Abstract
The ubiquitin proteasome system (UPS) is the major pathway for intracellular protein degradation in most organs, including the heart. UPS controls many fundamental biological processes such as cell cycle, cell division, immune responses, antigen presentation, apoptosis, and cell signaling. The UPS not only degrades substrates but also regulate activity of gene transcription at the post-transcription level. Emerging evidence suggests that impairment of UPS function is sufficient to cause a number of cardiac diseases, including heart failure, cardiomyopathies, hypertrophy, atrophy, ischemia-reperfusion, and atherosclerosis. Alterations in the expression of UPS components, changes in proteasomal peptidase activities and increased ubiquitinated and oxidized proteins have also been detected in diabetic cardiomyopathy (DCM).However, the pathophysiological role of the UPS in DCM has not been examined. Recently, in vitro and in vivo studies have proven highly valuable in assessing effects of various stressors on the UPS and, in some cases, suggesting a causal link between defective protein clearance and disease phenotypes in different cardiac diseases, including DCM. Translation of these findings to human disease can be greatly strengthened by corroboration of discoveries from experimental model systems using human heart tissue from well-defined patient populations. This review will summarize the general role of the UPS in different cardiac diseases, with major focus on DCM, and on recent advances in therapeutic development.
1. Overview of Ubiquitin Proteasome System:
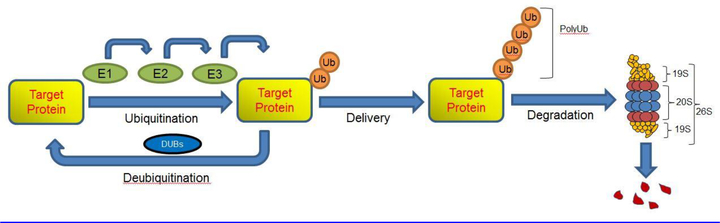
Over the past several decades, it has become increasingly apparent that protein degradation by the ubiquitin proteasome system (UPS) controls many fundamental biological processes such as cell cycle and cell division, immune responses and antigen presentation, apoptosis, and cell signaling [1]. In most mammalian cells, the UPS degrades more than 90% of proteins, ensuring that misfolded, oxidized, or damaged proteins, which possess an intrinsic toxicity, are degraded [1–3]. During the ubiquitination process, multiple ubiquitin proteins can be covalently attached to a target protein by ubiquitination enzymes. These ubiquitination enzymes include ubiquitin activating enzyme (E1), ubiquitin carrier protein (E2), and ubiquitin protein ligase (E3). Initially, E1 activates ubiquitin in an ATP-dependent manner, forming a thioester linkage between the carboxy-terminal glycine residue of ubiquitin and a cysteine in the active site of the E1 enzyme [4]. Activated ubiquitin is then transferred from an E1 enzyme to a cysteine residue of an E2 enzyme. E3 then catalyzes the final step of the ubiquitination process by transferring ubiquitin to lysine residues of targeted proteins, forming a polyubiquitin chain that earmarks the targeted proteins [1, 5]. Humans possess two E1 enzymes (UBA1 and UBA6), several dozen E2 enzymes, and several hundred E3 enzymes [6]. The specificity of the E3 enzymes determines the specific recognition of target proteins, providing selectivity in which proteins are targeted to the proteasome for degradation [7]. Upon ubiquitination, targeted protein is degraded by the 26S proteasome in an ATP-dependent manner [8]. The 26S proteasome is a complex consisting of a proteolytic core particle (20S proteasome) that is capped at both ends by 19S regulatory particles (19S regulatory complex). The 20S proteasome is a barrel-shaped complex comprised of four stacked rings and contains multiple catalytic centers in the chamber. The 19S proteasome recognizes a polyubiquitinated protein, unfolds it, liberates it from the polyubiquitin chain, and translocates the protein into the proteasome chamber for degradation (Figure 1). The ubiquitin molecules are recycled, and the peptides generated are used for antigen presentation or are degraded into amino acids that are recycled for new protein synthesis [9, 10].
Figure 1:
The ubiquitin-proteasome system (UPS) involves in polyubiquitination of the target protein prior to its degradation by the 26S proteasome. Polyubiquitination involves the combined action of three different enzymes: E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase). The eukaryotic 26S proteasome is a large, multicatalytic protein complex composed of the 19S regulatory complex and the 20S core. The damaged, misfolded, or mutant proteins are degraded by three major proteolytic activities. Figure adapted from [62].
2. UPS and cardiovascular diseases:
The UPS is the major pathway for intracellular protein degradation in most organs, including the heart. A functional UPS removes damaged or misfolded proteins that are critical for normal protein turnover and function [11–13]. Dysfunction of the UPS is rapidly gaining recognition as a potentially important mechanism involved in the pathogenesis of a number of cardiac diseases, including DCM [14, 15], and providing a strong rationale for further study of specific mechanisms of proteasome impairment and identification of specific targets for therapy. Given the consequentiality of these components in cardiac diseases, it is imperative to understand the effects of disease on the UPS itself.
2.1. UPS and Cardiac ischemia–reperfusion injury
Myocardial infarction remains an unsolved health problem, resulting in serious harm to human health. The ischemic myocardium is characterized by increased cardiomyocyte death, caused by necrosis and apoptosis [16]. Reperfusion is essential for myocardial salvage after myocardial infarction; however, it is often followed by additional myocardial injury, termed myocardial ischemia/reperfusion (I/R) injury [17]. The mechanisms of I/R injury involve the generation of reactive oxygen species, intracellular calcium overload, microvascular and endothelial dysfunction, altered myocardial metabolism, and concurrent activation of neutrophils, platelets, and complement [18]. Mice with myocardial I/R have been shown to have UPS function insufficiently, which is characterized by decreased proteosome activity and degraded proteins and which can lead apoptosis [19, 20]. It was shown recently that soluble receptor for advanced glycation end-products (sRAGE) inhibits I/R-induced apoptosis separately from blocking the interaction of RAGE with ligands [21]. The downstream of UPS function involves in regulating key apoptosis-related signaling pathways, such as MAPK, JNK, calcineurin, FOXO, and p53. sRAGE decreases the delayed-type hypersensitivity response in both RAGE−/− and RAGE+/+ mice in a similar manner, indicating that sRAGE not only purely blocks cell surface RAGE activation but also has other regulation pathways [22]. Inhibition of sRAGE on I/R-induced cardiomyocyte apoptosis is associated with activation and expression of proteasome, including improved proteasome activity and elevated β1i and β5i expression mediated by STAT3 activation [21]. However, further studies are required to demonstrate the concise mechanisms of sRAGE regulating the UPS following myocardial I/R.
The molecular chaperone machinery also plays a necessary role in protecting against myocardial infarction after I/R injury [23]. Previous literature revealed that CHIP, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardio protection after myocardial infarction in mice. CHIP also regulates the response to injury in the heart, where it is preferentially expressed, under pathophysiologically relevant conditions [24–26]. Mice lacking CHIP may be especially useful tool for analyzing how protective systems defend against myocardial damage, because CHIP coordinates a critical tier of the response to stress and protein damage in injured cells. The role of protein quality control mechanisms in protecting against cardiac injury needs to be reconsidered in order to find ways to rescue myocardium after I/R injury.
As discussed earlier, cardiomyocyte apoptosis plays a central role in the pathogenesis of myocardial disease and reactive oxygen species is critically responsible for mediating cardiomyocyte apoptosis in I/R injury. MDM2, a E3 ubiquitin ligase, has been shown to regulate the expression of the ARC (Apoptosis Repressor with Caspase recruitment domain) in I/R injury [27]. ARC is an anti-apoptotic protein that has been found to be abundantly present in terminally differentiated cells, such as cardiomyocytes. MDM2 directly accelerates ARC protein turnover via ubiquitination and proteasomal-dependent degradation. This activity requires a functioning MDM2 ring finger domain because the MDM2(C464A) mutant was unable to direct ARC degradation. Furthermore, ARC degradation requires MDM2. MDM2 knock-out fibroblasts showed defective ARC degradation that could be rescued by MDM2. Proteasomal inhibitors rescued bothMDM2 and H(2)O(2)-induced degradation of ARC and inhibited cardiomyocyte apoptosis [27].
