Abstract
Detection and characterization of DNA damage is essential for evaluating genotoxicity, monitoring DNA repair, developing biomarkers for exposures, and evaluating the efficacy of chemotherapies. These diverse applications for DNA damage measurements have spurred the continual development and refinement of methodologies for detecting, characterizing, and quantifying DNA damage from isolated DNA and in cells and tissues. Current damage detection methods cover a wide range of techniques from radiolabeling to mass spectrometry, and use of these techniques varies widely based on expense, expertise, and knowledge of adduct formation. More generalizable, easy-to-use methods for detecting and quantifying DNA damage are needed, and there has been an emergence of fluorescence-based methodologies to address this need. Developments in these fluorescence-based strategies are reviewed here.
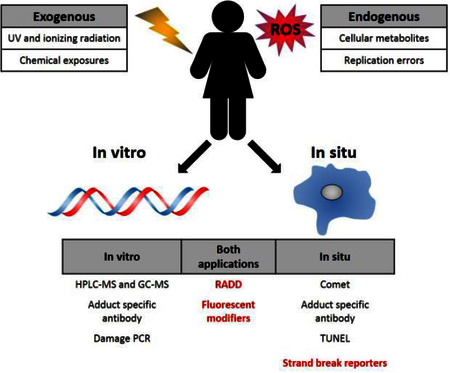
Graphical Abstract.
DNA adducts are induced endogenously via the production of reactive metabolic byproducts and during cellular propagation, and exogenously through environmental exposures to DNA damaging chemicals and radiation. The methods that require DNA purification are labeled in vitro, and methods capable of detecting adducts within a cellular context are identified as in situ.
Introduction
DNA damage is a natural consequence of daily life. Endogenous metabolism and exposures to environmental, dietary, and therapeutic chemicals, create modified DNA bases and strand breaks that if left unrepaired can produce mutations, chromosomal aberrations, and induce cell death. Detection, characterization, and quantification of these modifications, or adducts, and strand breaks provides important information about genotoxic exposures, their mechanistic actions, and cellular responses. These data are critical for risk assessment and for gauging the efficiency of DNA repair pathways, the effectiveness of chemopreventives, or the efficacy of chemotherapies [1–3].
Given these diverse applications, a large number of methods for the detection and quantification of DNA adducts have been developed. These methods range from the highly specific detection and quantification of DNA adducts using liquid chromatography and mass spectrometry (reviewed in [1,2,4]) to more broad spectrum detection of total DNA damage through polymerase chain reactions (PCR) [5,6].
An emerging tool in DNA damage detection are fluorescence-based strategies for detecting, characterizing, and quantifying DNA damage. Fluorescence methods offer sensitive and specific detection of damage and avoid the use of hazardous radiolabeled compounds. They are compatible with a large number of read-outs from single molecule imaging, which allows the heterogeneity of a system to be analyzed, to high content imaging with flow cytometry or microplate readers, which allows population level differences to be measured. These methods also allow spectrally distinct labeling strategies that may be multiplexed, increasing the potential throughput of experiments, and they are capable of measuring damage on isolated DNA and within cells and tissues.
The advantages of fluorescence-based methods are not new to DNA damage detection [1,3]. Antibody-based detection of damage in cells and tissues has been conducted for decades, typically using fluorophore-conjugated primary or secondary antibodies [1,7]. DNA intercalating dyes have been used to detect DNA strand breaks in single cell gel electrophoresis (SGCE) [8–10] or chromosomal instability by monitoring supercoiled DNA (the “halo” assay) [11] or formation of micronuclei [12]. However, new fluorescence-based DNA damage detection methodologies for use in vitro and in situ are emerging that provide sensitive and specific detection of damage, are compatible with single molecule and whole cell imaging, and can be coupled with genomic sequence analysis to better understand the damage context. Here, we will review these techniques and recent advances in these methodologies.
