimental or nonexistent. Patients would also be good candidates if they have a rare or neglected disease with no treatment options. Other candidates include those that have a medical condition such as an allergy or super sensitivity to the standard-of-care treatment. Cancer patients or others whose diseases become resistant to the standard treatment are also good candidates.
The particulars of a patient’s disease must be taken into consideration. Does the patient have a severe disease phenotype or secondary complications? Has the patient been afflicted with the illness for a prolonged period of time as in cancer or hereditary diseases? Has the patient exhausted all standard-of-care options and experimental treatments? Does the patient have a terminal condition? It is important to assess the health status and treatment regimen of the patient with these kinds of questions because this information will produce a better cost:benefit value. To this point, there is a worldwide effort to understand how lifelong environmental exposure, or the exposome, affects health and disease [22–24]. The individual’s microbiome, an internal environmental factor, is tightly intertwined with the exposome. The microbiome is widely appreciated for its ability to affect all aspects of human health and, when dysregulated, can be a disease initiator or modifier [25,26]. Each person’s exposome and microbiome is unique, and can thus introduce variables relating to drug metabolism, disease progression and treatment outcomes. The ‘unique disease principle’ has led to the field of molecular pathological epidemiology (MPE) which embraces heterogenous disease presentation with the understanding that the exposome can lead to differences in disease progression, morbidity and mortality [27,28]. MPE can help provide quantitative measures during the patient evaluation stage of PDS and has the potential to inform a precision medicine team about treatment options. To perform a personalized drug screen, obtaining viable patientderived samples including primary cells, iPS cells and their derivatives, engineered cells with patient-specific mutations or surgical tumor samples is crucial. Other sample types include isolated pathogens from patient tissues or bodily fluids.
Primary cells from patients
Although blood cells remain a convenient and useful source of patient cells, particularly for cell sorting, they are usually not sufficient to fulfill the need for PDS because they do not self-renew and are not suitable for imaging-based experiments. By contrast, dermal fibroblasts derived from patient skin biopsies are commonly used for diagnosis and study of genetic disease pathophysiology. Fibroblasts can be cultured and renewed for 20–30 passages, which is a necessary attribute for screening. Furthermore, the fibroblast monolayers can be easily imaged in phenotypic-based screens. Other types of patient primary cells are usually difficult to obtain or can only be obtained in small quantities for compound screening.
Tumor cells derived from patient surgical samples
Except for the small portion of blood-originating cancers, most cancer patients have solid tumors [29]. Surgically removed tumor samples are a good source of patient-derived cancer cells for drug screening [30]. Enzymatic digestion or mechanical dissociation are common methods used to dissociate cells from tumor samples but most isolated cancer cells either grow too slowly or cannot be passaged in normal culture conditions [30–32]. Cell immortalization using the SV40 oncogene [33] is a traditional method used to establish fast-growing cancer cell lines from primary tumor samples but, because the essential properties of the cells can change, they are not suitable for PDS. Several other methods have been reported to culture surgical sample-derived cancer cells without use of the traditional cell immortalization methods including the use of ROCK inhibitors in culture [34], transfecting patient-derived cells with the telomerase reverse transcriptase [35–37] and obtaining cancer stem cells from patient samples [38].
