SUMMARY
SF3B1 is recurrently mutated in chronic lymphocytic leukemia (CLL), but its role in the pathogenesis of CLL remain elusive. Here, we show that conditional expression of Sf3b1-K700E mutation in mouse B cells disrupts pre-mRNA splicing, alters cell development, and induces a state of cellular senescence. Combination with Atm deletion leads to the overcoming of cellular senescence and the development of CLL-like disease in elderly mice. These CLL-like cells show genome instability and dysregulation of multiple CLL-associated cellular processes, including deregulated B cell receptor (BCR) signaling, which we also identified in human CLL cases. Notably, human CLLs harboring SF3B1 mutations exhibit altered response to BTK inhibition. Our murine model of CLL thus provides insights into human CLL disease mechanisms and treatment.
Keywords: CLL, SF3B1, ATM, murine model, BCR signaling
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is a relatively indolent malignancy of mature CD19+ CD5+ B cells that is more prevalent in the elderly. Accumulating evidence from large-scale genetic characterizations of CLL has not only uncovered numerous recurrently somatically mutated genes in CLL (Landau et al., 2015; Puente et al., 2015; Wang et al., 2011) but has also suggested that CLL emerges from the stepwise acquisition of diverse mutations over time (Landau et al., 2015). Of these putative CLL drivers, thus far only the focal deletion of chromosome 13 [del(13q)] has been validated as a bona fide CLL driver based on the observation of the accumulation of pathognomonic clonal CD19+CD5+ cells in a mouse model harboring deletion of the miR-5/16 locus, contained within del(13q) in humans (Klein et al., 2010). Indeed, the functional effects of the vast majority of other individual CLL-associated events and how they cooperate together in the oncogenic process, as well as the minimum number of somatic events required to lead mature B cells towards a leukemic state, remain unknown.
SF3B1 is among the most frequently mutated genes in CLL. Recurrent mutations in SF3B1 commonly co-occur in CLL with focal deletion in chromosome 11 [del(11q)], a region that contains the essential DNA damage response gene ATM (Dohner et al., 2000). In CLL, SF3B1 mutation is often detected as a subclonal event, indicating that it tends to arise later in leukemic development and contribute to disease progression. Other lines of evidence, however, suggest that it can be also acquired early in the disease, as it has been implicated in clonal hematopoiesis (Jaiswal et al., 2014; Xie et al., 2014) and has been detected in the CLL precursor condition monoclonal B cell lymphocytosis (Ojha et al., 2014). To investigate the function of SF3B1 mutation, we established a conditional knock-in mouse model with B cell-restricted expression of Sf3b1-K700E mutation and examined how this mutation impacts the pathogenesis of CLL.
RESULTS
Conditional expression of heterozygous Sf3b1-K700E mutation in B cells leads to alternative RNA splicing
We generated a mouse model with conditional expression of the most commonly occurring SF3B1-K700E mutation in CLL from the endogenous Sf3b1 locus. To obtain B cell specific expression, the mouse line carrying the heterozygous Sf3b1-K700E allele (Sf3b1fl/+) was crossed with homozygous Cd19-Cre (Cd19-Cre+/+) mice, yielding both wild-type (WT, Cd19-Cre+/−Sf3b1+/+) and mutant mice (Sf3b1 MT, Cd19-Cre+/−Sf3b1fl/+) (Figure 1A). Although the floxed allele was present across tissues of the mouse (i.e. including T cells), the MT allele was specifically activated in CD19+ splenic B cells (Figure 1A). Targeted allele-specific quantitative RNA sequencing by pyrosequencing revealed that the MT allele was stably detected even in 24-month old mice and was expressed at a level similar to that of the WT allele (Figure 1B). Expression of the MT allele did not affect the SF3B1 protein expression in B cells, since both WT and Sf3b1MT B cells (as well as non-affected T cells from the same mice) expressed equivalent levels of SF3B1 protein (Figure 1C).
