Abstract
After more than ten years of accumulated efforts, genome-wide association studies (GWAS) have led to many findings, most of which have been deposited into the GWAS Catalog. Between GWAS’s inception and March 2017, the GWAS Catalog has collected 2,430 studies, 1,818 phenotypes, and 28,462 associated SNPs. We reclassified the psychology-related phenotypes into 198 reclassified phenotypes, which accounted for 472 studies and 6,632 SNPs. In total, 1,109 of the SNPs reached genome-wide significance. Of these, 133 were replicated for the same psychological trait in different studies. Another 379 SNPs were replicated within one original study. The SNPs rs2075650 and rs4420638 were linked to the most replications within a single reclassified phenotype; both were associated with Alzheimer’s disease (AD). Schizophrenia was associated with 76 SNPs. Alzheimer’s disease and schizophrenia were linked to many physical phenotypes, including cholesterol and body mass index, through common GWAS signals. Alzheimer’s disease also shared risk SNPs with age-related phenotypes such as age-related macular degeneration and longevity. Smoking-related SNPs were linked to lung cancer and respiratory function. Alcohol-related SNPs were associated with cardiovascular and digestive system phenotypes and disorders. Two separate studies also identified a shared risk SNP for bipolar disorder and educational attainment. This review revealed a list of reproducible SNPs worthy of future functional investigation. Additionally, by identifying SNPs associated with multiple phenotypes, we illustrated the importance of studying the relationships among phenotypes to resolve the nature of their causal links. The insights within this review will hopefully pave the way for future evidence-based genetic studies.
Introduction
The GWAS Catalog is a comprehensive database that archives genome-wide association studies (GWAS) investigating associations between single-nucleotide polymorphisms (SNPs) and a variety of phenotypes, ranging from psychiatric disorders (e.g. schizophrenia), to physical disorders, to other medical, physical, and psychological traits. GWAS feature data-driven, fairly objective designs and often use relatively large sample sizes, making them better-powered and less biased than most candidate-gene studies.
Motivation behind this review
The year 2017 marks the 10-year anniversary of psychiatric GWAS. While the first GWAS recorded by the GWAS Catalog was published in 2005 on the topic of age-related macular degeneration, A GWAS in the field of psychiatry was not published until a WTCCC-funded study in 2007. This study examined seven diseases, including bipolar disorder.1
Since then, genotyping costs have dropped dramatically and GWAS have become an essential step on the path towards uncovering genetic factors of phenotypes. Research concerning psychiatric disorders and relevant traits have accounted for a significant share of the accumulated GWAS findings and will be the focus of this review. Such findings are often selectively and mistakenly interpreted by the media, the public, clinicians, patients, and even some investigators in related fields. We feel the obligation to provide a scholastic, holistic, and stringent summary of the GWAS data. In doing this, we have identified SNPs that have been repeatedly reported in association with the same disorder and that should thus bear better confidence. We also hope to uncover hidden connections between disorders or traits by highlighting genomic regions that could be associated with multiple conditions.
Candidate genes have been studied for several decades in psychiatry. They have mostly included small sample sizes and often been underpowered. Some frequently-studied genes have been included in meta-analyses combining a large collection of studies. However, such meta-analyses have thus far reported disappointing results. Candidate genes have been found to be non-reproducible or insignificant upon further scrutiny, as in the case of two meta-analyses on bipolar disorder and schizophrenia candidate genes.2, 3 This article therefore focuses on GWAS signals. That being said, as GWAS produce more findings, it will likely be increasingly useful to compare results of candidate genes with GWAS signals. Furthermore, there will be a greater quantity of robust data from which to make more informed decisions about good candidate genes.
It should be noted that, as writing this summary requires some arbitrary decisions regarding inclusion criteria for disorders and traits and grouping of phenotypes, the reported results are somewhat swayed by bias. Its parameters should thus be refined and improved in future reviews.
Data
The data in this summary includes all studies recorded by the GWAS Catalog (https://www.ebi.ac.uk/gwas/) as of the date of download (March 30, 2017); the most recent study in the dataset was published on October 31st, 2016. The dataset includes literature sources, phenotype information, p-values, and identified SNPs, among many other pieces of data.
GWAS utilize stringent significance thresholds as a result of the multiple testing correction needed to account for millions of SNPs on the human genome. While the GWAS Catalog collected SNPs associated with p-values as large as 1 × 10−5 (with the exception of one result, which was linked to 2 × 10−5), the commonly used cut-off for genome-wide significance is typically p = 2 × 10−8 or 5 × 10−8.
The GWAS Catalog, as of the date of download, included a total of 2,430 papers, 1,818 phenotypes, and 28,462 SNPs. In order to redirect the focus of the analysis to psychiatry, we limited the set of phenotypes to a list of 198 reclassified psychological phenotypes, many of which are also directly related to psychiatric illness (Supplementary Table 1). In addition to retaining many phenotypes from the initial dataset, we added some broader phenotypes that combined similar traits in order to make parsing and comparison easier and more efficient. While many of the reclassified phenotypes are explicitly psychological in nature, we also included phenotypes that are not classically psychological but are likely related to or correlated with mental functioning. A total of 472 studies and 6,632 unique SNPs remained after phenotype reclassification and filtering. Out of these, 1,109 of the SNPs reached genome-wide significance. Roughly half of the significant SNPs were reproduced in some capacity (see section on reproducibility and Table 1).
Table 1.
Number of total and genome-wide significant SNPs in the entire GWAS Catalog and within the reclassified phenotypes. Also listed are the SNPs found in more than one study that reached genome-wide significance in at least one of the studies (Supplementary Table 2a) and those not in 2a but which are significant for multiple samples within the same study (Supplementary Table 2b).