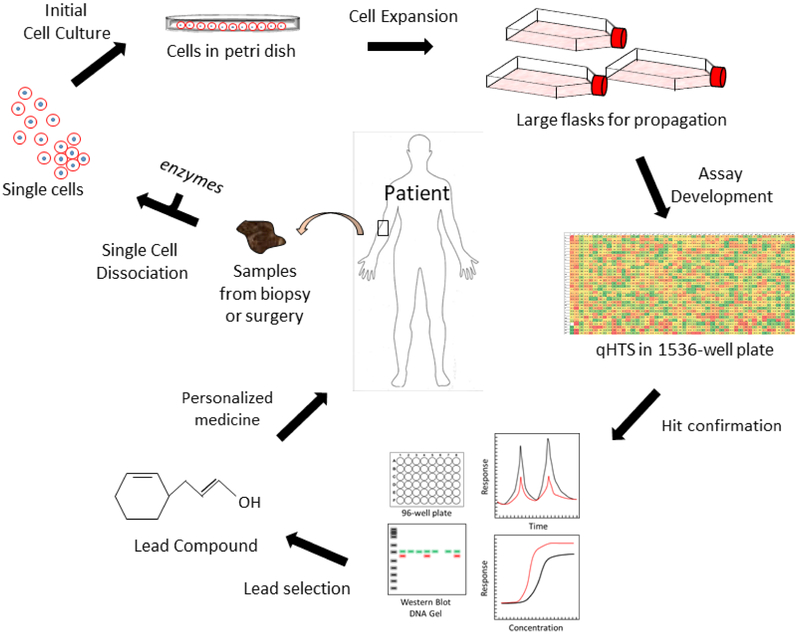
Furthermore, a diseased tissue might respond poorly to cell-dissociation protocols. For example, after dissociation the primary sample might not adhere well to plastic substrates along with decreased cell viability. Cells must be dissociated from the tissue using trypsin, collagenase or other digestion methods, and cultured in a flask or other plastic container with a suitable substrate such as poly-D-lysine, laminin, fibronectin or Matrigel™. Quality control must be performed on the cell culture to prevent contamination and to verify that the cells have the same genotype, as demonstrated by single tandem repeat (STR) profiling and phenotype as the cells from the primary tissue sample [39]. Figure 2 illustrates the different kinds of patient samples that can be obtained and cultured in a variety of ways for quantitative (q)HTS.
Figure 2.
Workflow for personalized medicine drug repurposing screen. A biopsy or sample is collected from the patient, possibly in the form of a removed tumor, skin punch, bacteria from blood urine or other bodily fluid. The sample is dissociated into single cells using a proteinase. The single cells are cultured in a smaller dish or flask before expansion. Once enough cells are collected, quantitative (q)HTS is performed and the hits are confirmed using biochemical assays and follow-up screens. Once the lead compound is selected, it is sent back to the clinic for use in the patient.
Assay development and execution
The laboratory conducting the PDS must also have a readily available, fully developed and robust assay in addition to an active drug-screening program. Generally, this also means that the labs need automated screening infrastructure. Although HTS assays are varied, an effective screening center should have high-content imaging machines, fluorescent and luminescent plate readers, high-throughput flow cytometry and high-throughput liquid-handling capabilities. A level of sophistication is also required for the center’s information technology infrastructure to handle the large quantities of data generated for each assay. Coupling the raw data with dedicated bioinformatics analysis allows for the team to expedite the delivery of meaningful data back to the clinic. The Assay Guidance Manual developed by NCATS and Eli Lilly is a continually revised and updated reference for the cutting-edge techniques and procedures required for PDS (https://www.ncbi.nlm.nih.gov/books/NBK53196).
Assays would preferably be developed with a primary or model cell line as closely related to the disease as possible to reduce assay artifacts and consequences. Researchers would need to collaborate with clinicians to select a drug library or a more narrow and focused selection of approved drugs for the screen. For example, approved cancer drugs and experimental drug candidates should be included in screens for cancer patients. If the mutations in the cancer and other genotype and/or phenotype information are known, compounds with known mechanisms of action to match the cancer profile should be prioritized for the screen. Depending on the disease and prognosis, there could be time to test non-FDA-approved compounds. Usually, time constraints must be considered because clinicians will need screening results quickly in the order of days to weeks.
Other information to consider includes the historical evidence of therapeutic efficacy, achievable blood plasma concentration (potency), potential side effects (tolerability) and available formulations for the proper route of administration. It will be essential to screen at clinically relevant concentrations of compounds to prevent a mismatch of dose and response between the in vitro and clinical data. Information about prior treatment regimens will be important for the team to assess whether a drug combination therapy should be developed. Assay design will vary significantly depending on these requirements. Researchers will need to move quickly to confirm the positive hits after the completion of the primary screen in a secondary screen with appropriate dose–response parameters and controls.