Molecular markers for in vivo detection of DNA damage
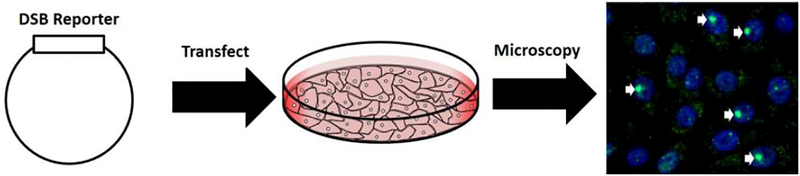
Antibodies against DNA adducts, like thymine dimers or 8-hydroxy-2’-deoxyguanosine (8-oxodG), or DNA repair proteins like Ku, XRCC1, or phosphorylated H2AX (γH2AX) have all been used to characterize DNA damage within isolated DNA and in cells [13–16]. However, all of these techniques rely on the generation of highly specific, high affinity antibodies for targets of interest. Given the natural affinity and specificity of DNA repair proteins for damage sites, a more direct detection strategy would be to use specific DNA repair or DNA binding proteins to directly characterize induced damage (Fig. 1).
Figure 1.
Overview of fluorescent reporters for DNA damage in vivo. Cells are transfected with a fluorescent reporter bound to a double strand break (DSB) binding protein, Gam or 53BP-1. Endogenous DSBs or exogenously induced DSBs can be detected via foci formation at the DSB site.
For example, the Gam protein from bacteriophage Mu recognizes double strand breaks (DBSs) generated during integration and protects the linear DNA from degradation [17,18]. This specific recognition of DNA ends led Shee et al. to fuse Gam to green fluorescent protein (Gam-GFP) and monitor the induction of double strand breaks in E. coli and mammalian systems [19]. In E. coli, specific detection of DSBs generated during replication, by I-SceI cleavage, and from genotoxic exposure was achieved with an estimated detection efficiency of 71–82% [19]. In mammalian systems, expressed Gam-GFP has been shown to detect laser and radiation-induced DSBs as well as failed mitotic events and shown to co-localized with phosphorylated H2AX (γH2AX) and 53BP-1 [19,20]. However, the detection efficiency in the mammalian systems was not as high. Competition with endogenous Ku proteins, to which Gam is related, likely contributes to this loss in detection efficiency [17,19]. Despite some limitations for mammalian systems, Gam-GFP provides specific and sensitive detection of in vivo DSBs and has been used to characterize DNA repair dynamics in E. coli [21,22]. Additionally, the competition with Ku in mammalian systems may provide another DNA repair linked application for this damage reporter, where Ku’s recruitment and role in nonhomologous end-joining (NHEJ) is dissected.
Similarly, another DSB reporter has also been developed for in vivo single-cell imaging based on truncating 53BP-1 (amino acids 1220–1711) and fusing it to the mApple fluorescent protein [23]. 53BP-1 recognizes DSBs and promotes NHEJ-mediated repair [24]. The truncated 53BP-1 was shown to accumulate at DSBs in a number of cell lines including breast, ovarian and neuronal cells, allowing study of the dynamics of strand break formation and repair after genotoxic exposures [23,25,26].
While these methodologies are still developing, engineered proteins may provide new reagents for the detection of strand breaks or DNA adducts in vivo. More work is needed to generate reagents compatible with single strand breaks (SSBs) and other DNA adducts, but in vivo imaging of induced damage would provide new insight into the genotoxicity of environmental and therapeutic chemicals and allow dynamics of DNA repair to be visualized.
Enzyme-mediated damage detection assays
Engineered protein reporters exploit the natural activities of DNA repair proteins, similarly a number of techniques have emerged utilizing DNA repair enzymes to excise and specifically label DNA damage sites [27–31]. In 2013, Lee et al. demonstrated that molecular combing of λ DNA molecules could be used to detect strand breaks induced by ultraviolet light (UV) [28]. Stretched DNA molecules were irradiated and visualized by fluorescent staining with the DNA intercalator, YOYO-1. Gaps in the YOYO-1 staining revealed DSBs in the DNA. Further damage characterization was carried out by coupling enzymatic processing by pyrimidine dimer glycosylase (PDG) with the incorporation of fluorescently-labeled dNTPs by DNA polymerase I to characterize the presence of single strand breaks (SSBs) and UV-induced base lesions [28].