MDM2 is also recognized as a negative regulator of the tumor suppressor p53 and thereby represents a critical upstream factor to determine whether cells enter the cell cycle or die by apoptosis [28]. In response to hypoxia, the protein level of p53 becomes elevated and mediates apoptosis in most cell systems [29]. Although adenoviral delivery of p53 was sufficient to promote apoptosis of isolated cardiomyocytes [30], in chronic coronary artery ligation and the Langendorff ischemia/reperfusion models, endogenous p53 was dispensable for apoptotic cell death [31]. Toth et al. showed that isolated cardiac myocytes overexpressing MDM2 acquired resistance to hypoxia/reoxygenation-induced cell death. Conversely, inactivation of MDM2 by a peptide inhibitor resulted in elevated p53 levels and promoted hypoxia/reoxygenation-induced apoptosis. [32]. Consistent with this, decreased expression of MDM2 in a genetic mouse model was accompanied by reduced functional recovery of the left ventricles determined with the Langendorff ex vivo model of ischemia/reperfusion. Overall these finding suggest that MDM2 may serve as a novel cardiac gene therapy target.
2.2. UPS and cardiac hypertrophy
Cardiac hypertrophy is key compensatory mechanism acting in response to pressure or volume overload, involving some alterations in signaling transduction pathways and transcription factors-regulation. These changes result in enhanced proteins’ synthesis leading to Left Ventricular Hypertrophy (LVH) [33].
Emerging evidences suggests that the UPS also attends to the cells’ growth, favoring protein’ synthesis, subsequently caused LVH [33]. Several recent reports have identified E3 ubiquitin ligases that exert anti-hypertrophic activity [4]. These E3s are Muscle-atrophy F-box (Atrogin1) and muscle-specific ring finger-1 (MuRF1) that are key mediators of skeletal muscle atrophy and found exclusively in skeletal muscle and the heart [34]. Further studies identified that these proteins have E3 ubiquitin ligase activity, and play a pathophysiology role in cardiac hypertrophy [35, 36]. Both Atrogin1 and MuRF1 inhibit cardiac hypertrophy by interacting with important hypertrophic signaling pathways [37].
Atrogin1 interacts with calcineurin and inhibits cardiac hypertrophy. An Atrogin1–calcineurin complex associates with Skp1, Cul1, and Roc1 to assemble an SCF (Atrogin1) complex with ubiquitin ligase activity [35]. Further, calcineurin dephosphorylates NFAT resulting in the transcription of hypertrophy-associated genes [35].
The MuRF1 is critical in regulating both pathological and physiological cardiac hypertrophy in vivo [38]. Mice globally lacking the striated muscle-specific MuRF1 (MuRF1−/−) exhibit an exaggerated physiological hypertrophy in response to exercise and an exaggerated pathological hypertrophy in response to pressure overload. This is attributed to MuRF1’s ability to inhibit serum response factor (SRF) and insulin-like growth factor-1 (IGF-1) signaling pathways, in part, through its targeted inhibition of cJun [39, 40]. MuRF1 also has a more direct role in regulating cardiac muscle mass by targeting proteasome dependent degradation of sarcomere proteins [41, 42]. MuRF1 interacts directly with cardiac troponin I, cardiac myosin binding protein-c (cMyBP-c), and myosin heavy chain, to direct their subsequent poly-ubiquitination and proteasome-dependent degradation [43, 44]. This regulation of sarcomere degradation explains why MuRF1−/− mice have resistance to cardiac atrophy and a limited ability to regress upon unloading after the induction of pressure overload-induced cardiac hypertrophy [45]. By regulating both the indirect signaling processes that activate cardiac hypertrophy and directly targeting sarcomere proteins for degradation, MuRF1 regulates the heart’s response to external stress that lead to disease. The muscle-specific ubiquitin ligase MuRF1 has also recently been shown in vivo to specifically inhibit PPARαactivity, but not PPARβ/δ or PPARγ1 activity [46]. In contrast to the previous studies on nuclear receptors, MuRF1 was found to inhibit PPARα in cardiomyocytes through mono-ubiquitination that enhances nuclear export without promoting PPARα degradation [46]. Complementary to these studies, the closely related muscle-specific ubiquitin ligase MuRF2 inhibited PPARγ1 [47], whereas MuRF3 inhibited PPARβ activity in vivo by mono-ubiquitination [48]. Recently it was found that MuRF1 inhibits thyroid receptor T3-induced physiological cardiac hypertrophy in vivo [39]. MuRF1 directly interacts with a specific thyroid receptor to mediate its posttranslational modification and inhibit its activity. Recent evidence has also begun to delineate the role of MDM2 in the context of cardiac hypertrophy. MDM2 overexpression promotes cell survival and attenuates hypertrophy [37]. MDM2 down-regulates the sarcomeric protein T-cap (telethonin) through the proteasomal pathway, an effect that is inhibited by p14/ARF expression [32]. This proteasomal degradation of T-cap, which inhibits cardiac hypertrophy, occurs in a ubiquitin-independent manner by a mechanism yet to be defined [49]. These findings suggest that the UPS is essential for protection against apoptosis and depression of proteasome activities in cardiac hypertrophy.
The MuRF1 protein also inhibits pathologic cardiac hypertrophy by attenuating PKCε activity and translocation [50]. Upon stimulation with phenylephrine or PMA to induce hypertrophy in cardiomyocytes, MuRF1 interacts with the receptor for activated protein kinase C (RACK1) and blocks PKCε translocation (and activity) to focal adhesion kinases. Blocking PKCε signaling results in the inhibition of focal adhesion formation and downstream ERK1/2 signaling. It has also been shown that cardiac troponin I (cTnI) is targeted for ubiquitin-dependent proteasomal degradation through interaction with MuRF1 by its RING finger domain [44]. Increased MuRF1 expression correlates with reduced cTnI and reduced indices of contractility, indicating that MuRF1 may play a key role in cardiac physiology beyond its antihypertrophic effects [40].
2.3. UPS and cardiac atrophy
Cardiac atrophy is seen in mechanical unloading of the heart, such as that seen after the placement of the left ventricular assist device (LVAD) [51]. Atrophic remodeling of the heart is associated with simultaneous activation of regulators of both proteolysis and protein synthesis. Much less is known about the UPS in cardiac atrophy, likely because this state is physiologically less relevant. Studies using gene expression profiling have shown that transcript levels of ubiquitin B (UbB), the ubiquitin conjugating enzyme (E2–14 kDa), the ubiquitin ligases muscle atrophy F-box protein (Mafbx)/Atrogin-1, and MuRF-1, as well as the proteasomal subunit PSMB4, are increased in skeletal muscle atrophy induced by nutrient deprivation, unloading, diabetes, uremia, and cancer[52, 53].Recent study also confirmed that UbB is an important marker for cardiac remodeling and a potential target to modulate UPS-mediated cardiac remodeling. [4, 54].
2.4. UPS and cardiomyopathies
Cardiomyopathies are intrinsic heart muscle diseases characterized by adverse ventricular and atrial remodeling and systolic and/or diastolic dysfunction. Types of cardiomyopathy includes ischemic cardiomyopathy, which results from previous myocardial infarction(s), and/or ongoing coronary ischemia and hypertrophic cardiomyopathy (HCM), which is the most common inherited cardiomyopathy (population frequency, 1:500); HCM is characterized by cardiac hypertrophy and diastolic dysfunction that typically develops over many years, and can be complicated by heart failure, arrhythmias, and sudden cardiac death [55, 56]. The majority of HCM cases are linked to autosomal dominant mutations in cardiac sarcomere genes, with a small percentage of others associated with mutations in Z-disc, metabolic, and Ca2+handling genes [57–59]. However, to reach any conclusions, it is necessary to find distinct features of different cardiomyopathies in patients from whom tissue were obtained [11, 60, 61].
A major proteasome functional insufficiency has also been shown in familial dilated cardiomyopathies [62, 63]. In the pure form of familial dilated cardiomyopathies, more than 12 identified genes encode different components of cardiomyocytes, including sarcomeric proteins, lamin A/C, and cytoskeletal proteins such as AB-crystallin (CryAB) and desmin [62, 63]. In failing hearts of patients with familial dilated cardiomyopathies, researchers have observed marked accumulation of ubiquitinated proteins as well as up-regulation of several UPS components, concomitant with elevated levels of pro-apoptotic p53 [64]. Researchers have also frequently found Desmin aggregates in human heart failure studies that clearly indicate that UPS dysfunction could be a causative factor in the development of dilated cardiomyopathies [65].