| Total # of SNPs | # of Genome-wide Significant SNPs | # of Replicated SNPs | |
|---|---|---|---|
| Entire GWAS Catalog | 28,462 | 8,740 | — |
| Reclassified Psychiatry-Related Phenotypes | 6,632 | 1,109 | Total 512, including 133 (replicated across studies) + 379 (replicated in the same study) |
Finally, the dataset included a broad range of samples. The paper with the smallest sample size among the psychiatry-related studies was a study on clozapine-induced cytotoxicity that used 90 European ancestry lymphoblast cell lines.4 The largest sample size included samples from 549,935 individuals as part of a study on depression, neuroticism, and subjective well-being.5
Reproducibility
As in any other scientific discipline, reproducibility is a critical indication of the strength of genetic findings. Here, we identified SNPs that have been associated with the same (or very similar) reclassified disorders or traits in two or more studies. If the specific risk allele of the SNPs of interest were reported consistently for each study, we also included the allele (denoted by a hyphen and the base letter following the name of the SNP). We excluded non-identical reclassified “compound phenotypes” (phenotypes that are a combination of multiple disorders or traits) if they only matched with reclassified phenotypes that were part of the compound. For example, if the only reclassified phenotypes found to be significant for a SNP were schizophrenia and the compound phenotype autism, bipolar disorder, or schizophrenia, the SNP was not considered to be replicated. However, a SNP found to be related to psychosis and mood in two different studies would be considered replicated. Furthermore, we required at least one of the findings corresponding to the phenotype and SNP to reach genome-wide significance (p ≤ 2 × 10−8). All further replicated findings for the same SNP and the same or very similar reclassified phenotype met the catalog’s p-value cut-off of p ≤ 2 × 10−5. In Supplementary Table 2a, we listed the 133 SNPs meeting the above criteria (excluding replication samples within a single paper) for reclassified phenotypes. Supplementary Table 2b lists 379 SNPs that were significantly replicated in samples within a single paper but were not listed in Supplementary Table 2a.
We consider the 133 SNPs replicated by two or more separate publications arguably more reliable than the 379 SNPs replicated by the same publications, so our discussion focuses on the former. The SNPs rs2075650 and rs4420638 were linked to the most replications within a single reclassified phenotype. Both are brain eQTL (see Supplementary Table 2a). rs2075650 was reported in association with Alzheimer’s disease in eight different studies6–13 and in association with cognitive decline in one study.14 rs4420638 was linked to Alzheimer’s disease in six studies15–20 and cognitive decline in one study.21
rs2075650 is located in an intron of the TOMM40 gene, a mitochondria membrane protein. It is a brain eQTL SNP for the gene PPM1N and many other genes (though not for APOE4 or TOMM40). The G-allele for rs2075650 was reported to confer an odds ratio of two in a small case-control study.22 This SNP is also associated with hippocampal atrophy. However, there is question as to whether it carries an independent AD risk. One study posits that rs2075650’s relationship with AD is more attributable to its modest linkage disequilibrium with rs429358.23 rs429358 is on the fourth exon of APOE and, along with rs7412, determines the APOE haplotype, where the variant ε4 is the strongest risk factor for AD. Similarly, another SNP, rs429358, was associated with AD in three studies (Supplementary Table 2a). The SNP rs4420638 is located downstream of the APOC1 gene. rs4420638’s association with AD could also be related to its linkage disequilibrium with rs429358 and so is likely also not an entirely independent risk locus.24, 25 rs4420638 is a brain eQTL SNP for non-APOE genes, according to BRAINEAC. The evidence regarding GWAS signal linkage disequilibrium and the link between GWAS signals and brain eQTL may merit further investigation to clarify issues of causality.
Narcolepsy and nicotine-related phenotypes were also linked to highly replicated SNPs. They were associated with the SNPs rs115415526–29 and rs1051730,30–33 respectively, each in four different studies. Additionally, rs1154155 was identified in three studies in association with narcolepsy27, 28, 34 and in one study in association with narcolepsy with cataplexy.29 rs1154155 is mapped to the downstream region of TRAJ10, a gene in the TRAJ (T cell receptor alpha joining) gene cluster. rs1154155 is an eQTL SNP for mir208a in white matter. rs1051730 is a coding synonymous variant in CHRNA3 but is also an eQTL SNP for CHRNA5 in brain. Both CHRNA3 and CHRNA5 are strong candidates for nicotine dependence.
Schizophrenia was the disease with by far the largest number of SNPs replicated within-phenotype —76— though many of these replications came from the same two studies, which shared samples.35, 36 Educational attainment accounted for eleven replicated SNPs, and phenotypes including the term “Alzheimer’s” were associated with 24 replicated SNPs.
While, at face value, the aforementioned findings could suggest strong genetic underpinnings of Alzheimer’s disease and schizophrenia, it should be noted that some topics have been given more attention than others. For example, the search term “GWAS Alzheimer’s disease” returns about four times as many results in Google Scholar as “GWAS educational attainment.”
The fact that some studies drew from the same samples as other studies must also be taken into account. For example, only four publications from two research groups (from the Netherlands and the UK) have studied educational attainment,37–40 and many of the samples from the four studies either were shared or came from the same biobank. Because recruiting huge samples is difficult, it is common for consortia to publish several studies on the same continuously growing sample. Thus, not all of the SNPs we recorded should be considered independently replicated. Going forward, it will be important for researchers to be cognizant of the limited validity of many reported GWAS findings.
Table 1 summarizes the quantity of SNPs according to criteria of significance, phenotype class, and replication status.
Reproducible GWAS Signals as Brain eQTL
We entered the 133 reproduced GWAS signals into the UK Brain Expression Consortium’s (UKBEC) expression quantitative trait loci (eQTL) database (http://www.braineac.org/). Among the 133 SNPs, a total of 78 SNPs (59%) exceeded the threshold cut-off required to reach significance (0.05/133 replicated SNPs = 3.8×10−4, see Supplementary Table 2a). Of these, the SNP rs17693963 (associated with schizophrenia, bipolar disorder, or schizoaffective disorder41, 42) was the strongest brain eQTL and was associated with expression of the gene ZNF389 in white matter (p = 2.8×10−10), among other brain regions. GTEx data also reported rs17693963 as an eQTL SNP in many tissues (including brain) and as being associated with several genes. The second strongest brain eQTL was rs950169 (associated with schizophrenia35, 36). This SNP was also associated with SCAND2 in cerebellum (p = 1.4×10−8), hippocampus (p = 2.2×10−8), and other brain regions. GTEx also confirmed rs950169 as a universal eQTL in many tissue types and as being associated with multiple genes.
Cross-Phenotype Analysis
GWAS can help draw attention to psychiatric disorders that may share genomic risk factors. The literature with the greatest impact on this topic has largely pointed to shared genetics between schizophrenia and other psychiatric conditions, perhaps most notably bipolar disorder, intellectual disability, and autism spectrum disorder, based on polygenic risk scores and shared rare variants.36, 43–45 In this analysis, we again focused on individual SNPs recorded in the GWAS Catalog, which primarily includes common variants. For cases in which phenotypes were redundant, they were sometimes summarized or combined (e.g. “hematocrit” and “red blood cell traits,” when linked to the same SNP, were condensed to “red blood cell traits”).