Concluding remarks and perspectives for personalized drug screening
As improvements in in vitro cell culture techniques and new compound screening assays are disseminated and adopted by researchers, personalized drug screening approaches to identify effective therapeutics for individual patients will have greater opportunities in clinical settings. As mentioned above, patient-derived cells and samples are needed to maintain conditions pathologically and phenotypically similar to the disease in patients – this is a crucial issue. Advances in 3D organoid cultures are accelerating, providing better in vitro cell models that mimic body tissues, mimic pathophysiology (such as tumor morphology and microenvironment) and can also provide insights about drug penetration into tissue [40,41]. However, there are challenges when working at the cutting-edge of drug discovery. Many organoids are derived from stem cells, but stem cell generation and reprogramming demand a significant amount of time and resources. Further development of technologies for convenient stem cell and organoid models are needed for better in vitro models for PDS.
At the present time, the monolayer cell culture is still useful for certain cases of PDS despite the points discussed above. Another alternative to monolayer cell culture is to develop an ex vivo model of the diseased tissue by isolating a larger sample and creating tissue slices 40–100 μm in thickness that could be cultured for an extended period of time to conduct drug screening [42,43]. Tissue slices have more similarity of structure and morphology to human tissues and are relatively easier to culture in vitro, whereas organoids are more-complex in nature and require advanced cell culture techniques. As samples become more complex, analysis of the massive amounts of data becomes more challenging. Nevertheless, data from advanced models such as these should be more clinically relevant than more-traditional methods such as the monolayer primary cell culture. More information on ex vivo preparations can also be found in an excellent review by Meijer et al. [42].
For cancer therapeutic development, circulating tumor cells (CTCs) provide an interesting option for clinicians and researchers alike. Indeed, a method for enumerating CTCs has been developed in the past decade and approved by the FDA as a liquid cancer biopsy [44,45]. Acquiring these cells is less invasive than surgical biopsy and they could have the potential to be enriched and expanded for drug repurposing screens. As these methods become more sophisticated and technology enables more-rapid development and analysis, personalized drug screening will be readily considered by clinicians as a therapeutic approach for intractable disease. In addition, combining the results of PDS and personal genome analysis brings two powerful technologies together to advance the future development of precision medicine.
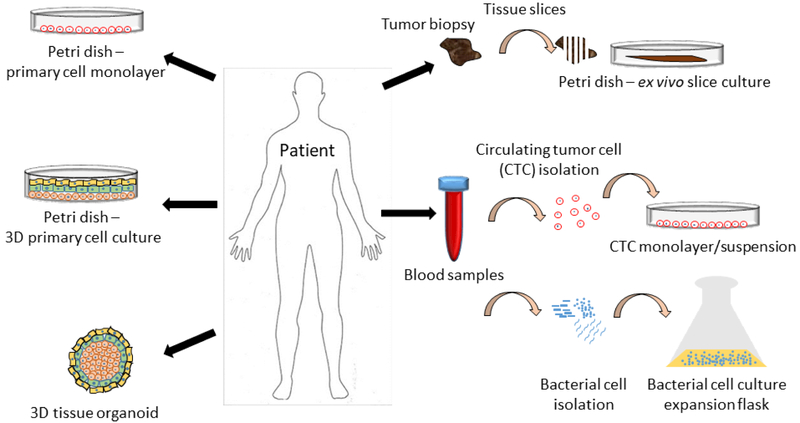
Figure 3.
Patient sample cell culture options for a personalized drug repurposing screen. Samples from patients can vary in scope and complexity. Samples could be in the form of skin fibroblasts used to grow a monolayer of cells in culture. Multiple cell types could be isolated for a more complex 3D layered culture system with one cell type growing with or on top of anothere cell type. Multiple cell types could be combined together to form 3D spheroids or organoids. A tumor could be resected and used for ex vivo tissue slice culture. Circulating tumor cells could be isolated from blood samples and cultured in a monolayer or suspension. Finally, bacteria could be isolated from blood, urine or other bodily fluids and expanded in large culture flasks.