Activation of autophagy is another potential consequence of UPS inhibition [66]. While the mechanisms by which these two pathways interact are not fully understood, multiple lines of evidence implicate p62 as a key linker protein and sensor of proteotoxic stress. p62 is a multifunctional adaptor protein that binds K63-linked ubiquitin chains and also directly binds to microtubule-associated protein 1A/1B-light chain 3, which could promote autophagosome formation [66]. In summary, proteasome dysfunction in human heart failure may have profound downstream effects on diverse pathways that actively contribute to cardiomyopathies.
2.5. UPS and atherosclerosis
Atherosclerosis is a chronic inflammatory disease accompanied by the intimal thickening of systemic arterial walls [67]. Rupture of vulnerable plaques as well as thrombotic/embolic occlusion and arterial narrowing can cause lethal ischemic disease, including acute coronary syndromes and myocardial infarction [68]. Like other cardiovascular diseases, dysfunctional proteasomes can also cause atherosclerosis [69]. An atherosclerosis study in high fat diet-fed pigs identified that proteasomal activity was elevated in coronary arteries during the progression of atherosclerosis. Furthermore, this study confirmed that chronic proteasome inhibition is associated with increased oxidative stress, impairment in coronary endothelium-dependent vasorelaxation, and intimal thickening, resembling and aggravating the vascular effects of traditional cardiovascular risk factors such as hypercholesterolemia [70]. Inhibition of proteasome activity is also associated with in determining atherosclerotic plaque instability [70]. Herrmann et al. 2007 examined the long-term administration of the proteasome inhibitor MLN-273 in pigs fed a high cholesterol diet and identified the elevation of oxidative stress and exacerbation of atherosclerosis [70]. Wlick et al. (2012) also reported that low-dose proteasome inhibition exerts antioxidative and anti-inflammatory effects and attenuates development of atherosclerotic lesions in low-density lipoprotein receptor–deficient mice [71]. These studies clearly suggest that treatment with low doses of proteasome inhibitors exerts favorable anti-inflammatory and antioxidative effects in vascular cells.
Role of ubiquitin proteasome systemin diabetic cardiomyopathy
3.1. Diabetic cardiomyopathy; an overview
Diabetic cardiomyopathy (DCM) is a disorder of the heart independent of hypertension and coronary arterial disease; the condition results in wide range of structural abnormalities to the myocardium, and eventually leads to left ventricular hypertrophy (LVH), diastolic and systolic dysfunction, or a combination of all three [72]. Hyperglycemia, increased ROS, and perhaps other altered metabolic factors present in diabetes can cause protein glycation, oxidation, and cross-linking [37], with accumulation of these abnormal proteins in the heart contributing to the development of diabetic cardiomyopathy. However, it is unknown if these abnormal protein accumulations also contribute to DCM. An in-depth study of proteasome alteration in diabetic hearts has not yet occurred, but both increased and decreased proteasome function were implicated in diabetes [73]. It appears that hyperglycemia and hyperinsulinemia can increase UPS function, which may be responsible for endothelial damage and atherosclerosis as well as muscle wasting in diabetes [74]. Since increased ROS is a major pathogenic factor in diabetes, it is conceivable that oxidative modification of proteasome subunits may play a role in altering proteasome function in diabetes [72–74].
More than forty years have passed since DCM was discovered, but the mechanisms responsible for the development of DCM remain largely unknown despite comprehensive research. Recently, research has focused on gene transcriptions to elucidate mechanisms of disease progression. Several transcription factors (TFs) were reported to play vital roles in the development of DCM [75–77]. UPS not only degrades substrates, but also regulates activity of gene transcription at the post-transcription level [78]. Therefore it could be possible that UPS may regulate activity of gene transcription into the development of DCM. Additionally, the UPS is associated with DCM by its contribution to insulin resistance and insulin deficiency, as it degrades the insulin receptor and the insulin receptor substrate and regulates insulin gene transcription. Moreover, UPS are involved in cardiac apoptosis, hypertrophy, and fibrosis which are the main cardiac structural changes associated with diabetes [37]. This suggests that the UPS act as a fundamental factor of DCM [37]. Up until now, even though many molecular mechanisms have been proposed for DCM, little attention is paid to the roles of the UPS in DCM [79]. Therefore, in this section we will summarize the general role of UPS in DCM.
3.2. UPS regulates p53 signaling pathway in DCM
p53 is a well-known proapoptotic protein in the heart [64, 80], and high levels of p53 are found in several types of heart diseases, including DCM [80]. Besides proapoptosis, p53 also damages the heart by increasing mitochondrial oxygen consumption and ROS generation [80]. In the heart, three E3s target p53 as their substrates: MDM2, its homologue MDM4, and CHIP [81]. Through degradation of p53, the three E3s inhibit myocyte apoptosis, thus protecting the heart from stressors including hyperglycemia and diabetes [82]. However, p53 also shows antiapoptotic activity through activating the Bcl-2 family [83]. Mice deficient in p53 exhibit an increased risk of cardiac rupture after myocardial infarction [84], indicating a beneficial role of p53 in the heart. However, it must be remembered that p53 inhibition is accompanied by a higher risk of cancer. In addition, the E3s regulate apoptosis by modulating the activities of caspases, which play essential roles in apoptosis.
3.3. UPS regulates vital transcription factors in DCM
New molecular methods and knockout animal models have gained deeper insight in the mechanism of DCM at the transcription level. Currently several TFs, including PPARα, Nrf-2 and FoxOs, have been identified as playing major roles in DCM and as having their activities regulated by the UPS, providing support for the hypothesis that the UPS plays a causal role in DCM. PPARα was the first identified TF that is related to the genesis of DCM [85]. It was found that expression of PPARα target genes involved in cardiac fatty acid uptake and oxidation pathways was increased in MHC-PPAR mice. Surprisingly, the expression of genes involved in glucose transport and utilization was reciprocally repressed in MHC-PPAR hearts. Two types of ubiquitin ligases, MG53 and MuRF-1, act as upstream regulators of PPARα [85]. MG53 positively upregulates the expression level of PPARα, thus triggering a cascade of events that leads to DCM. Additionally, in vitro loss- and gain-of-function studies firmly have established PPARα as downstream to MG53 [85]. In contrast, MuRF1 inhibits PPARα activity by monoubiquitination and facilitates its nuclear export, thus blocking PPARα-induced gene transcription [46].
Another TF involved in DCM is nuclear factor (NF) erythroid 2-related factor 2(Nrf-2), which regulates expression of a set of antioxidant proteins (such as NAD(P)H quinone oxidoreductase 1, glutathione S-transferase, and heme oxygenase-1) thus protecting the heart in oxidative stress. Under baseline conditions, Nrf-2 is combined with Kelch-like ECH-associated protein 1 (Keap1), which acts as a substrate adaptor of the cullin3-Keap1-E3 ubiquitin ligase complex. MG-132, a proteasome inhibitor, can upregulate Nrf2-mediated antioxidative function and downregulate NF-κB-mediated inflammation [86]. A previous study suggests that MG-132 has a therapeutic effect on diabetic cardiomyopathy in OVE26 diabetic mice, possibly through the upregulation of Nrf2-dependent antioxidative function and downregulation of NF-κB-mediated inflammation. With the exception of preventing Nrf-2 degradation, modulation activity of Keap1 is another way to enhance Nrf-2 level. It enables Nrf-2 release from the cullin3 ubiquitin ligase complex [87]; thus, it is a promising target to protect the heart from diabetes.