We identified the SNPs and SNP risk alleles mapped to distinct phenotypes (e.g. excluding matches between single phenotypes and compound phenotypes encapsulating that phenotype and excluding very similar phenotypes). The cross-phenotype analysis required a more stringent threshold cut-off than the reproducibility analysis because, unlike in the case of reproducibly, this analysis needed to account for all examined phenotypes. We thus required that the associations for all phenotypes reach genome-wide significance (p ≤ 2 × 10−8).
Because our focus is on psychiatry, we only summarized findings concerning SNPs linked to at least one psychological trait, though we also included all significant findings for non-psychiatric/psychological phenotypes also associated with the same SNPs. The SNPs associated with multiple phenotypes are plotted in a network in Figure 1.
Figure 1.
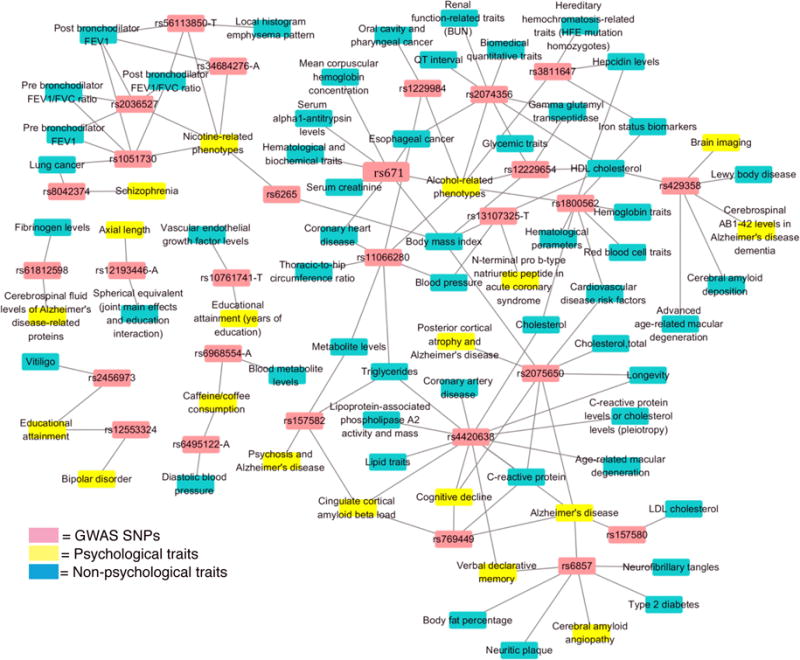
SNP-Phenotype Network. All the SNPs (in red) showed a significant association (p ≤ 2 × 10−8) with psychological/psychiatric phenotypes (in yellow) and at least one other psychological/psychiatric phenotype or a non-psychological phenotype (in turquoise).
Addictive Behavior and Substance Use
Many significant SNPs were mapped to the use of various substances, a phenotype category that has been measured in both controls and those affected by addiction. Four SNPs were associated with both nicotine-related phenotypes and phenotypes related to respiration. rs1051730 was linked to smoking behavior30–33 and nicotine dependence30 (combined into “nicotine-related phenotypes”), post bronchodilator FEV1, post bronchodilator FEV1/FVC ratio, pre bronchodilator FEV1, pre bronchodilator FEV1/FVC ratio, and lung cancer;46, 47 rs2036527 was related to smoking behavior,48 post bronchodilator FEV1, post bronchodilator FEV1/FVC ratio, pre bronchodilator FEV1, and pre bronchodilator FEV1/FVC ratio;49 rs34684276-A was associated with post bronchodilator FEV1, post bronchodilator FEV1/FVC ratio,49 and nicotine dependence;50 and rs56113850-T was associated with nicotine metabolite ratio in current smokers,51 local histogram emphysema pattern,52 post bronchodilator FEV1, and post bronchodilator FEV1/FVC ratio.49 While these connections between smoking and respiratory phenotypes are meaningful, there exists the strong possibility that respiratory changes are the product of smoking and that these SNPs are not organic genetic causes of lung cancer, etc.
Additionally, rs6265 was associated with both body mass index53 and smoking behavior.31
Alcohol use also shared common risk SNPs with many other disorders and traits, particularly those related to the circulatory and digestive systems. One meta-analysis of “maxdrinks,” an alcohol-related phenotype measuring the greatest number of drinks that an individual has ever consumed in a 24-hour period, included the results of two studies.54 While neither of the two studies yielded a statistically significant association for rs1229984, the SNP achieved a p-value of 2.04 × 10−8 for the meta-analysis. Two other studies, one on alcohol dependence55 and the other on the effects of alcohol and smoking on esophageal cancer risk,56 found evidence for rs1229984 as a risk SNP. Further, this SNP was found to be linked to oral cavity and pharyngeal cancer.57 rs671 was associated with drinking behavior,58 coronary heart disease,59 alcohol consumption (maxi-drinks) and response to alcohol consumption (flushing) in small samples of Han Chinese participants,60 esophageal cancer,56 serum alpha1-antitrypsin levels,61 body mass index,62 mean corpuscular hemoglobin concentration,63 serum creatinine,64 and hematological and biochemical traits.63 A study assessing Han Chinese drinkers and nondrinkers65 identified rs11066280, a SNP also associated with blood pressure phenotypes,66–68 esophageal cancer,69 coronary heart disease,70 thoracic-to-hip circumference ratio,71 metabolite levels,72 and triglycerides,73 in other studies. The SNP rs1800562 was associated with transferrin glycosylation,74 which is relevant to alcohol consumption, as well as with iron status biomarkers,75–78 hematological parameters,79 hepcidin levels,80 cardiovascular disease risk factors,81 cholesterol,82, 83 hemoglobin,84, 85 and red blood cell traits.84, 86 rs12229654 was linked to body mass index,62 glycemic traits,87 HDL cholesterol, gamma glutamyl transpeptidase,72 and alcohol consumption.88 rs2074356 was linked to alcohol consumption,88 QT interval,89, 90 gamma glutamyl transpeptidase, HDL cholesterol,72 esophageal cancer,69 glycemic traits,87 biomedical quantitative traits,91 and renal function-related traits (BUN),64 and rs3811647 was linked to hepcidin levels,80 iron status biomarkers,75, 77, 92 alcohol consumption (transferrin glycosylation),74 and hereditary hemochromatosis-related traits (HFE mutation homozygotes).93
Coffee consumption94, 95 held a genome-wide association with diastolic blood pressure96 at rs6495122-A and with blood metabolite levels97 at rs6968554-A.