Highlights:
Personalized drug screening (PDS) using patient-derived cells and approved drugs predicts drug efficacy in vitro before drug treatment in patients
This ‘bedside to bench and back to patient’ practice is a collaborative effort between basic researchers and clinicians
Personalized drug screening combined with personalized genomic analysis can contribute greatly to development of future precision medicine
The infrastructure and technologies need to be further developed for implementation of personalized drug screening in the clinical setting
Acknowledgments
This work was funded by the National Institutes of Health Intramural Research Program at the National Center for Advancing Translational Sciences (to W.Z.). The funding sources granted a large scope to the work that could be undertaken. This funding support had a very broad input into the study design, in the collection, analysis and interpretation of data, in the writing of the report and in the decision to submit the paper for publication. Dr DeeAnn Visk edited the manuscript for grammar, spelling, clarity and compliance with the journal style. K.G. and C.C. prepared the figures. K.G., C.C. and W.Z. wrote the manuscript. R.E.M, N.M., Y.C., K.C. and J.K.P. were responsible for carrying out the glioblastoma experiments, N.S., D-T.N. and C.C. analyzed the glioblastoma data. W.Z. conceived the idea for the manuscript.
Footnotes
Teaser: Examples of personalized medicine are discussed in this review, detailing the therapeutic benefits with drug-repurposing screens.
Conflicts of Interest
The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peck RW (2018) Precision medicine is not just genomics: the right dose for every patient. Ann. Rev. Pharmacol. Toxicol. 58, 105–122 [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA et al. (2017) Bedside back to bench: building bridges between basic and clinical genomic research. Cell 169, 6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verbelen M et al. (2017) Cost-effectiveness of pharmacogenetic-guided treatment: are we there yet? Pharmacogenomics J. 17, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips KA et al. (2013) The economic value of personalized medicine tests: what we know and what we need to know. Genet. Med. 16, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips KA et al. (2001) Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA 286, 2270–2279 [DOI] [PubMed] [Google Scholar]
- 6.Zhou L and Rupa AP (2018) Categorization and association analysis of risk factors for adverse drug events. Enr. J. Clin. Pharmacol. 74, 389–404 [DOI] [PubMed] [Google Scholar]
- 7.Zheng W et al. (2018) Drug repurposing screens and synergistic drug- combinations for infectious diseases. Br. J. Pharmacol. 175, 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan H et al. (2012) Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med. 367, 1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohwieler M et al. (2017) Stem cell-derived organoids to model gastrointestinal facets of cystic fibrosis. United European Gastroenterology J. 5, 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksson R et al. (2011) Impact of therapy on quality of life, neurocognitive function and their correlates in glioblastoma multiforme: a review. J. Neuro Oncol. 104, 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stupp R et al. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 [DOI] [PubMed] [Google Scholar]
- 12.Lee J et al. (2006) Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum- cultured cell lines. Cancer Cell 9, 391–403 [DOI] [PubMed] [Google Scholar]
- 13.Cornejo-Juarez P et al. (2015) The impact of hospital-acquired infections with multi drug-resistant bacteria in an oncology intensive care unit. Int. J. Infect. Dis. 31,31–34 [DOI] [PubMed] [Google Scholar]
- 14.Lim C et al. (2016) Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. eLife 5, el8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prestinaci F et al. (2015) Antimicrobial resistance: a global multifaceted phenomenon. Pathogens Global Health 109, 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins A et al. (2001) Bacterial antibiotic resistance In eLS, John Wiley & Sons [Google Scholar]
- 17.Sun W et al. (2016) Rapid antimicrobial susceptibility test for identification of new therapeutics and drug combinations against multidrug-resistant bacteria. Emerging Microbes Amp Infections 5, el16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutting GR (2014) Cystic fibrosis genetics: from molecular understanding to clinical application. Nat. Rev. Genet. 16, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekkers JF et al. (2016) Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 8, 344ra384. [DOI] [PubMed] [Google Scholar]