Forkhead box (other) transcriptional factors (FoxOs) are important TFs in the development of DCM, and their levels are consistently high in the diabetic heart [88–90]. FOXOs are responsible for regulating the ubiquitin-proteasomal pathway, as well as the autophagy-lysosomal pathway. The ubiquitin-proteasomal pathway is activated by an absence of nutrients, thus leading to proteasome-dependent protein degradation [91]. FOXOs consist of a highly conserved forkhead/winged helix DNA-binding domain, which encompasses the most common 110 amino acids of the FOXO family and embodies 3a, 3b, and 2 winged helices, facilitating its DNA binding [92]. FOXO1 and FOXO3 are expressed globally, and FOXO1 isoform is abundantly located in hepatic, fatty tissue and pancreatic β cells [93]. Post-translational modifications (PTM) such as phosphorylation, acetylation, ubiquitination, arginine methylation, and O-glycosylation [94] are known to determine the FOXO1 nuclear transit and transcriptional activity [95]. These modifications can either enhance or reduce the FOXO1 transcriptional activity, as determined by the upstream target and/or the sites concerned. AKT phosphorylates FOXO1 facilitates its nuclear transit, which consecutively decreases the transcriptional function of FOXO1 [96]. However, several other kinases like mitogen-activated protein kinases (also known as JNKs), cyclin-dependent kinase 2 and nuclear factor κB (NFκB) kinase are also involved in FOXO1 phosphorylation [97]. The nuclear compartmentalization and transcriptional function of FOXO1 can also be modified by other PTM like acetylation, ubiquitination, glycosylation and methylation [98]. Disturbances of myocardial glucose and lipid metabolism are initial events in DCM [88]. During insulin resistance, pyruvate dehydrogenase kinase 4 (PDK4) is known to inhibit glucose oxidation by blocking pyruvate from undergoing mitochondrial oxidation through phosphorylating the E1 moiety of pyruvate dehydrogenase complex [99]. Enhanced expression of FOXO1 downstream target gene PDK4 gene is also observed in high fat and obese animal models of insulin resistance, which adversely regulate insulin actions [100]. FOXO1 can modulate glucose metabolism in adult cardiomyocytes in insulin resistance and diabetic conditions by inhibiting glucose oxidation preceded by PDK4 activation, subsequently altering the substrate preference for fatty acid and lactate [101]. These findings collectively suggest that FOXO1 is a key factor responsible for anomaly of glucose and lipid metabolic pathways in insulin resistance.
3.4. UPS regulates TGFβ pathway in DCM.
Myocardial fibrosis and collagen deposition are the main structural changes observed in DCM. It is known that transforming growth factor-β (TGFβ) plays a causal role in cardiac fibrosis [102], and the Smad proteins are signaling transducers downstream from TGFβ receptors. Three families of Smad proteins have been identified: receptor-activated Smad2 and Smad3(R-Smads), whose nuclear translocation can activate transcription of a select set of profibrosis genes; common partnerSmad4; and inhibitory Smad6 and Smad7 (I-Smads). Smad7 is a key regulator of TGFβ signaling through negative feedback loops [103]. It forms a stable complex with the TGFβ receptor, thus inhibiting phosphorylation and nuclear translocation ofR-Smads [104]. Smad7 also inhibits TGFβ signaling in the nucleus by interacting with transcription repressors such as histone deacetylases or disrupting formation of the R-Smads-DNA complex [105]. Overexpression of Smad7 in primary cardiac myofibroblasts significantly reduces collagen synthesis, concomitant with increased expression of metalloproteinase-2 [106]. Three members of the E3s mediate degradation of Smad proteins. The Smad ubiquitination regulatory factor 1 (Smurf1) and factor 2 (Smurf2) bind to TGF receptors through Smad7 [106–108] and degrade the TGFβ receptor, thus blocking profibrosis signaling. Arkadia, another E3 ubiquitin ligase, induces the ubiquitination and degradation of Smad7 and its repressors c-Ski and SnoN, thus enhancing TGFβ signaling [109, 110]. In Otsuka Long-Evans Tokushima Fatty rats that typically showed hyperglycemia after 18 weeks of age [111], cardiac expression of TGFβ receptorwas increased at 15 weeks of age, concomitant with enhanced collagen deposition [111]. Thus, interventions targeting Smurf1or Smurf2 are promising strategies to block cardiac fibrosis in the early stage of diabetes.
3.5. UPS regulates apoptotic pathways in DCM
Myocardial apoptosis is a deeply controlled self-protective process in response to severe stress. It is increasingly clear that the UPS regulates cardiac apoptosis via inhibitors of apoptosis protein (IAPs). IAPs were initially identified in viruses by their ability to inhibit apoptosis in infected cells [112]. The IAPs have a conserved RING finger motif at the COOH terminus that has E3 ubiquitin ligase activity. To date, a growing number of IAPs have been identified, including three in the heart: two cellular IAPs (cIAP1 and cIAP2) and an X-linked IAP (XIAP). Each of these IAPs has a specific target: cIAP1 targetscaspase-3 and caspase-9, cIAP2 targets caspase-3 and caspase-7[113], and XIAP targets caspase-3 [114], which accounts for rapid degradation of active caspase-3 in a proteasome-dependent manner [115]. Overexpression of XIAP protects cell function and decreases the number of human islets required to reverse hyperglycemia in the diabetic mice [116]. XIAP also protects the heart from ischemic injury, as evidenced by smaller infarct size, reduced apoptosis, and decreased level of cleaved caspase-3 [111]. However, in-vivo evidence is needed to confirm the role of IAPs in diabetes induced myocardial apoptosis.
3.6. UPS regulates calcium sensing receptor in DCM.
Calcium sensing receptor (CaSR) is a G protein-coupled receptors that is widely expressed in prokaryotic and eukaryotic cells. Early study found that the function of CaSR is to regulate the release of parathyroid hormone and maintain the body’s calcium homeostasis [117]. Furthermore, CaSR is also closely related to cell differentiation, proliferation, migration, apoptosis, brain development, and wound healing [118, 119]. Previously, it was found that CaSR is involved in myocardial ischemia reperfusion injury, myocardial infarction, and pulmonary hypertension [120, 121]. Moreover, recently it was observed that the cardiac function and the expression of CaSR were decreased in the rat diabetic myocardium [122]. Reports suggest that high glucose (HG) decreased the expression of CaSR, mitochondrial fusion proteins (Mfn1, Mfn2), cell gap junction related proteins (Cx43, β-catenin, Ncadherin), and intracellular ATP concentration. In contrast, HG increased extracellular ATP concentration, the expression of gp78, mitochondrial fission proteins (Fis1, Drp1), and the ubiquitination levels of Mfn1, Mfn2, and Cx43. Moreover, CaSR agonist and gp78-siRNA significantly reduced the stated changes [123]. Taken together, these results suggest that HG induces myocardial energy metabolism disorder via a reduction in CaSR expression and activation of the gp78-ubiquitin proteasome system. Stimulation of CaSR significantly attenuates HG-induced abnormal myocardial energy metabolism, indicating that CaSR may hold promise as a potential therapeutic target for DCM.
4. Therapeutic potentials of the UPS
The UPS has a central position in maintaining cellular homeostasis. Because of its involvement in a wide variety of important regulatory processes many proteins involved in the ubiquitination, deubiquitination, or ubiquitin-mediated degradation of proteins are considered to be potential drug targets. New research tools are rapidly being developed to make novel drug inhibitors that target different UPS steps.