According to research that has been conducted thus far, SNPs mapped to the use of substances overlap almost exclusively with SNPs related to physical, non-psychological phenotypes, which could either reflect the physical consequences of excessive substance use or indicate common genetic predispositions. Surprisingly, there were no significant associations between substance use phenotypes and psychiatric disorders that are frequently comorbid with substance abuse.
Educational Attainment
Findings pertaining to educational attainment also appeared multiple times in cross-phenotype analysis. The term “educational attainment” had different implications depending on the study. For example, “educational attainment,” in some research, indicated years of education, while other research measured “educational attainment” according to performance on a reasoning task. rs10761741-T has been identified as a SNP risk allele for vascular endothelial growth factor levels98 and years of education.37 In the case of the former, the T allele at this locus seemed to indicate greater epinephrine-induced platelet aggregation and higher circulating VEGF levels. Serum VEGF levels have been reported as being associated with prefrontal cortex volume in schizophrenia patients.99 Therefore, there is a possibility that this SNP might be related to educational attainment as a result of its contribution to brain structure.
rs2456973 was associated with vitiligo,100 a skin disorder, and educational attainment.37 There is no obvious connection between the two phenomena.
One meta-analysis reported rs12193446-A as the strongest SNP risk allele for refractive error × education interaction.101 Another study also revealed rs12193446-A as the SNP allele most strongly determining ocular axial length.102 Similarly, a study of a rural population in India reported a positive correlation between ocular axial length and education; the same result was found in two other studies—one with a Chinese population and one with an elderly White population.103–105
The SNP rs12553324 was found to be associated with level of educational attainment40 and, in a separate study, was also identified as an area of interest for bipolar disorder.106 Glahn et al., in a non-genetic 2006 study predating both of the former, found that bipolar patients had fewer years of educational attainment.107 On the contrary, when measuring academic achievement in terms of performance rather than years in school, MacCabe et al. found somewhat mixed results.108 While there was a relationship between bipolar disorder and low grades, there was also a very increased rate of eventual bipolar disorder diagnosis amongst those with high grades, compared to those with average grades. A genetic study reported a positive genetic correlation, achieved through linkage disequilibrium score regression, between bipolar disorder and years of education.109 One paper studying polygenic risk score results, which represent combined contribution of many common SNPs to risk, found that high risk score of bipolar disorder and schizophrenia are predictive of high educational attainment.110 There appears to be a relationship between bipolar disorder and educational attainment; the specific nature of this link should be further studied.
Alzheimer’s Disease
Several studies mapped common SNPs to Alzheimer’s disease and many other phenotypes.6–12, 14–16, 21, 23, 25, 73, 81–83, 111–146 More than one SNP was mapped to both Alzheimer’s disease and each of the following traits:
Verbal declarative memory
Cingulate cortical amyloid beta load (covariate in one study)
C-reactive proteins
Longevity
Cholesterol
Cognitive decline
Age-related macular degeneration
Triglycerides
Given that both Alzheimer’s disease and these phenotypes are largely related to aging, the findings are not surprising. Inflammation, as it relates to Alzheimer’s, could also be an interesting further topic of study because of inflammation’s link to C-reactive protein.
Schizophrenia
Schizophrenia-focused GWAS findings also attained cross-phenotype significance. In particular, rs13107325-T was linked to schizophrenia,35 as well as to body mass index,117, 147, 148 HDL cholesterol,82, 83 blood pressure,149, 150 and N-terminal pro b-type natriuretic peptide in acute coronary syndrome.151 Some of these findings are congruent with research that has found higher rates of cardiovascular diseases in schizophrenia.152, 153 rs8042374 was associated with schizophrenia35, 36 and lung cancer,154 a finding that is not incredibly surprising given the high rates of smoking in schizophrenics.155
While some SNPs associated with both schizophrenia only and cohorts with schizophrenia and other disorders (e.g. autism, bipolar disorder) were excluded from the results due to the complications of linking a compound trait to a single finding, the high volume of SNPs associated with such combined effects suggests heretofore uncovered genetic commonalities between schizophrenia and other psychiatric disorders. Although polygenic analysis indicates that bipolar and schizophrenia share genetic risk factors, they did not share any significant individual SNP associations according to our analysis. This may be related to imbalance in sample sizes. Schizophrenia GWAS typically have much larger sample sizes than bipolar disorder GWAS. Still, there was some evidence for shared GWAS signals between the two disorders. For example, the SNP rs1006737 in the CACNA1C gene was linked to schizophrenia, with a p of 2 × 10−8, but with bipolar disorder at p = 7 × 10−8.42 Thus, the bipolar association did not meet the significance threshold for our analysis. However, both schizophrenia and bipolar disorder were associated with several different SNPs reaching genome-wide significance in CACNA1C.
Supplementary Table 3 summarizes the cross-phenotype SNPs and their associated phenotypes, with the phenotypes that were later transformed into reclassified phenotypes listed under the name of the reclassified phenotypes.
Limitation of the current review
We acknowledge that there are many areas of GWAS that were not deeply addressed within this paper. In writing this review, we relied primarily on the data as it was presented in the GWAS Catalog. Certain statistical parameters such as effect size could not be easily summarized and compared, as not all studies used the same criteria. Also, as the GWAS Catalog includes only genome-wide association studies, our analysis did not take candidate gene studies into account, meaning that our points of comparison were not exhaustive.
The present review is intended to serve as a broad summary and analysis of psychiatric GWAS. We believe that future studies and reviews that approach GWAS from a different angle or that focus on finer statistical details, even going so far as to scrutinize raw data from individuals, could lead to interesting and important findings.
Conclusion
GWAS research has yielded many discoveries in both psychiatry and physical pathology. Further research and improvement in big data parsing methods will be an imperative part of fully legitimizing the field, but there already exist strong bases for forming theories about genome-wide associations.
Most psychiatric disorders appear to share specific risk loci with physical disorders or phenotypes instead of other psychiatric disorders or psychological traits. This could perhaps be because many psychiatric disorders are characterized by a rather wide assortment of sub-phenotypes, some of which may be more linked to physical phenotypes from a biological perspective. Perhaps most notably, GWAS research on substance use has led to many associations with physical traits, which may harbor implications for addiction research.
More validation is needed
As shown in Table 1, only about half of the psychological genome-wide significant associations have been validated, either in an independent study or in a replication sample. Because of the possibility of false discovery, the likelihood of a GWAS signal being a true marker of the tested phenotype holds fairly limited promise prior to replication.