Zirkin et al. refined and extended this technique to detect abasic sites, uracils, oxidative, and UV-induced DNA lesions from DNA isolated from treated cells or human tissues. Isolated DNA was treated prior to combing with an enzymatic cocktail, PreCR®, which contains bacterial DNA glycosylases, EndoIV, EndoVIII, Fpg, T4 PDG, UDG, and Bst DNA polymerase [32]. DNA damage is recognized and excised by these enzymes and damage sites are detected by the incorporation of fluorescently-labeled nucleotides by the DNA polymerase. Modified versions of this assay, with different enzyme cocktails, have been utilized to detect oxidative DNA damage in viral genomes [27] and alcohol-induced DNA damage in the E. coli genome [29].
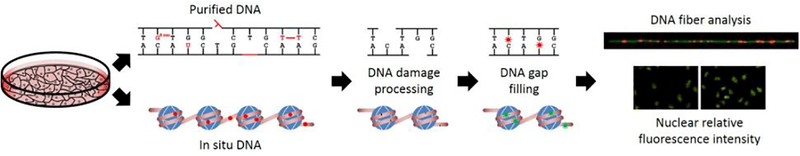
Recently, a further modification of this assay, Repair Assisted Damage Detection (RADD) was developed with a refined enzyme cocktail for the detection of abasic sites, uracils, strand breaks, and oxidative and UV-induced DNA lesions within cells (Fig. 2) [31].
Figure 2.
Overview of enzyme-mediated DNA damage detection strategies that can be performed on purified DNA or on fixed cells. First, DNA repair enzymes process the DNA damage and create the DNA end chemistry needed for DNA polymerase to fill the gap with a fluorescently tagged nucleotide. Damage sites are imaged and quantified by counting the number of lesions per length of DNA or through measurement of the fluorescence intensity of the nuclear area.
While the RADD in cell assay allows relative quantification of DNA damage [31], the DNA combing strategy can detect very low levels of endogenous DNA damage 0–10 labels per Mbp and significant increases ~ 1–1.5 fold in damage after genotoxic exposures [27–30]. Additionally, since the DNA context is preserved and damage sites are labeled, these techniques are compatible with optical sequence mapping [27–29] or next generation sequencing of isolated damage fragments (similar to [33,34]), allowing new insight into genome damage susceptibility.
These DNA repair enzyme-mediated damage detection strategies offer significant improvements over other enzyme detection strategies such as terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) [35–37]. TUNEL has been employed to detect DNA strand breaks, typically in the context of apoptosis, but also in some cases DNA adducts across a variety of biological samples [38–40]. However, TUNEL is highly specific for DNA strand breaks with 3’-OH ends, limiting the utility for detecting different types of DNA damage. While several TUNEL modifications have emerged extending its ability to detect other DNA ends (i.e., 3’-PO4) or improving DNA damage detection by incorporating FPG to excise oxidative DNA adducts [41], the repair enzyme-mediated strategies outlined here offer more robust strategies for detecting specific classes of DNA damage or a broad spectrum of DNA damage [30,31]. The only limitation to these techniques are the specificity of known DNA repair enzymes, and engineering of enzymes for specific lesions would open new opportunities to study damage and its sequence context.
Fluorescent modification of DNA adducts
The intrinsic fluorescence of some DNA adducts and fluorescent conjugates DNA adducts by HPLC techniques [1,2]. While these fluorescent strategies are limited in their applications, they highlight that covalent modification of DNA adducts with fluorescent molecules could enhance detection and quantification. To this end, new synthetic strategies have been developed for chemical modifiers that specifically and covalently bind some DNA adducts. These modifiers may be fluorophores, but they could also be functional groups, such as azides, alkynes, amines, or other functional groups compatible with fluorescent labeling.