4.1. Proteasome inhibition
In recent years, several types of low-molecular weight inhibitors of the proteasome have been identified that can readily enter cells and selectively inhibit the proteolytic function of the proteasome complex [124]. Bortezomib (Velcadei) is a novel dipeptide boronic acid that is the first proteasome inhibitor to have progressed to clinical trials. Bortezomib has now been approved by the U.S. Food and Drug Administration (FDA) and the European Agency for the Evaluation of Medicinal Products (EMEA) for the treatment of multiple myeloma patients. Therefore, most of the data related to clinical studies and side effects were obtained from cancer studies. The risks that are associated with bortezomib therapy include new or worsening peripheral neuropathy, orthostatic hypotension, gastrointestinal adverse events and thrombocytopenia. Moreover, the acute development or exacerbation of congestive heart failure has been seen in patients with risk factors for, or pre-existing, heart disease [125]. Although there seems to be some risk of cardiovascular side effects when proteasome inhibitors are applied as cancer treatment intermittently for several weeks, it has been proposed that proteasome inhibitors might have beneficial effects. These effects are due to their potential anti-inflammatory properties when administered for a short time immediately after acute ischemic events, such as myocardial infarction or stroke. Based on animal studies, it has been proposed that proteasome inhibitors might have the potential to attenuate reperfusion injury and might work synergistically with current myocardial infarction–reperfusion therapies [126]. Existing thrombolytic agents have a narrow window of time for therapeutic application. The hope for the future is to increase this period using proteasome inhibitors, which would expand the patient population that could receive thrombolytic therapy [127, 128]. The current treatment for acute myocardial infarction is percutaneous coronary intervention (PCI), but this treatment is limited to hospitals with cardiac surgery departments. Therefore, thrombolytic therapy, while not the favored option, is the treatment of choice when PCI cannot be preformed immediately. As such, expanding the range of patients who can receive this treatment is critical for increasing the survival rate of acute myocardial infarction [129]. The anti-inflammatory action of proteasome inhibitors might also offer an opportunity for tolerance in duction in transplant recipients. It has been demonstrated in vitro that proteasome inhibitors could suppress the proliferation and induce the apoptosis of activated T cells. This finding suggests that such inhibitors could be used as a novel category of immunosuppressants in blocking allograft rejection. Indeed, in a mouse heterotopic heart allograft rejection model, the proteasome inhibitor dipeptide boronic acid prolonged heart allograft survival from 7 to 35 days [35]. Initial reports on the effects of proteasome inhibitors indicate that proteasome inhibition might also be an effective therapeutic strategy for the reduction of restenosis after balloon angioplasty of coronary arteries and stent implantation. In a balloon-injury model of the rat carotid artery, local administration of the proteasome inhibitor MG132 effectively reduced neointima formation, which was associated with strong antiproliferative and proapoptotic effects on vascular smooth muscle cells and reduced infiltration of macrophages [130]. Although these proof-of-concept experiments have established novel and interesting therapeutic options for the treatment of cardiac diseases, ongoing and future clinical studies are needed to confirm the experimental findings obtained with proteasome inhibitors in the clinical setting.
4.2. Deubiquitinating enzymes
The process of ubiquitination can be reversed by Deubiquitinating enzymes (DUBs). DUBs either trim or completely remove ubiquitin chains from ubiquitinated proteins. DUBs demonstrate specificity for both substrates as well particular ubiquitin chain types [131]. DUBs have key functions in the regulation of important cellular functions. By reversing ubiquitination, DUBs affect the function, localization, or stability of proteins. Recently it was found that the reduced ubiquitin specific protease 14(USP14) deubiquitinase activity resulted in lower expression levels of the myocardial hypertrophy specific marker β-MHC, and subsequently led to decreased GSK-3β phosphorylation [132]. Furthermore, this finding indicated that USP14 might represent a novel therapeutic target for cardiac hypertrophy treatment.
Another DUB named Cyclindromatosis (CYLD) is also associated with mediating cardiac maladaptive remodeling and dysfunction [133]. CYLD was originally identified as a gene mutated in familial cylindromatosis, a genetic condition that predisposes individuals tothe development of tumors of skin appendages, termed cylindroma [134]. Subsequent studies have revealed that CYLD regulates diverse physiological processes, ranging from cell cycle progression and immune response to spermatogenesis and osteoclastogenesis, and also plays a key role in the pathogenesis of cancer, lung fibrosis, and inflammatory bowel disease [134, 135] Knockout of CYLD improved survival rate and alleviated cardiac hypertrophy, fibrosis, apoptosis, oxidative stress, and dysfunction in mice that were subjected to sustained pressure overload induced by transverse aortic construction [133]. This data suggests that CYLD may act as a potential therapeutic target in cardiac protection.
4.3. 11S proteasome targeted therapy
The 11S-activated proteasome is generally believed to mediate protein degradation in a ubiquitin- and ATP-independent manner [7]. The 11S proteasome can be formed by either PA28α and PA28β hetero heptamers (α3β4 or α4β3) or by PA28γ in homo heptamers (γ7) [136]. Using a mouse model with enhanced proteasome function by cardiac-specific overexpression of PA28α, it has been found that restoration of UPS function attenuates cardiac dysfunction [19]. This novel finding suggests that insufficient proteasome function contributes to the pathogenesis of diabetic cardiomyopathy. PA28α overexpression improves UPS function in diabetic hearts. It has been reported that overexpression of PA28α upregulates 11S proteasome abundance by stabilizing PA28β, leading to increased association of 11S proteasomes with 20S proteasomes [136, 137]. The 20S proteasomes are capable of degrading denatured, non-ubiquitinated proteins in an ATP-independent manner [138] and the 11S proteasomes upregulate 20S proteolytic activity by binding to the 20S α-ring subunits and facilitating gate opening of the 20S proteasome [139]. Therefore, it is plausible that under pathological conditions such as diabetes, cells may resort to the 11S–20S proteasomes for bulk degradation of damaged proteins, and as a consequence of PA28αOE, the increased 11S proteasomes protect cardiomyocytes against diabetes-induced proteotoxicity. In support of this notion, the 11S proteasomes were reported to be strongly induced by oxidative stress, in turn promoting the degradation of oxidized proteins [140]. It has been shown that the diabetic PA28αOE hearts displayed a significant reduction in diabetes-induced cardiac fibrosis and apoptosis, which, however, remained higher than in nondiabetic hearts [19]. Furthermore, the findings of this study indicate that proteasome function in cardiomyocytes, in combination with other strategies (e.g., elevation of autophagy, antioxidant,anti-inflammatory), could be more effective in limiting diabetic cardiac injury.
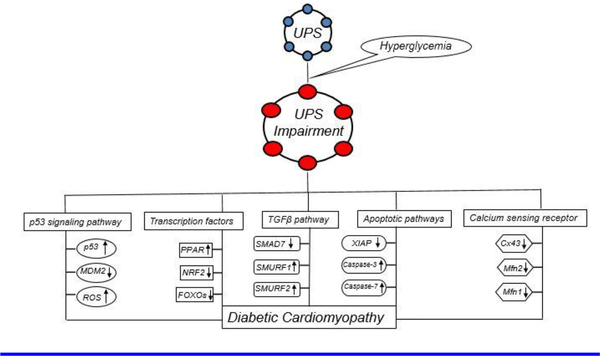
5. Conclusion:
The UPS is a quality control system in the mammalian body that eliminates potentially harmful abnormal proteins. In the heart, the UPS plays an important role in intracellular pathways such as transcriptional regulation, apoptosis, and inflammation. Hyperglycemia-induced Impairment of the UPS alters these pathways and leads to structural damage, such as fibrosis and hypertrophy. Together all of these changes lead to DCM (Figure 2). The studies presented in this review highlight the important signaling molecules and pathways regulated by the UPS that lead to cardiac diseases. Targeting these signaling molecules and pathways will potentially enable the development of future therapeutic options.
Figure 2:
The hyperglycemia induced UPS impairment dysregulates several signaling pathways that leads to DCM. Figure adopted from [37].
7. Background and Translational Significance:
UPS impairment is important mechanism involved and related to the number of cardiac diseases. In this review we highlighted the important signaling pathways that altered in cardiac diseases and related to UPS dysfunction. Targeting these pathways may be useful for better therapeutic approach in cardiac diseases.