Heterogeneity issues across studies
Any discrepancies between individual studies must be taken into consideration as confounding variables. For example, studies included a broad range of sample sizes, some of which only included a single racial or ethnic group; these studies should be replicated within different populations. It should also be noted that some studies drew from the same consortia, meaning that many samples likely shared participants. Additionally, some of the papers in the database were meta-analyses of other GWAS, which creates another opportunity for sample overlaps.
Differentiating causal associations from indirect correlational relationships
In assessing the data, it is important to beware of tempting but potentially false causal conclusions. For example, rs1051730 was mapped to smoking and some related physical conditions, including lung cancer. In order to find evidence that the listed physical health risks are governed by the same genomic regions (rather than just being a result of smoking behavior), it would be important to compare smokers to non-smokers, a variable that was not stringently controlled with respect to the loci of interest.
Despite these caveats, the relationships drawn between non-pathological phenotypes and psychiatric disorders is an important area of study. The findings linking the two types of traits promote the idea that physical phenotypes may eventually be able to serve as valid biomarkers of psychopathology. Going forward, it will be important to make efforts to distinguish between shared genetic roots and phenotypes that co-occur but which are not the results of a common genetic cause.
Future Directions
Revealing the functionality of GWAS-associated SNPs and resolving the causal nature of the relationship between SNPs and their associated traits should be one of the major objectives of post-GWAS conducted in the coming years. The CRISPR/Cas9 editing of risk alleles in induced pluripotent stem cells (iPSC),156 as well as gene knockdown and knockout studies, promise to bring exciting new insights to the discipline of GWAS.
The study of genetic relationships between various phenotypes is a vast new field of research with possibilities that will only increase as the rate of research accelerates and the GWAS database grows. Such research is giving us a new perspective on the etiology and pathology of disorders.
GWAS, while still in its infancy, has led to a prodigious quantity of publications and areas of study. We hope the insights gleaned in this investigation will help to guide future research in psychiatric genomics by highlighting worthwhile areas of investigation, ultimately enhancing our understanding of psychiatric disorders.
Supplementary Material
Supplementary Table 1. List of Phenotypes Included in the Review and Their Corresponding Reclassifications: A list of all the initial phenotypes (left column) and the 198 reclassified phenotypes into which some of them were transformed (corresponding cell in the right column). The reclassified phenotypes reflect the important themes in the psychiatric GWAS studies.
Supplementary Table 2a. Replicated GWAS Findings from Independent Publication-or SNP alleles found in more than one publication for the same or very similar reclassified phenotypes along with the number of studies reporting the SNP.
Supplementary Table 2b. Replicated GWAS Findings from the Same Publications: SNPs or SNP alleles found in both initial and replication samples in one paper studying a reclassified phenotype but not replicated across publications.
Supplementary Table 3. SNPs Associated with Multiple Phenotypes: A list of SNPs or SNP alleles with p-values of ≤2 × 10−8 replicated across phenotypes, including at least one significant association with a psychiatric phenotype; the reclassified phenotypes are included in place of the original phenotypes when applicable, and redundant or very similar non-reclassified phenotypes are summarized or omitted.
Acknowledgments
We thank Dr. Elliot S. Gershon for valuable discussions and comments.
Footnotes
Conflict of interest.
The authors declare no conflict of interest.
Supplementary information is available at MP’s website.
References
- 1.Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seifuddin F, Mahon PB, Judy J, Pirooznia M, Jancic D, Taylor J, et al. Meta-analysis of genetic association studies on bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(5):508–518. doi: 10.1002/ajmg.b.32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O’Donovan MC, et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015;20(5):555–562. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de With SA, Pulit SL, Wang T, Staal WG, van Solinge WW, de Bakker PI, et al. Genome-wide association study of lymphoblast cell viability after clozapine exposure. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(2):116–122. doi: 10.1002/ajmg.b.32287. [DOI] [PubMed] [Google Scholar]
- 5.Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48(6):624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson PT, Estus S, Abner EL, Parikh I, Malik M, Neltner JH, et al. ABCC9 gene polymorphism is associated with hippocampal sclerosis of aging pathology. Acta Neuropathol. 2014;127(6):825–843. doi: 10.1007/s00401-014-1282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Palma E, Bustos BI, Villaman CF, Alarcon MA, Avila ME, Ugarte GD, et al. Overrepresentation of glutamate signaling in Alzheimer’s disease: network-based pathway enrichment using meta-analysis of genome-wide association studies. PLoS One. 2014;9(4):e95413. doi: 10.1371/journal.pone.0095413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinzen EL, Need AC, Hayden KM, Chiba-Falek O, Roses AD, Strittmatter WJ, et al. Genome-wide scan of copy number variation in late-onset Alzheimer’s disease. J Alzheimers Dis. 2010;19(1):69–77. doi: 10.3233/JAD-2010-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 12.Naj AC, Beecham GW, Martin ER, Gallins PJ, Powell EH, Konidari I, et al. Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 2010;6(9):e1001130. doi: 10.1371/journal.pgen.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, et al. Genome-wide association study of CSF biomarkers Abeta1-42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76(1):69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies G, Harris SE, Reynolds CA, Payton A, Knight HM, Liewald DC, et al. A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing. Mol Psychiatry. 2014;19(1):76–87. doi: 10.1038/mp.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold C, Hooli BV, Mullin K, Liu T, Roehr JT, Mattheisen M, et al. Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease with OSBPL6, PTPRG, and PDCL3. Mol Psychiatry. 2016;21(11):1608–1612. doi: 10.1038/mp.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, et al. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 18.Webster JA, Myers AJ, Pearson JV, Craig DW, Hu-Lince D, Coon KD, et al. Sorl1 as an Alzheimer’s disease predisposition gene? Neurodegener Dis. 2008;5(2):60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- 19.Kamboh MI, Barmada MM, Demirci FY, Minster RL, Carrasquillo MM, Pankratz VS, et al. Genome-wide association analysis of age-at-onset in Alzheimer’s disease. Mol Psychiatry. 2012;17(12):1340–1346. doi: 10.1038/mp.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coon KD, Dunckley TL, Stephan DA. A generic research paradigm for identification and validation of early molecular diagnostics and new therapeutics in common disorders. Mol Diagn Ther. 2007;11(1):1–14. doi: 10.1007/BF03256218. [DOI] [PubMed] [Google Scholar]