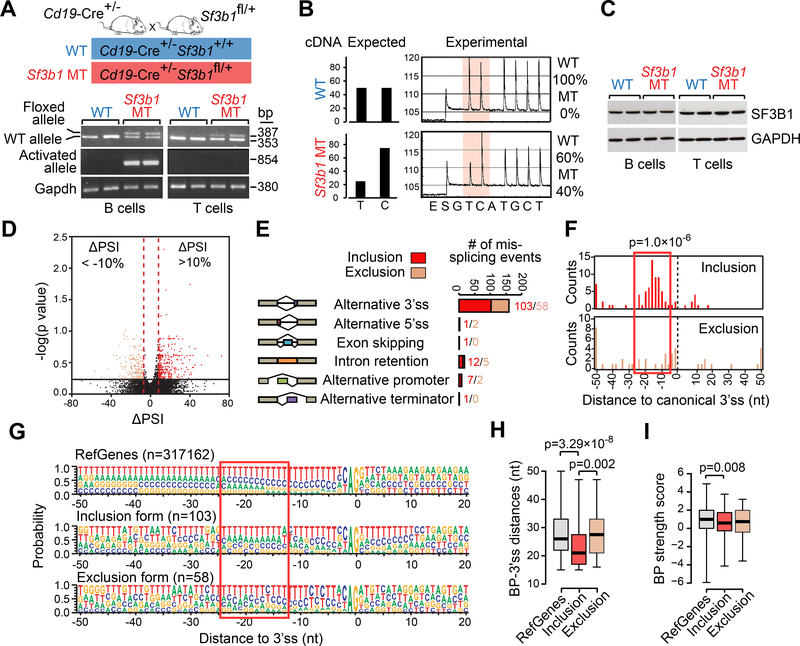
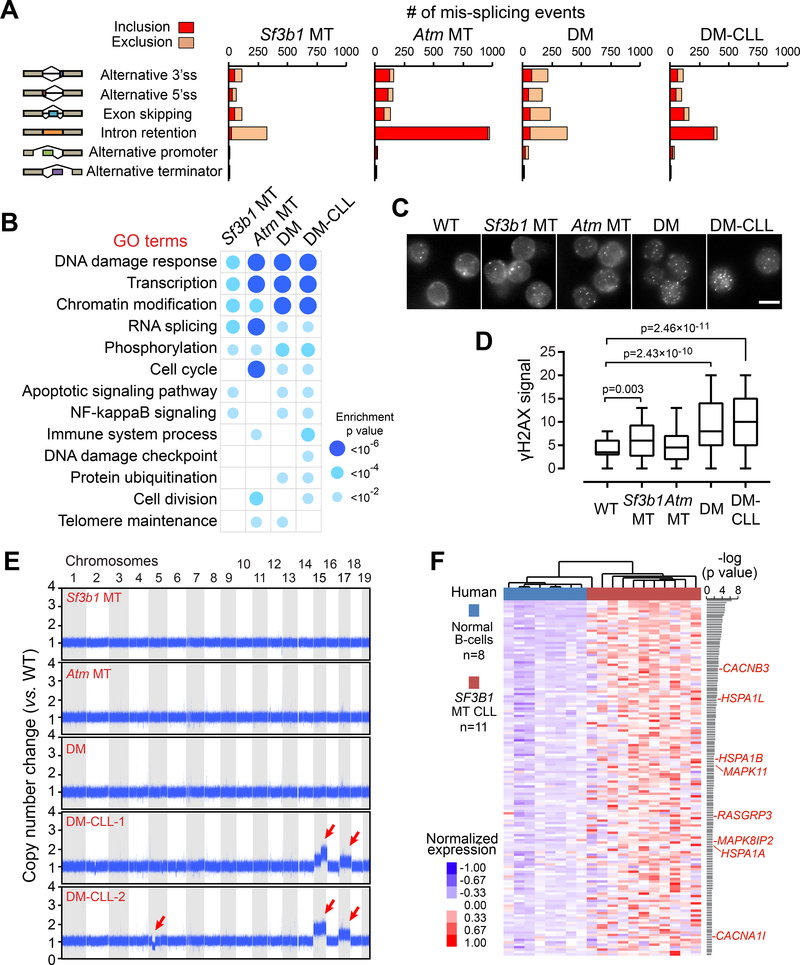
Figure 1. Conditional expression of Sf3b1-K700E in mouse B cells.
(A) PCR of genomic DNA from B and T cells from mice with WT or MT Sf3b1 to detect the floxed allele and the activated Sf3b1-K700E allele. (B) The percentages of WT or MT Sf3b1 alleles from pyrosequencing profiles in B cells are shown. (C) Western blot of SF3B1 in B cells and T cells with WT and MT Sf3b1 are shown. Two biological replicates are shown for each group. (D) Volcano plot shows ΔPSI versus log10 (p value) of all splicing changes identified by JuncBASE. Events with |ΔPSI|>10% and p<0.05 were considered significant. (E) Different categories of mis-splicing events in MT versus WT cells are shown. Events with ΔPSI>10% were defined as inclusion and events with ΔPSI<−10% were defined as exclusion in MT compared to WT cells. (F) Histogram shows the distance between the alternative and canonical 3’ss. The 0 point defines the position of the canonical 3’ss. (G) Sequence motifs around all RefGene 3’ss, MT inclusion 3’ss and MT exclusion 3’ss are shown. The height of each letter indicates the probability that nucleotide is used at that position. The red box highlights the region with heightened usage of adenosine upstream of the inclusion 3’ss. (H) The distance between the predicted branch point and the corresponding 3’ss are shown. The 0 point defines the position of the 3’ss. (I) The strength of the branch point associated with different groups of 3’ss are shown. In H and I, center lines show the means; box limits indicate the 25th and 75th percentiles and whiskers extend to minimum and maximum values. See also Figures S1 and S2, and Table S1.