6. Acknowledgement:
This work was supported by grants from the National Institutes of Health (HL111278). All authors have read the journal’s policy on conflicts of interest. The authors do not have any conflict of interest. All authors have also read the journal’s authorship agreement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8. References:
- [1].Pagan J, Seto T, Pagano M, Cittadini A. Role of the ubiquitin proteasome system in the heart. Circ Res. 2013;112:1046–58. [DOI] [PubMed] [Google Scholar]
- [2].Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–11. [DOI] [PubMed] [Google Scholar]
- [3].Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–4. [DOI] [PubMed] [Google Scholar]
- [4].Willis MS, Patterson C. Into the heart: the emerging role of the ubiquitin-proteasome system. J Mol Cell Cardiol. 2006;41:567–79. [DOI] [PubMed] [Google Scholar]
- [5].Zhong JL, Huang CZ. Ubiquitin proteasome system research in gastrointestinal cancer. World J Gastrointest Oncol. 2016;8:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jung T, Catalgol B, Grune T. The proteasomal system. Mol Aspects Med. 2009;30:191–296. [DOI] [PubMed] [Google Scholar]
- [7].Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. [DOI] [PubMed] [Google Scholar]
- [8].Hershko A The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture). Angew Chem Int Ed Engl. 2005;44:5932–43. [DOI] [PubMed] [Google Scholar]
- [9].Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haas AL, Bright PM. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J Biol Chem. 1987;262:345–51. [PubMed] [Google Scholar]
- [11].Day SM. The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am J Physiol Heart Circ Physiol. 2013;304:H1283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Flegal KM, Kit BK, Orpana H, Graubard BI. Association of all-cause mortality with overweight and obesity using standard body mass index categories: a systematic review and meta-analysis. JAMA. 2013;309:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kriegenburg F, Ellgaard L, Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2012;279:532–42. [DOI] [PubMed] [Google Scholar]
- [14].Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–24. [DOI] [PubMed] [Google Scholar]
- [15].Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eefting F, Rensing B, Wigman J, Pannekoek WJ, Liu WM, Cramer MJ, et al. Role of apoptosis in reperfusion injury. Cardiovasc Res. 2004;61:414–26. [DOI] [PubMed] [Google Scholar]
- [17].Takemura G, Nakagawa M, Kanamori H, Minatoguchi S, Fujiwara H. Benefits of reperfusion beyond infarct size limitation. Cardiovasc Res. 2009;83:269–76. [DOI] [PubMed] [Google Scholar]
- [18].Luqman N, Sung RJ, Wang CL, Kuo CT. Myocardial ischemia and ventricular fibrillation: pathophysiology and clinical implications. Int J Cardiol. 2007;119:283–90. [DOI] [PubMed] [Google Scholar]
- [19].Li J, Ma W, Yue G, Tang Y, Kim IM, Weintraub NL, et al. Cardiac proteasome functional insufficiency plays a pathogenic role in diabetic cardiomyopathy. J Mol Cell Cardiol. 2017;102:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Portbury AL, Ronnebaum SM, Zungu M, Patterson C, Willis MS. Back to your heart: ubiquitin proteasome system-regulated signal transduction. J Mol Cell Cardiol. 2012;52:526–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guo CX, Jiang X, Zeng XJ, Wang HX, Li HH, Du FH, et al. Soluble receptor for advanced glycation endproducts protects against ischemia/reperfusion-induced myocardial apoptosis via regulating the ubiquitin proteasome system. Free Radic Biol Med. 2016;94:17–26. [DOI] [PubMed] [Google Scholar]
- [22].Liliensiek B, Weigand MA, Bierhaus A, Nicklas W, Kasper M, Hofer S, et al. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J Clin Invest. 2004;113:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang C, Xu Z, He XR, Michael LH, Patterson C. CHIP, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. Am J Physiol Heart Circ Physiol. 2005;288:H2836–42. [DOI] [PubMed] [Google Scholar]
- [24].Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McClellan AJ, Frydman J. Molecular chaperones and the art of recognizing a lost cause. Nat Cell Biol. 2001;3:E51–3. [DOI] [PubMed] [Google Scholar]
- [26].Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Foo RS, Chan LK, Kitsis RN, Bennett MR. Ubiquitination and degradation of the anti-apoptotic protein ARC by MDM2. J Biol Chem. 2007;282:5529–35. [DOI] [PubMed] [Google Scholar]
- [28].Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- [29].Hammond EM, Giaccia AJ. The role of p53 in hypoxia-induced apoptosis. Biochem Biophys Res Commun. 2005;331:718–25. [DOI] [PubMed] [Google Scholar]
- [30].Regula KM, Kirshenbaum LA. p53 activates the mitochondrial death pathway and apoptosis of ventricular myocytes independent of de novo gene transcription. J Mol Cell Cardiol. 2001;33:1435–45. [DOI] [PubMed] [Google Scholar]
- [31].Bialik S, Geenen DL, Sasson IE, Cheng R, Horner JW, Evans SM, et al. Myocyte apoptosis during acute myocardial infarction in the mouse localizes to hypoxic regions but occurs independently of p53. J Clin Invest. 1997;100:1363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Toth A, Nickson P, Qin LL, Erhardt P. Differential regulation of cardiomyocyte survival and hypertrophy by MDM2, an E3 ubiquitin ligase. J Biol Chem. 2006;281:3679–89. [DOI] [PubMed] [Google Scholar]
- [33].Cacciapuoti F Role of ubiquitin-proteasome system (UPS) in left ventricular hypertrophy (LVH). Am J Cardiovasc Dis. 2014;4:1–5. [PMC free article] [PubMed] [Google Scholar]
- [34].Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–8. [DOI] [PubMed] [Google Scholar]
- [35].Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Maejima Y, Usui S, Zhai P, Takamura M, Kaneko S, Zablocki D, et al. Muscle-specific RING finger 1 negatively regulates pathological cardiac hypertrophy through downregulation of calcineurin A. Circ Heart Fail. 2014;7:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bai T, Wang F, Mellen N, Zheng Y, Cai L. Diabetic cardiomyopathy: role of the E3 ubiquitin ligase. Am J Physiol Endocrinol Metab. 2016;310:E473–83. [DOI] [PubMed] [Google Scholar]
- [38].Parry TL, Desai G, Schisler JC, Li L, Quintana MT, Stanley N, et al. Fenofibrate unexpectedly induces cardiac hypertrophy in mice lacking MuRF1. Cardiovasc Pathol. 2016;25:127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wadosky KM, Rodriguez JE, Hite RL, Min JN, Walton BL, Willis MS. Muscle RING finger-1 attenuates IGF-I-dependent cardiomyocyte hypertrophy by inhibiting JNK signaling. Am J Physiol Endocrinol Metab. 2014;306:E723–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mearini G, Gedicke C, Schlossarek S, Witt CC, Kramer E, Cao P, et al. Atrogin-1 and MuRF1 regulate cardiac MyBP-C levels via different mechanisms. Cardiovasc Res. 2010;85:357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, et al. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117:2486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci U S A. 2004;101:18135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Willis MS, Rojas M, Li L, Selzman CH, Tang RH, Stansfield WE, et al. Muscle ring finger 1 mediates cardiac atrophy in vivo. Am J Physiol Heart Circ Physiol. 2009;296:H997–H1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rodriguez JE, Liao JY, He J, Schisler JC, Newgard CB, Drujan D, et al. The ubiquitin ligase MuRF1 regulates PPARalpha activity in the heart by enhancing nuclear export via monoubiquitination. Mol Cell Endocrinol. 2015;413:36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].He J, Quintana MT, Sullivan J, T LP, T JG, Schisler JC, et al. MuRF2 regulates PPARgamma1 activity to protect against diabetic cardiomyopathy and enhance weight gain induced by a high fat diet. Cardiovasc Diabetol. 2015;14:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Quintana MT, He J, Sullivan J, Grevengoed T, Schisler J, Han Y, et al. Muscle ring finger-3 protects against diabetic cardiomyopathy induced by a high fat diet. BMC Endocr Disord. 2015;15:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tian LF, Li HY, Jin BF, Pan X, Man JH, Zhang PJ, et al. MDM2 interacts with and downregulates a sarcomeric protein, TCAP. Biochem Biophys Res Commun. 2006;345:355–61. [DOI] [PubMed] [Google Scholar]