- 21.De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS, et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol Aging. 2012;33(5):1017e1011–1015. doi: 10.1016/j.neurobiolaging.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS One. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10(4):686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyholt DR, Yu CE, Visscher PM. On Jim Watson’s APOE status: genetic information is hard to hide. Eur J Hum Genet. 2009;17(2):147–149. doi: 10.1038/ejhg.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry. 2007;68(4):613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Wang Z, Liu Y, Tian H, Yao X, Zhang J. Development and characterization of 11 polymorphic microsatellite markers in Tapiscia sinensis (Staphyleaceae) Appl Plant Sci. 2013;1(12) doi: 10.3732/apps.1300051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hor H, Kutalik Z, Dauvilliers Y, Valsesia A, Lammers GJ, Donjacour CE, et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat Genet. 2010;42(9):786–789. doi: 10.1038/ng.647. [DOI] [PubMed] [Google Scholar]
- 28.Hallmayer J, Faraco J, Lin L, Hesselson S, Winkelmann J, Kawashima M, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41(6):708–711. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toyoda H, Miyagawa T, Koike A, Kanbayashi T, Imanishi A, Sagawa Y, et al. A polymorphism in CCR1/CCR3 is associated with narcolepsy. Brain Behav Immun. 2015;49:148–155. doi: 10.1016/j.bbi.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobacco Genetics C. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han F, Faraco J, Dong XS, Ollila HM, Lin L, Li J, et al. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet. 2013;9(10):e1003880. doi: 10.1371/journal.pgen.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goes FS, McGrath J, Avramopoulos D, Wolyniec P, Pirooznia M, Ruczinski I, et al. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet B Neuropsychiatr Genet. 2015;168(8):649–659. doi: 10.1002/ajmg.b.32349. [DOI] [PubMed] [Google Scholar]
- 36.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533(7604):539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111(38):13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151) Mol Psychiatry. 2016;21(6):758–767. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sleiman P, Wang D, Glessner J, Hadley D, Gur RE, Cohen N, et al. GWAS meta analysis identifies TSNARE1 as a novel Schizophrenia / Bipolar susceptibility locus. Sci Rep. 2013;3:3075. doi: 10.1038/srep03075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Schizophrenia Working Group of the Psychiatric Genomics C et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2014;19(9):1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Schizophrenia C. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Cai T, Jiang Y, Chen H, He X, Chen C, et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry. 2016;21(2):290–297. doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A Genome-wide Association Study of Lung Cancer Identifies a Region of Chromosome 5p15 Associated with Risk for Adenocarcinoma. Am J Hum Genet. 2011;88(6):861. doi: 10.1016/j.ajhg.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40(12):1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL, et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lutz SM, Cho MH, Young K, Hersh CP, Castaldi PJ, McDonald ML, et al. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet. 2015;16:138. doi: 10.1186/s12863-015-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hancock DB, Reginsson GW, Gaddis NC, Chen X, Saccone NL, Lutz SM, et al. Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. doi: 10.1038/tp.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel YM, Park SL, Han Y, Wilkens LR, Bickeboller H, Rosenberger A, et al. Novel Association of Genetic Markers Affecting CYP2A6 Activity and Lung Cancer Risk. Cancer Res. 2016;76(19):5768–5776. doi: 10.1158/0008-5472.CAN-16-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castaldi PJ, Cho MH, San Jose Estepar R, McDonald ML, Laird N, Beaty TH, et al. Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190(4):399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 54.Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, et al. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet. 2013;132(10):1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, et al. Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet. 2013;132(6):657–668. doi: 10.1007/s00439-013-1281-8. [DOI] [PubMed] [Google Scholar]
- 56.Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137(5):1768–1775. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 57.McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, Byrnes G, et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7(3):e1001333. doi: 10.1371/journal.pgen.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, et al. Confirmation of ALDH2 as a Major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J. 2011;75(4):911–918. doi: 10.1253/circj.cj-10-0774. [DOI] [PubMed] [Google Scholar]
- 59.Takeuchi F, Yokota M, Yamamoto K, Nakashima E, Katsuya T, Asano H, et al. Genome-wide association study of coronary artery disease in the Japanese. Eur J Hum Genet. 2012;20(3):333–340. doi: 10.1038/ejhg.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, et al. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014;165B(2):103–110. doi: 10.1002/ajmg.b.32213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Setoh K, Terao C, Muro S, Kawaguchi T, Tabara Y, Takahashi M, et al. Three missense variants of metabolic syndrome-related genes are associated with alpha-1 antitrypsin levels. Nat Commun. 2015;6:7754. doi: 10.1038/ncomms8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang JY, et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23(20):5492–5504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42(3):210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 64.Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44(8):904–909. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X, Lu X, Wang L, Chen S, Li J, Cao J, et al. Common variants at 12q24 are associated with drinking behavior in Han Chinese. Am J Clin Nutr. 2013;97(3):545–551. doi: 10.3945/ajcn.112.046482. [DOI] [PubMed] [Google Scholar]
- 66.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43(6):531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet. 2015;24(3):865–874. doi: 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47(11):1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y, et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet. 2011;43(7):679–684. doi: 10.1038/ng.849. [DOI] [PubMed] [Google Scholar]
- 70.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44(8):890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cha S, Park AY, Kang C. A Genome-Wide Association Study Uncovers a Genetic Locus Associated with Thoracic-to-Hip Ratio in Koreans. PLoS One. 2015;10(12):e0145220. doi: 10.1371/journal.pone.0145220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011;43(10):990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- 73.Lu X, Huang J, Mo Z, He J, Wang L, Yang X, et al. Genetic Susceptibility to Lipid Levels and Lipid Change Over Time and Risk of Incident Hyperlipidemia in Chinese Populations. Circ Cardiovasc Genet. 2016;9(1):37–44. doi: 10.1161/CIRCGENETICS.115.001096. [DOI] [PubMed] [Google Scholar]