- 20.Cholon DM and Gentzsch M (2018) Recent progress in translational cystic fibrosis research using precision medicine strategies. J. Cystic Fibrosis 17, S52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekkers JF et al. (2013) A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939. [DOI] [PubMed] [Google Scholar]
- 22.Rattray NJW et al. (2018) Beyond genomics: understanding exposotypes through metabolomics. Human Genomics 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warth B et al. (2017) Exposome-scale investigations guided by global metabolomics, pathway analysis, and cognitive computing. Anal. Chem. 89, 11505–11513 [DOI] [PubMed] [Google Scholar]
- 24.Kim KN and Hong YC (2017) The exposome and the future of epidemiology: a vision and prospect. Environ. Health Toxicol. 32, e2017009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cani PD (2018) Human gut microbiome: hopes, threats and promises. Gut doi: http://dx.doi.Org/10.l136/gutjnl-2018-316723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon Y (2016) Microbiome-linked crosstalk in the gastrointestinal exposome towards host health and disease. Pediatric Gastroenterology Hepatology Nutrition 19, 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamada T et al. (2017) Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J. Gastroenterol. 52, 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogino S et al. (2018) Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 67, 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolston KVI (2017) Infections in cancer patients with solid tumors: a review. Infect. Dis. Ther. 6, 69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra A et al. (2013) Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 31, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naipal KAT et al. (2015) Attenuated XPC expression is not associated with impaired DNA repair in bladder cancer. PLoS One 10, e0126029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volovitz I et al. (2016) A non-aggressive, highly efficient, enzymatic method for dissociation of human brain-tumors and brain-tissues to viable single-cells. BMC Neurosci. 17, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbard K and Ozer HL (1999) Mechanism of immortalization. Age 22, 65–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilson SG et al. (2015) ROCK inhibition facilitates in vitro expansion of glioblastoma stem-like cells. PLoS One 10, e0132823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neelima PS et al. (2009) Effect of human telomerase reverse transcriptase transfection on differentiation in BeWo choriocarcinoma cells. Reproductive Biomedicine 18, 838–849 [DOI] [PubMed] [Google Scholar]
- 36.Gu Y et al. (2004) A telomerase-immortalized primary human prostate cancer clonal cell line with neoplastic phenotypes. Int. J. Oncol. 25, 1057–1064 [PubMed] [Google Scholar]
- 37.Janik K et al. (2016) Efficient and simple approach to in vitro culture of primary epithelial cancer cells. Biosci. Rep. 36, e00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmini G et al. (2016) Establishment of cancer stem cell cultures from human conventional osteosarcoma../. Visualized Experiments 116, 53884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bornman DM et al. (2012) Short-read, high-throughput sequencing technology for STR genotyping. BioTechniques 2012, 1–6 [PMC free article] [PubMed] [Google Scholar]
- 40.Dutta D et al. (2017) Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 23, 393–410 [DOI] [PubMed] [Google Scholar]
- 41.Liu F et al. (2016) Drug discovery via human-derived stem cell organoids. Front. Pharmacol. 7, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meijer TG et al. (2017) Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Science 3, FSO190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roife D et al. (2016) Ex vivo testing of patient-derived xenografts mirrors the clinical outcome of patients with pancreatic ductal adenocarcinoma. Clin. Cancer Res 22, 6021–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Albuquerque A et al. (2012) Prognostic and predictive value of circulating tumor cell analysis in colorectal cancer patients. J. Transl. Med. 10, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L et al. (2016) Promise and limits of the CellSearch® platform for evaluating pharmacodynamics in circulating tumor cells (CTC). Semin. Oncol. 43, 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]