- [50].Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, et al. Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol. 2004;167:1147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kinoshita M, Takano H, Taenaka Y, Mori H, Takaichi S, Noda H, et al. Cardiac disuse atrophy during LVAD pumping. ASAIO Trans. 1988;34:208–12. [PubMed] [Google Scholar]
- [52].Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. [DOI] [PubMed] [Google Scholar]
- [53].Temparis S, Asensi M, Taillandier D, Aurousseau E, Larbaud D, Obled A, et al. Increased ATPubiquitin-dependent proteolysis in skeletal muscles of tumor-bearing rats. Cancer Res. 1994;54:5568–73. [PubMed] [Google Scholar]
- [54].Razeghi P, Sharma S, Ying J, Li YP, Stepkowski S, Reid MB, et al. Atrophic remodeling of the heart in vivo simultaneously activates pathways of protein synthesis and degradation. Circulation. 2003;108:2536–41. [DOI] [PubMed] [Google Scholar]
- [55].Ho CY. Hypertrophic cardiomyopathy in 2012. Circulation. 2012;125:1432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–20. [DOI] [PubMed] [Google Scholar]
- [57].Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. [DOI] [PubMed] [Google Scholar]
- [58].Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–10. [DOI] [PubMed] [Google Scholar]
- [59].Hipp AA, Heitkamp HC, Rocker K, Dickhuth HH. Hypertrophic cardiomyopathy--sports-related aspects of diagnosis, therapy, and sports eligibility. Int J Sports Med. 2004;25:20–6. [DOI] [PubMed] [Google Scholar]
- [60].Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;124:2761–96. [DOI] [PubMed] [Google Scholar]
- [61].Jessup M, Abraham WT, Casey DE, Feldman AM, Francis GS, Ganiats TG, et al. 2009 focused update: ACCF/AHA Guidelines for the Diagnosis and Management of Heart Failure in Adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation. 2009;119:1977–2016. [DOI] [PubMed] [Google Scholar]
- [62].Schlossarek S, Carrier L. The ubiquitin-proteasome system in cardiomyopathies. Curr Opin Cardiol. 2011;26:190–5. [DOI] [PubMed] [Google Scholar]
- [63].Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovasc Res. 2010;85:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Birks EJ, Latif N, Enesa K, Folkvang T, Luong le A, Sarathchandra P, et al. Elevated p53 expression isassociated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovasc Res. 2008;79:472–80. [DOI] [PubMed] [Google Scholar]
- [65].Heling A, Zimmermann R, Kostin S, Maeno Y, Hein S, Devaux B, et al. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circ Res. 2000;86:846–53. [DOI] [PubMed] [Google Scholar]
- [66].Lamark T, Johansen T. Autophagy: links with the proteasome. Curr Opin Cell Biol. 2010;22:192–8. [DOI] [PubMed] [Google Scholar]
- [67].Ross R Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340:115–26. [DOI] [PubMed] [Google Scholar]
- [68].Libby P, Aikawa M. Stabilization of atherosclerotic plaques: new mechanisms and clinical targets. Nat Med. 2002;8:1257–62. [DOI] [PubMed] [Google Scholar]
- [69].Herrmann J, Lerman LO, Lerman A. On to the road to degradation: atherosclerosis and the proteasome. Cardiovasc Res. 2010;85:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Herrmann J, Saguner AM, Versari D, Peterson TE, Chade A, Olson M, et al. Chronic proteasome inhibition contributes to coronary atherosclerosis. Circ Res. 2007;101:865–74. [DOI] [PubMed] [Google Scholar]
- [71].Wilck N, Fechner M, Dreger H, Hewing B, Arias A, Meiners S, et al. Attenuation of early atherogenesis in low-density lipoprotein receptor-deficient mice by proteasome inhibition. Arterioscler Thromb Vasc Biol. 2012;32:1418–26. [DOI] [PubMed] [Google Scholar]
- [72].Severson DL. Diabetic cardiomyopathy: recent evidence from mouse models of type 1 and type 2 diabetes. Can J Physiol Pharmacol. 2004;82:813–23. [DOI] [PubMed] [Google Scholar]
- [73].Li YF, Wang X. The role of the proteasome in heart disease. Biochim Biophys Acta. 2011;1809:141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Marfella R, DA M, Di Filippo C, Siniscalchi M, Sasso FC, Ferraraccio F, et al. The possible role of the ubiquitin proteasome system in the development of atherosclerosis in diabetes. Cardiovasc Diabetol. 2007;6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bai Y, Cui W, Xin Y, Miao X, Barati MT, Zhang C, et al. Prevention by sulforaphane of diabetic cardiomyopathy is associated with up-regulation of Nrf2 expression and transcription activation. J Mol Cell Cardiol. 2013;57:82–95. [DOI] [PubMed] [Google Scholar]
- [76].Battiprolu PK, Hojayev B, Jiang N, Wang ZV, Luo X, Iglewski M, et al. Metabolic stress-induced activation of FoxO1 triggers diabetic cardiomyopathy in mice. J Clin Invest. 2012;122:1109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rubler S, Dlugash J, Yuceoglu YZ, Kumral T, Branwood AW, Grishman A. New type of cardiomyopathy associated with diabetic glomerulosclerosis. Am J Cardiol. 1972;30:595–602. [DOI] [PubMed] [Google Scholar]
- [78].Liu F, Song R, Feng Y, Guo J, Chen Y, Zhang Y, et al. Upregulation of MG53 induces diabetic cardiomyopathy through transcriptional activation of peroxisome proliferation-activated receptor alpha. Circulation. 2015;131:795–804. [DOI] [PubMed] [Google Scholar]
- [79].Bugger H, Abel ED. Molecular mechanisms of diabetic cardiomyopathy. Diabetologia. 2014;57:66071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Nakamura H, Matoba S, Iwai-Kanai E, Kimata M, Hoshino A, Nakaoka M, et al. p53 promotes cardiac dysfunction in diabetic mellitus caused by excessive mitochondrial respiration-mediated reactive oxygen species generation and lipid accumulation. Circ Heart Fail. 2012;5:106–15. [DOI] [PubMed] [Google Scholar]
- [81].Naito AT, Okada S, Minamino T, Iwanaga K, Liu ML, Sumida T, et al. Promotion of CHIP-mediated p53 degradation protects the heart from ischemic injury. Circ Res. 2010;106:1692–702. [DOI] [PubMed] [Google Scholar]
- [82].van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–73. [DOI] [PubMed] [Google Scholar]
- [83].Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17:631–6. [DOI] [PubMed] [Google Scholar]
- [84].Zhang Y, Kohler K, Xu J, Lu D, Braun T, Schlitt A, et al. Inhibition of p53 after acute myocardial infarction: reduction of apoptosis is counteracted by disturbed scar formation and cardiac rupture. J Mol Cell Cardiol. 2011;50:471–8. [DOI] [PubMed] [Google Scholar]
- [85].Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chen J, Regan RF. Increasing expression of heme oxygenase-1 by proteasome inhibition protects astrocytes from heme-mediated oxidative injury. Curr Neurovasc Res. 2005;2:189–96. [DOI] [PubMed] [Google Scholar]
- [87].Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem Res Toxicol. 2008;21:705–10. [DOI] [PubMed] [Google Scholar]
- [88].Kandula V, Kosuru R, Li H, Yan D, Zhu Q, Lian Q, et al. Forkhead box transcription factor 1: role in the pathogenesis of diabetic cardiomyopathy. Cardiovasc Diabetol. 2016;15:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Storz P Forkhead homeobox type O transcription factors in the responses to oxidative stress.Antioxid Redox Signal. 2011;14:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wang J, Song Y, Wang Q, Kralik PM, Epstein PN. Causes and characteristics of diabetic cardiomyopathy. Rev Diabet Stud. 2006;3:108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Milan G, Romanello V, Pescatore F, Armani A, Paik JH, Frasson L, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun. 2015; 6:6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Obsil T, Obsilova V. Structural basis for DNA recognition by FOXO proteins. Biochim Biophys Acta. 2011;1813:1946–53. [DOI] [PubMed] [Google Scholar]
- [93].Kitamura T, Nakae J, Kitamura Y, Kido Y, Biggs WH 3rd, Wright CV, et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J Clin Invest. 2002;110:1839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14–3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–45. [DOI] [PubMed] [Google Scholar]
- [95].Maiese K, Hou J, Chong ZZ, Shang YC. A fork in the path: Developing therapeutic inroads with FoxO proteins. Oxid Med Cell Longev. 2009;2:119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:290527. [DOI] [PubMed] [Google Scholar]