- 74.Kutalik Z, Benyamin B, Bergmann S, Mooser V, Waeber G, Montgomery GW, et al. Genome-wide association study identifies two loci strongly affecting transferrin glycosylation. Hum Mol Genet. 2011;20(18):3710–3717. doi: 10.1093/hmg/ddr272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benyamin B, McRae AF, Zhu G, Gordon S, Henders AK, Palotie A, et al. Variants in TF and HFE explain approximately 40% of genetic variation in serum-transferrin levels. Am J Hum Genet. 2009;84(1):60–65. doi: 10.1016/j.ajhg.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benyamin B, Ferreira MA, Willemsen G, Gordon S, Middelberg RP, McEvoy BP, et al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet. 2009;41(11):1173–1175. doi: 10.1038/ng.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McLaren CE, Garner CP, Constantine CC, McLachlan S, Vulpe CD, Snively BM, et al. Genome-wide association study identifies genetic loci associated with iron deficiency. PLoS One. 2011;6(3):e17390. doi: 10.1371/journal.pone.0017390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5:4926. doi: 10.1038/ncomms5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, Teumer A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41(11):1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Traglia M, Girelli D, Biino G, Campostrini N, Corbella M, Sala C, et al. Association of HFE and TMPRSS6 genetic variants with iron and erythrocyte parameters is only in part dependent on serum hepcidin concentrations. J Med Genet. 2011;48(9):629–634. doi: 10.1136/jmedgenet-2011-100061. [DOI] [PubMed] [Google Scholar]
- 81.Middelberg RP, Ferreira MA, Henders AK, Heath AC, Madden PA, Montgomery GW, et al. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med Genet. 2011;12:123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41(11):1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin X, Li J, Yin G, Zhao Q, Elias D, Lykkesfeldt AE, et al. Integrative analyses of gene expression and DNA methylation profiles in breast cancer cell line models of tamoxifen-resistance indicate a potential role of cells with stem-like properties. Breast Cancer Res. 2013;15(6):R119. doi: 10.1186/bcr3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Go MJ, Hwang JY, Kim YJ, Hee Oh J, Kim YJ, Heon Kwak S, et al. New susceptibility loci in MYL2, C12orf51 and OAS1 associated with 1-h plasma glucose as predisposing risk factors for type 2 diabetes in the Korean population. J Hum Genet. 2013;58(6):362–365. doi: 10.1038/jhg.2013.14. [DOI] [PubMed] [Google Scholar]
- 88.Baik I, Cho NH, Kim SH, Han BG, Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr. 2011;93(4):809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- 89.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46(8):826–836. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41(4):399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 92.Pichler I, Minelli C, Sanna S, Tanaka T, Schwienbacher C, Naitza S, et al. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum Mol Genet. 2011;20(6):1232–1240. doi: 10.1093/hmg/ddq552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Tayrac M, Roth MP, Jouanolle AM, Coppin H, le Gac G, Piperno A, et al. Genome-wide association study identifies TF as a significant modifier gene of iron metabolism in HFE hemochromatosis. J Hepatol. 2015;62(3):664–672. doi: 10.1016/j.jhep.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 94.Amin N, Byrne E, Johnson J, Chenevix-Trench G, Walter S, Nolte IM, et al. Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol Psychiatry. 2012;17(11):1116–1129. doi: 10.1038/mp.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Coffee, Caffeine Genetics C. Cornelis MC, Byrne EM, Esko T, Nalls MA, et al. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol Psychiatry. 2015;20(5):647–656. doi: 10.1038/mp.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41(6):677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi SH, Ruggiero D, Sorice R, Song C, Nutile T, Vernon Smith A, et al. Six Novel Loci Associated with Circulating VEGF Levels Identified by a Meta-analysis of Genome-Wide Association Studies. PLoS Genet. 2016;12(2):e1005874. doi: 10.1371/journal.pgen.1005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pillai A, Howell KR, Ahmed AO, Weinberg D, Allen KM, Bruggemann J, et al. Association of serum VEGF levels with prefrontal cortex volume in schizophrenia. Mol Psychiatry. 2016;21(5):686–692. doi: 10.1038/mp.2015.96. [DOI] [PubMed] [Google Scholar]
- 100.Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44(6):676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fan Q, Verhoeven VJ, Wojciechowski R, Barathi VA, Hysi PG, Guggenheim JA, et al. Meta-analysis of gene-environment-wide association scans accounting for education level identifies additional loci for refractive error. Nat Commun. 2016;7:11008. doi: 10.1038/ncomms11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng CY, Schache M, Ikram MK, Young TL, Guggenheim JA, Vitart V, et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am J Hum Genet. 2013;93(2):264–277. doi: 10.1016/j.ajhg.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nangia V, Jonas JB, Sinha A, Matin A, Kulkarni M, Panda-Jonas S. Ocular axial length and its associations in an adult population of central rural India: the Central India Eye and Medical Study. Ophthalmology. 2010;117(7):1360–1366. doi: 10.1016/j.ophtha.2009.11.040. [DOI] [PubMed] [Google Scholar]
- 104.Wong TY, Foster PJ, Johnson GJ, Seah SK. Education, socioeconomic status, and ocular dimensions in Chinese adults: the Tanjong Pagar Survey. Br J Ophthalmol. 2002;86(9):963–968. doi: 10.1136/bjo.86.9.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee KE, Klein BE, Klein R, Quandt Z, Wong TY. Association of age, stature, and education with ocular dimensions in an older white population. Arch Ophthalmol. 2009;127(1):88–93. doi: 10.1001/archophthalmol.2008.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landen M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25(15):3383–3394. doi: 10.1093/hmg/ddw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Glahn DC, Bearden CE, Bowden CL, Soares JC. Reduced educational attainment in bipolar disorder. J Affect Disord. 2006;92(2–3):309–312. doi: 10.1016/j.jad.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 108.MacCabe JH, Lambe MP, Cnattingius S, Sham PC, David AS, Reichenberg A, et al. Excellent school performance at age 16 and risk of adult bipolar disorder: national cohort study. Br J Psychiatry. 2010;196(2):109–115. doi: 10.1192/bjp.bp.108.060368. [DOI] [PubMed] [Google Scholar]
- 109.Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DC, Ritchie SJ, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry. 2016;21(11):1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18(7):953–955. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 111.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41(1):47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Feulner TM, Laws SM, Friedrich P, Wagenpfeil S, Wurst SH, Riehle C, et al. Examination of the current top candidate genes for AD in a genome-wide association study. Mol Psychiatry. 2010;15(7):756–766. doi: 10.1038/mp.2008.141. [DOI] [PubMed] [Google Scholar]