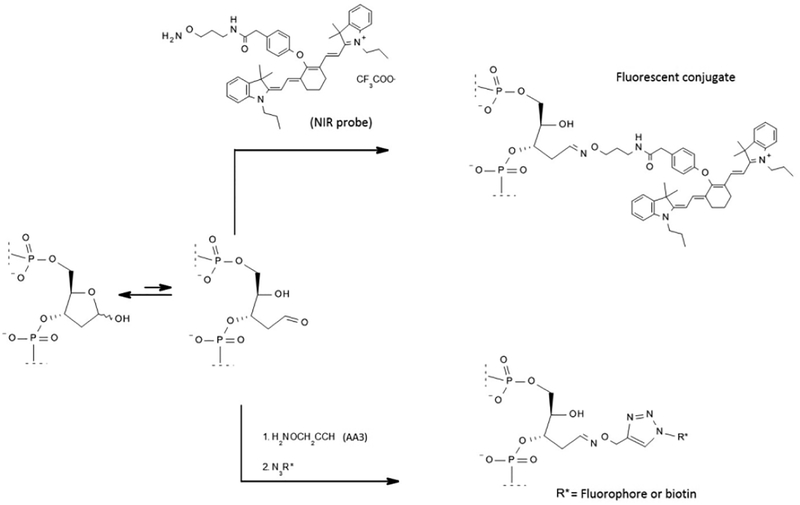
Fluorophore conjugation directly to DNA adducts is less common, though a near-infrared (NIR) fluorescent probe has been developed for in vitro and in situ imaging of abasic sites [42] (Fig. 3). This probe binds abasic sites with higher affinity than the two current detection reagents for abasic sites, aldehyde-reactive probe (ARP, alkoxyamine) or methoxyamine (MX). Sensitivity of the probe was demonstrated by quantifying chemotherapy-induced abasic sites in DNA isolated from colorectal cancer cells [42]. While this probe has potential in vivo applications, the cell permeability was not demonstrated and inhibition of AP-endonuclease 1 (APE1) above ARP and MX levels was observed [42]. With resolution of these issues, this reagent may be a new analytical tool for assessing abasic sites in vivo and in vitro.
Figure 3.
Two different approaches for fluorescent modification of DNA adducts. Here, an absaic site is modified directly with the NIR probe (top, [42]) or with AA3, a modified alkoxyamine (bottom, [44]). Labeling of the AA3-conjugated molecule is achieved by the addition of an azide-conjugated fluorophore or biotin (bottom, [44]).
Abasic site detection is more commonly conducted through reactions of the abasic sites with ARP modified with biotin, which reacts with the open form of deoxyribose sugar forming an oxime. The biotinylated abasic site can then be readily detected. However, ARP has numerous drawbacks, including competition with endogenous biotin, low reactivity, and production of side products [43]. An improved abasic detection reagent, AA3, has been synthesized, which addresses these shortcomings by functionalizing the abasic site with an alkyne group, which can be subsequently labeled via Click chemistry (Fig. 3) [44]. This functionalization allows users to dictate the fluorescent molecules or functional groups attached to the abasic site. AA3 also inhibits the activity of APE1, but the inhibition is similar to MX [44].
A reagent for interstrand crosslink detection has also been generated by attaching an alkyne group to psoralen, 8-propargyloxypsoralen (8-POP). Psoralen generates interstrand crosslinks when activated by UV light, the 8-POP derivative generates interstrand crosslinks and labels them with an alkyne group, offering fluorophore compatible labeling strategies for the detection of DNA damage in vivo and in vitro
Direct labeling of DNA damage sites with fluorophores or chemical modifiers that facilitate fluorescent labeling offer new strategies for the detection and quantification of DNA damage, particularly of abasic sites and interstrand crosslinks, which are difficult to detect. While the efficiency of these reactions have not been adequately measured, these tools offer new labeling opportunities that overcome antibody affinity and dissociation issues due to their covalent nature. However, users need to be aware of potential genotoxic side effects of these agents and their competition with endogenous repair proteins.