- [97].Martinez SC, Tanabe K, Cras-Meneur C, Abumrad NA, Bernal-Mizrachi E, Permutt MA. Inhibition ofFoxo1 protects pancreatic islet beta-cells against fatty acid and endoplasmic reticulum stress-induced apoptosis. Diabetes. 2008;57:846–59. [DOI] [PubMed] [Google Scholar]
- [98].Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–63. [DOI] [PubMed] [Google Scholar]
- [99].Puthanveetil P, Wang Y, Wang F, Kim MS, Abrahani A, Rodrigues B. The increase in cardiac pyruvate dehydrogenase kinase-4 after short-term dexamethasone is controlled by an Akt-p38-forkhead box other factor-1 signaling axis. Endocrinology. 2010;151:2306–18. [DOI] [PubMed] [Google Scholar]
- [100].Majer M, Popov KM, Harris RA, Bogardus C, Prochazka M. Insulin downregulates pyruvate dehydrogenase kinase (PDK) mRNA: potential mechanism contributing to increased lipid oxidation in insulin-resistant subjects. Mol Genet Metab. 1998;65:181–6. [DOI] [PubMed] [Google Scholar]
- [101].Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–36. [DOI] [PubMed] [Google Scholar]
- [102].Leask A TGFbeta, cardiac fibroblasts, and the fibrotic response. Cardiovasc Res. 2007;74:207–12. [DOI] [PubMed] [Google Scholar]
- [103].Itoh S, ten Dijke P. Negative regulation of TGF-beta receptor/Smad signal transduction. Curr Opin Cell Biol. 2007;19:176–84. [DOI] [PubMed] [Google Scholar]
- [104].Li JH, Zhu HJ, Huang XR, Lai KN, Johnson RJ, Lan HY. Smad7 inhibits fibrotic effect of TGF-Beta on renal tubular epithelial cells by blocking Smad2 activation. J Am Soc Nephrol. 2002;13:1464–72. [DOI] [PubMed] [Google Scholar]
- [105].Zhang S, Fei T, Zhang L, Zhang R, Chen F, Ning Y, et al. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol. 2007;27:4488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wang B, Omar A, Angelovska T, Drobic V, Rattan SG, Jones SC, et al. Regulation of collagen synthesis by inhibitory Smad7 in cardiac myofibroblasts. Am J Physiol Heart Circ Physiol. 2007;293:H1282–90. [DOI] [PubMed] [Google Scholar]
- [107].He X, Gao X, Peng L, Wang S, Zhu Y, Ma H, et al. Atrial fibrillation induces myocardial fibrosis through angiotensin II type 1 receptor-specific Arkadia-mediated downregulation of Smad7. Circ Res. 2011;108:164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang B, Hao J, Jones SC, Yee MS, Roth JC, Dixon IM. Decreased Smad 7 expression contributes to cardiac fibrosis in the infarcted rat heart. Am J Physiol Heart Circ Physiol. 2002;282:H1685–96. [DOI] [PubMed] [Google Scholar]
- [109].Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, et al. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 2003;22:6458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H, et al. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J Biol Chem. 2007;282:20492–501. [DOI] [PubMed] [Google Scholar]
- [111].Kawano K, Hirashima T, Mori S, Saitoh Y, Kurosumi M, Natori T. Spontaneous long-term hyperglycemic rat with diabetic complications. Otsuka Long-Evans Tokushima Fatty (OLETF) strain. Diabetes. 1992;41:1422–8. [DOI] [PubMed] [Google Scholar]
- [112].Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239–52. [DOI] [PubMed] [Google Scholar]
- [113].Zhao D, Sun Y, Wei X, Liang H, Zhao L, Dong X, et al. cIAP1 attenuates shear stress-induced hBMSC apoptosis for tissue-engineered blood vessels through the inhibition of the mitochondrial apoptosis pathway. Life Sci. 2015;137:81–8. [DOI] [PubMed] [Google Scholar]
- [114].Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J Biol Chem. 2000;275:26661–4. [DOI] [PubMed] [Google Scholar]
- [115].Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A. 2001;98:8662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Emamaullee JA, Rajotte RV, Liston P, Korneluk RG, Lakey JR, Shapiro AM, et al. XIAP overexpression in human islets prevents early posttransplant apoptosis and reduces the islet mass needed to treat diabetes. Diabetes. 2005;54:2541–8. [DOI] [PubMed] [Google Scholar]
- [117].Hendy GN, Canaff L. Calcium-sensing receptor, proinflammatory cytokines and calcium homeostasis. Semin Cell Dev Biol. 2016;49:37–43. [DOI] [PubMed] [Google Scholar]
- [118].Milara J, Mata M, Serrano A, Peiro T, Morcillo EJ, Cortijo J. Extracellular calcium-sensing receptor mediates human bronchial epithelial wound repair. Biochem Pharmacol. 2010;80:236–46. [DOI] [PubMed] [Google Scholar]
- [119].Tharmalingam S, Hampson DR. The Calcium-Sensing Receptor and Integrins in Cellular Differentiation and Migration. Front Physiol. 2016;7:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Guo J, Li HZ, Wang LC, Zhang WH, Li GW, Xing WJ, et al. Increased expression of calcium-sensing receptors in atherosclerosis confers hypersensitivity to acute myocardial infarction in rats. Mol Cell Biochem. 2012;366:345–54. [DOI] [PubMed] [Google Scholar]
- [121].Jiang CM, Han LP, Li HZ, Qu YB, Zhang ZR, Wang R, et al. Calcium-sensing receptors induce apoptosis in cultured neonatal rat ventricular cardiomyocytes during simulated ischemia/reperfusion. Cell Biol Int. 2008;32:792–800. [DOI] [PubMed] [Google Scholar]
- [122].Bai SZ, Sun J, Wu H, Zhang N, Li HX, Li GW, et al. Decrease in calcium-sensing receptor in the progress of diabetic cardiomyopathy. Diabetes Res Clin Pract. 2012;95:378–85. [DOI] [PubMed] [Google Scholar]
- [123].Wang Y, Gao P, Wei C, Li H, Zhang L, Zhao Y, et al. Calcium sensing receptor protects high glucoseinduced energy metabolism disorder via blocking gp78-ubiquitin proteasome pathway. Cell Death Dis. 2017;8:e2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Herrmann J, Ciechanover A, Lerman LO, Lerman A. The ubiquitin-proteasome system in cardiovascular diseases-a hypothesis extended. Cardiovasc Res. 2004;61:11–21. [DOI] [PubMed] [Google Scholar]
- [125].Zolk O, Schenke C, Sarikas A. The ubiquitin-proteasome system: focus on the heart. Cardiovasc Res. 2006;70:410–21. [DOI] [PubMed] [Google Scholar]
- [126].Bao J, Sato K, Li M, Gao Y, Abid R, Aird W, et al. PR-39 and PR-11 peptides inhibit ischemiareperfusion injury by blocking proteasome-mediated I kappa B alpha degradation. Am J Physiol Heart Circ Physiol. 2001;281:H2612–8. [DOI] [PubMed] [Google Scholar]
- [127].Di Napoli M, Papa F. The proteasome system and proteasome inhibitors in stroke: controlling the inflammatory response. Curr Opin Investig Drugs. 2003;4:1333–42. [PubMed] [Google Scholar]
- [128].Di Napoli M, Papa F. MLN-519. Millennium/PAION. Curr Opin Investig Drugs. 2003;4:333–41. [PubMed] [Google Scholar]
- [129].Aversano T, Aversano LT, Passamani E, Knatterud GL, Terrin ML, Williams DO, et al. Thrombolytic therapy vs primary percutaneous coronary intervention for myocardial infarction in patients presenting to hospitals without on-site cardiac surgery: a randomized controlled trial. JAMA. 2002; 287: 1943–51. [DOI] [PubMed] [Google Scholar]
- [130].Meiners S, Laule M, Rother W, Guenther C, Prauka I, Muschick P, et al. Ubiquitin-proteasome pathway as a new target for the prevention of restenosis. Circulation. 2002;105:483–9. [DOI] [PubMed] [Google Scholar]
- [131].D’Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–40. [DOI] [PubMed] [Google Scholar]
- [132].Liu N, Chai R, Liu B, Zhang Z, Zhang S, Zhang J, et al. Ubiquitin-specific protease 14 regulates cardiac hypertrophy progression by increasing GSK-3beta phosphorylation. Biochem Biophys Res Commun. 2016;478:1236–41. [DOI] [PubMed] [Google Scholar]
- [133].Wang H, Lai Y, Mathis BJ, Wang W, Li S, Qu C, et al. Deubiquitinating enzyme CYLD mediates pressure overload-induced cardiac maladaptive remodeling and dysfunction via downregulating Nrf2. J Mol Cell Cardiol. 2015;84:143–53. [DOI] [PubMed] [Google Scholar]
- [134].Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, et al. CYLD negatively regulates transforming growth factor-beta-signalling via deubiquitinating Akt. Nat Commun. 2012;3:771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Li J, Powell SR, Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. FASEB J. 2011;25:883–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem. 2003;278:311–8. [DOI] [PubMed] [Google Scholar]
- [139].Odiete O, Konik EA, Sawyer DB, Hill MF. Type 1 diabetes mellitus abrogates compensatory augmentation of myocardial neuregulin-1beta/ErbB in response to myocardial infarction resulting in worsening heart failure. Cardiovasc Diabetol. 2013;12:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432:585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]