- 113.Antunez C, Boada M, Gonzalez-Perez A, Gayan J, Ramirez-Lorca R, Marin J, et al. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer’s disease. Genome Med. 2011;3(5):33. doi: 10.1186/gm249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li J, Zhang Q, Chen F, Yan J, Kim S, Wang L, et al. Genetic Interactions Explain Variance in Cingulate Amyloid Burden: An AV-45 PET Genome-Wide Association and Interaction Study in the ADNI Cohort. Biomed Res Int. 2015;2015:647389. doi: 10.1155/2015/647389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kristiansson K, Perola M, Tikkanen E, Kettunen J, Surakka I, Havulinna AS, et al. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 2012;5(2):242–249. doi: 10.1161/CIRCGENETICS.111.961482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hollingworth P, Sweet R, Sims R, Harold D, Russo G, Abraham R, et al. Genome-wide association study of Alzheimer’s disease with psychotic symptoms. Mol Psychiatry. 2012;17(12):1316–1327. doi: 10.1038/mp.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dorajoo R, Li R, Ikram MK, Liu J, Froguel P, Lee J, et al. Are C-reactive protein associated genetic variants associated with serum levels and retinal markers of microvascular pathology in Asian populations from Singapore? PLoS One. 2013;8(7):e67650. doi: 10.1371/journal.pone.0067650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schott JM, Crutch SJ, Carrasquillo MM, Uphill J, Shakespeare TJ, Ryan NS, et al. Genetic risk factors for the posterior cortical atrophy variant of Alzheimer’s disease. Alzheimers Dement. 2016;12(8):862–871. doi: 10.1016/j.jalz.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19(3):351–357. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53(3):1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ramirez A, van der Flier WM, Herold C, Ramonet D, Heilmann S, Lewczuk P, et al. SUCLG2 identified as both a determinator of CSF Abeta1-42 levels and an attenuator of cognitive decline in Alzheimer’s disease. Hum Mol Genet. 2014;23(24):6644–6658. doi: 10.1093/hmg/ddu372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10(9):e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. 439e431–432. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302(1):37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ligthart S, Vaez A, Hsu YH, Inflammation Working Group of the CC, Pmi Wg XCP, LifeLines Cohort S et al. Bivariate genome-wide association study identifies novel pleiotropic loci for lipids and inflammation. BMC Genomics. 2016;17:443. doi: 10.1186/s12864-016-2712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371(9611):483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010;30(11):2264–2276. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keller M, Schleinitz D, Forster J, Tonjes A, Bottcher Y, Fischer-Rosinsky A, et al. THOC5: a novel gene involved in HDL-cholesterol metabolism. J Lipid Res. 2013;54(11):3170–3176. doi: 10.1194/jlr.M039420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grallert H, Dupuis J, Bis JC, Dehghan A, Barbalic M, Baumert J, et al. Eight genetic loci associated with variation in lipoprotein-associated phospholipase A2 mass and activity and coronary heart disease: meta-analysis of genome-wide association studies from five community-based studies. Eur Heart J. 2012;33(2):238–251. doi: 10.1093/eurheartj/ehr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Suchindran S, Rivedal D, Guyton JR, Milledge T, Gao X, Benjamin A, et al. Genome-wide association study of Lp-PLA(2) activity and mass in the Framingham Heart Study. PLoS Genet. 2010;6(4):e1000928. doi: 10.1371/journal.pgen.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132(6–7):324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 137.Deelen J, Beekman M, Uh HW, Broer L, Ayers KL, Tan Q, et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum Mol Genet. 2014;23(16):4420–4432. doi: 10.1093/hmg/ddu139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Diabetes Genetics Initiative of Broad Institute of H, Mit LU, Novartis Institutes of BioMedical R. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 139.Debette S, Ibrahim Verbaas CA, Bressler J, Schuur M, Smith A, Bis JC, et al. Genome-wide studies of verbal declarative memory in nondemented older people: the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Biol Psychiatry. 2015;77(8):749–763. doi: 10.1016/j.biopsych.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kauwe JS, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL, et al. Genome-wide association study of CSF levels of 59 alzheimer’s disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet. 2014;10(10):e1004758. doi: 10.1371/journal.pgen.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Vries PS, Chasman DI, Sabater-Lleal M, Chen MH, Huffman JE, Steri M, et al. A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum Mol Genet. 2016;25(2):358–370. doi: 10.1093/hmg/ddv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu Y, Day FR, Gustafsson S, Buchkovich ML, Na J, Bataille V, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cook JP, Morris AP. Multi-ethnic genome-wide association study identifies novel locus for type 2 diabetes susceptibility. Eur J Hum Genet. 2016;24(8):1175–1180. doi: 10.1038/ejhg.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78(2):256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ridker PM, Pare G, Parker A, Zee RY, Danik JS, Buring JE, et al. Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am J Hum Genet. 2008;82(5):1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang C, Pierce BL. Genetic susceptibility to accelerated cognitive decline in the US Health and Retirement Study. Neurobiol Aging. 2014;35(6):1512e1511–1518. doi: 10.1016/j.neurobiolaging.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 147.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Felix JF, Bradfield JP, Monnereau C, van der Valk RJ, Stergiakouli E, Chesi A, et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet. 2016;25(2):389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43(10):1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.International Consortium for Blood Pressure Genome-Wide Association S. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johansson A, Eriksson N, Lindholm D, Varenhorst C, James S, Syvanen AC, et al. Genome-wide association and Mendelian randomization study of NT-proBNP in patients with acute coronary syndrome. Hum Mol Genet. 2016;25(7):1447–1456. doi: 10.1093/hmg/ddw012. [DOI] [PubMed] [Google Scholar]
- 152.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92(2):197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nasrallah HA, Meyer JM, Goff DC, McEvoy JP, Davis SM, Stroup TS, et al. Low rates of treatment for hypertension, dyslipidemia and diabetes in schizophrenia: data from the CATIE schizophrenia trial sample at baseline. Schizophr Res. 2006;86(1–3):15–22. doi: 10.1016/j.schres.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 154.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.de Leon J, Diaz FJ. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophr Res. 2005;76(2–3):135–157. doi: 10.1016/j.schres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 156.Bassett AR. Editing the genome of hiPSC with CRISPR/Cas9: disease models. Mamm Genome. 2017;28(7–8):348–364. doi: 10.1007/s00335-017-9684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. List of Phenotypes Included in the Review and Their Corresponding Reclassifications: A list of all the initial phenotypes (left column) and the 198 reclassified phenotypes into which some of them were transformed (corresponding cell in the right column). The reclassified phenotypes reflect the important themes in the psychiatric GWAS studies.
Supplementary Table 2a. Replicated GWAS Findings from Independent Publication-or SNP alleles found in more than one publication for the same or very similar reclassified phenotypes along with the number of studies reporting the SNP.
Supplementary Table 2b. Replicated GWAS Findings from the Same Publications: SNPs or SNP alleles found in both initial and replication samples in one paper studying a reclassified phenotype but not replicated across publications.
Supplementary Table 3. SNPs Associated with Multiple Phenotypes: A list of SNPs or SNP alleles with p-values of ≤2 × 10−8 replicated across phenotypes, including at least one significant association with a psychiatric phenotype; the reclassified phenotypes are included in place of the original phenotypes when applicable, and redundant or very similar non-reclassified phenotypes are summarized or omitted.