Conclusions
With continual improvements in optics and microscopy and the affordability of laser emitting diodes, fluorescence-based methodologies have significantly expanded. Further, single molecule techniques and fluorescence reading devices have become cheaper, smaller, and more readily available. These advancements have improved sequencing technologies and will likely significantly improve the detection and quantification of DNA damage. Here, we reviewed emerging fluorescence-based techniques for the detection and quantification of DNA damage. While these techniques are not as well-known as mass spectrometry, HPLC, or radiolabeling for adduct detection, they offer new tools for damage characterization that are compatible with other characterization methods, such as live cell imaging and microirradiation [19,31], genomic sequencing [27–29], and are capable of being multiplexed with the analysis of other DNA modifications, such as epigenetic marks [30]. While the sensitivity and detection efficiency of these fluorescence-based techniques still require improvement compared to well accepted adduct detection methods (Table 1), overall these offer new opportunities to characterize genotoxicity and examine DNA repair.
Table 1:
Comparison of established DNA adduct detection methods with reviewed emerging fluorescence-based methods.
| Established methods | Maximum Sensitivity | Total DNA required (μg) | Citations |
| HPLC-ECD/fluorescence | 1 in 108 | 1–100 | [1,2] |
| Immunoassay | 1 in 108 | 1–200 | [1,2] |
| Radio labeled compounds | 1 in 108 | 1–100 | [1,2] |
| 32P-post-labelling | 1 in 1010 | 1–10 | [1,2] |
| LC-MS and GC-MS | 1 in 108 | 1–100 | [2,7] |
| AMS | 1 in 1012 | 1–2000 | [1,2] |
| Emerging methods | Maximum Sensitivity | Total DNA required (μg) | Citations |
| Strand break reporter | 1 in 105* | NA | [19,23] |
| In vitro RADD | 1 in 105* | 0.1–1 | [27–30] |
| In vivo RADD | ND | NA | [31] |
| Fluorescent modifiers | ND | 0.1–1 | [42,44] |
estimated from Mbp measurements
Highlights.
DNA adducts are important biological markers of exposure and DNA repair
Fluorescence-based methods offer new opportunities for assessing DNA adducts
Fluorescent-tagging of DNA repair proteins allow in vivo damage measurements
Labeled dNTPs inserted at damage sites by repair enzymes also allow damage detection
Specific DNA adducts are labeled using covalent binding to chemical modifiers
Fluorescent methods are improving the detection and quantification of DNA damage
Acknowledgements
NH and NRG are supported by the National Institutes of Health [ES023813]. The authors would like to thank Dr. Yuval Ebenstein for providing the image of combed DNA, and Dr. Marie Migaud for comments and assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Phillips DH, Farmer PB, Beland FA, Nath RG, Poirier MC, Reddy MV, Turteltaub KW: Methods of DNA adduct determination and their application to testing compounds for genotoxicity. Environ Mol Mutagen 2000, 35:222–233. [DOI] [PubMed] [Google Scholar]
- •2.Brown K: Methods for the detection of DNA adducts. Methods Mol Biol 2012, 817:207–230.Review of the methodology for the most commonly used DNA adduct detection schemes.
- 3.Figueroa-Gonzalez G, Perez-Plasencia C: Strategies for the evaluation of DNA damage and repair mechanisms in cancer. Oncol Lett 2017, 13:3982–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •4.Balbo S, Turesky RJ, Villalta PW: DNA adductomics. Chem Res Toxicol 2014, 27:356–366.Review of DNA adduct formation and detection with particular focus on mass spectrometry and recent advances in that field.
- 5.Wang W, Scheffler K, Esbensen Y, Eide L: Quantification of DNA Damage by Real-Time qPCR. Methods Mol Biol 2016, 1351:27–32. [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer GP, Denissenko MF, Tang MS: PCR-based approaches to adduct analysis. Toxicol Lett 1998, 102–103:447–451. [DOI] [PubMed] [Google Scholar]
- •7.Hwa Yun B, Guo J, Bellamri M, Turesky RJ: DNA adducts: Formation, biological effects, and new biospecimens for mass spectrometric measurements in humans. Mass Spectrom Rev 2018.Review of newer methodologies for DNA adduct detection by mass spectrometry.
- 8.Ostling O, Johanson KJ: Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun 1984, 123:291–298. [DOI] [PubMed] [Google Scholar]
- 9.Collins AR: The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol 2004, 26:249–261. [DOI] [PubMed] [Google Scholar]
- 10.Sykora P, Witt KL, Revanna P, Smith-Roe SL, Dismukes J, Lloyd DG, Engelward BP, Sobol RW: Next generation high throughput DNA damage detection platform for genotoxic compound screening. Sci Rep 2018, 8:2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roti Roti JL, Wright WD: Visualization of DNA loops in nucleoids from HeLa cells: assays for DNA damage and repair. Cytometry 1987, 8:461–467. [DOI] [PubMed] [Google Scholar]
- 12.Fenech M: The micronucleus assay determination of chromosomal level DNA damage. Methods Mol Biol 2008, 410:185–216. [DOI] [PubMed] [Google Scholar]
- 13.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y: GammaH2AX and cancer. Nat Rev Cancer 2008, 8:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM: Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol 2003, 81:123–129. [DOI] [PubMed] [Google Scholar]
- 15.Gassman NR, Wilson SH: Micro-irradiation tools to visualize base excision repair and single-strand break repair. DNA Repair (Amst) 2015, 31:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britton S, Coates J, Jackson SP: A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 2013, 202:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.d’Adda di Fagagna F, Weller GR, Doherty AJ, Jackson SP: The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep 2003, 4:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akroyd J, Symonds N: Localization of the gam gene of bacteriophage mu and characterisation of the gene product. Gene 1986, 49:273–282. [DOI] [PubMed] [Google Scholar]
- ••19.Shee C, Cox BD, Gu F, Luengas EM, Joshi MC, Chiu LY, Magnan D, Halliday JA, Frisch RL, Gibson JL, et al. : Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife 2013, 2:e01222.Gam-GFP is developed for the detection of double strand breaks in bacterial and mammalian systems.
- •20.Pedersen RS, Karemore G, Gudjonsson T, Rask MB, Neumann B, Heriche JK, Pepperkok R, Ellenberg J, Gerlich DW, Lukas J, et al. : Profiling DNA damage response following mitotic perturbations. Nat Commun 2016, 7:13887.Application of Gam-GFP as a strand break sensor in mammalian cells.
- •21.Xia J, Chen LT, Mei Q, Ma CH, Halliday JA, Lin HY, Magnan D, Pribis JP, Fitzgerald DM, Hamilton HM, et al. : Holliday junction trap shows how cells use recombination and a junction-guardian role of RecQ helicase. Sci Adv 2016, 2:e1601605.Application of Gam-GFP to study DNA repair dynamics in bacteria.
- 22.Pedersen IB, Helgesen E, Flatten I, Fossum-Raunehaug S, Skarstad K: SeqA structures behind Escherichia coli replication forks affect replication elongation and restart mechanisms. Nucleic Acids Res 2017, 45:6471–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••23.Yang KS, Kohler RH, Landon M, Giedt R, Weissleder R: Single cell resolution in vivo imaging of DNA damage following PARP inhibition. Sci Rep 2015, 5:10129.Development and use of truncated 53BP-1 mApple as an in vivo double strand break marker.
- 24.Panier S, Boulton SJ: Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 2014, 15:7–18. [DOI] [PubMed] [Google Scholar]
- 25.Miller MA, Askevold B, Yang KS, Kohler RH, Weissleder R: Platinum compounds for high-resolution in vivo cancer imaging. ChemMedChem 2014, 9:1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •26.Moruno-Manchon JF, Koellhoffer EC, Gopakumar J, Hambarde S, Kim N, McCullough LD, Tsvetkov AS: The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging (Albany NY) 2017, 9:1957–1970.Application of 53BP-1 mApple to monitor DNA damage in mammalian cells.
- ••27.Lee J, Kim Y, Lim S, Jo K: Single-molecule visualization of ROS-induced DNA damage in large DNA molecules. Analyst 2016, 141:847–852.DNA fiber analysis with an enzyme-mediated damage detection cocktail for damage measurements.
- •28.Lee J, Park HS, Lim S, Jo K: Visualization of UV-induced damage on single DNA molecules. Chem Commun (Camb) 2013, 49:4740–4742.Development of DNA fiber assay for examining induced DNA damage on coverslips.
- 29.Kang Y, Lee J, Kim J, Oh Y, Kim D, Lee J, Lim S, Jo K: Analysis of alcohol-induced DNA damage in Escherichia coli by visualizing single genomic DNA molecules. Analyst 2016, 141:4326–4331. [DOI] [PubMed] [Google Scholar]
- ••30.Zirkin S, Fishman S, Sharim H, Michaeli Y, Don J, Ebenstein Y: Lighting up individual DNA damage sites by in vitro repair synthesis. J Am Chem Soc 2014, 136:7771–7776.Extension of DNA fiber assay for the detection of DNA damage from DNA isolated from treated cells or from human samples using a comprehensive damage cocktail.
- ••31.Holton NW, Ebenstein Y, Gassman NR: Broad spectrum detection of DNA damage by Repair Assisted Damage Detection (RADD). DNA Repair (Amst) 2018, 66–67:42–49.Application of enzyme-mediated DNA damage detection in cells, demonstrating in situ DNA detection within chromatin context.
- 32.Diegoli TM, Farr M, Cromartie C, Coble MD, Bille TW: An optimized protocol for forensic application of the PreCR Repair Mix to multiplex STR amplification of UV-damaged DNA. Forensic Sci Int Genet 2012, 6:498–503. [DOI] [PubMed] [Google Scholar]
- •33.Hu J, Adar S, Selby CP, Lieb JD, Sancar A: Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev 2015, 29:948–960.Genome-wide sequencing of damage sites.
- •34.Hu J, Lieb JD, Sancar A, Adar S: Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc Natl Acad Sci U S A 2016, 113:11507–11512.Genome-wide sequencing of damage sites.
- 35.Didenko VV: In situ labeling of DNA breaks and apoptosis by T7 DNA polymerase. Methods Mol Biol 2011, 682:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornsby PJ, Didenko VV: In situ ligation: a decade and a half of experience. Methods Mol Biol 2011, 682:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loo DT: TUNEL assay. An overview of techniques. Methods Mol Biol 2002, 203:21–30. [DOI] [PubMed] [Google Scholar]
- 38.Didenko VV, Ngo H, Baskin DS: Early necrotic DNA degradation: presence of blunt-ended DNA breaks, 3’ and 5’ overhangs in apoptosis, but only 5’ overhangs in early necrosis. Am J Pathol 2003, 162:1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dierendonck JH: DNA damage detection using DNA polymerase I or its Klenow fragment. Applicability, specificity, limitations. Methods Mol Biol 2002, 203:81–108. [DOI] [PubMed] [Google Scholar]
- 40.Otsuki Y, Ito Y: Quantitative differentiation of both free 3’ OH and 5’ OH DNA ends using terminal transferase-based labeling combined with transmission electron microscopy. Methods Mol Biol 2002, 203:41–54. [DOI] [PubMed] [Google Scholar]
- 41.Baskin DS, Widmayer MA, Sharpe MA: Quantification of DNase type I ends, DNase type II ends, and modified bases using fluorescently labeled ddUTP, terminal deoxynucleotidyl transferase, and formamidopyrimidine-DNA glycosylase. Biotechniques 2010, 49:505–512. [DOI] [PubMed] [Google Scholar]
- ••42.Condie AG, Yan Y, Gerson SL, Wang Y: A Fluorescent Probe to Measure DNA Damage and Repair. PLoS One 2015, 10:e0131330.Synthesis and characterization of a fluorescent abasic site reporter.
- 43.Bennett SE, Kitner J: Characterization of the aldehyde reactive probe reaction with AP-sites in DNA: influence of AP-lyase on adduct stability. Nucleosides Nucleotides Nucleic Acids 2006, 25:823–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••44.Wei S, Shalhout S, Ahn YH, Bhagwat AS: A versatile new tool to quantify abasic sites in DNA and inhibit base excision repair. DNA Repair (Amst) 2015, 27:9–18.Synthesis and characterization of improved abasic site detection reagent.