We and others have previously reported that aberrant 3’ splice site (ss) selection is the predominant splicing defect associated with SF3B1 mutation (Alsafadi et al., 2016; Darman et al., 2015; Ferreira et al., 2014; Wang et al., 2016). In a re-analysis of RNA-sequencing (RNA-seq) data generated from samples from 37 CLL cases (Wang et al., 2016), we observed that the majority of alternative 3’ss events associated with MT SF3B1 were inclusion events, with preferential use of a cryptic 3’ss (‘inclusion alt 3’ss’, ΔPSI> 10%), rather than exclusion events (‘exclusion alt 3’ ss’, ΔPSI<−10%) (Figure S1A). The inclusion alt 3’ss were enriched within the 10 to 20 nucleotides (nt) upstream of the corresponding canonical 3’ss (Figure S1B). Heightened usage of adenosine was observed upstream of the inclusion alt 3’ss (Figure S1C), suggesting the presence of motifs within branch point (BP) sequences for the corresponding canonical 3’ss and/or within alternative BP sequences for the cryptic 3’ss.
RNA-seq analysis of splenic B cells derived from WT and Sf3b1 MT mice (n=3 each, 3-month of age) revealed altered RNA splicing in a manner similar to human CLL with SF3B1 mutation. Specifically, the majority (161 of 192 [84%]) of mis-spliced events in Sf3b1 MT mice (|ΔPSI|>10%, p<0.05, per the tool JuncBASE (Brooks et al., 2011)) involved alternative 3’ ss usage (Figure 1D-E and Table S1). Similar to human data, Sf3b1 MT B cells had more inclusion (n=103) rather than exclusion (n=58) alternative 3’ss events compared to WT cells (Figure 1E). Moreover, inclusion alt 3’ss tended to cluster 10 to 20 nucleotides upstream of the canonical 3’ss (Figure 1F), with high adenosine usage in its upstream motif (Figure 1G), as observed in human CLLs. For both human and mouse, the distance between 3’ss and its BP (predicted by BPfinder (Corvelo et al., 2010)) was shorter for inclusion alt 3’ss compared to both exclusion alt 3’ss and RefGene canonical 3’ss (Figure 1H and Figure S1D). Although sequences of the BPs were similar amongst different groups (Figure S1E), the strength of the BPs was consistently lower for inclusion alt 3’ss compared to canonical RefGene 3’ss (Figure S1F and Figure 1I). Thus, in line with prior studies (Alsafadi et al., 2016; Darman et al., 2015), mutated Sf3b1 promotes altered recognition of BPs compared to WT cells.
RNA-seq data confirmed that the MT allele of Sf3b1 is expressed at a similar level as the WT allele (Figure S2A). Using quantitative PCR, we validated the cryptic 3’ss detected by RNA-seq analysis in several genes (Figure S2B-C). Moreover, as shown in Figure S2D-E, we observed no discernible change in expression or subcellular localization of SF3B1 or other U2 snRNP components in Sf3b1 MT B cells, and the assembly of functional 17S and 12S U2 snRNP complexes was normal (Figure S2F). Together, these data suggest that we have successfully generated a mouse model with B cell-restrictive expression of Sf3b1-K700E mutation present at physiologic levels, which functions in a manner similar to human CLL.
Expression of Sf3b1-K700E causes defects of B cell function
To investigate the impact of Sf3b1-K700E expression on B cell development and function, we characterized the B cell compartments from the peripheral blood, marrow, peritoneal cavity and spleen of WT and Sf3b1 MT mice using immunophenotypic markers (Shay and Kang, 2013). As shown in Figure 2A, the total number of splenocytes was significantly reduced in Sf3b1 MT mice versus WT mice (12-month old) as a result of significant reduction in B cells but not in non-B cells (i.e. CD3+ T cells). Spleen weight was also decreased in Sf3b1 MT mice versus WT mice (Figure S3A), although the average overall body weight and the number of total bone marrow mononuclear cells (BMCs) were unaffected (Figure S3B-C). Moreover, as shown in Figure 2B, the percentages of splenic T2 and T3 transitional B cells were decreased in Sf3b1 MT mice versus WT mice. In contrast, both the percentage and absolute number of marginal zone (MZ) B cells were increased in Sf3b1 MT mice (Figure 2B-C). No significant difference in B cell subset composition between WT and Sf3b1 MT mice in bone marrow (BM) fractions (Figure S3D-F) or peritoneal cavity (Figure S3G-H) was observed. With up to 24-months of observation, the percentages of T, B, and macrophage subpopulations in peripheral blood remained similar between WT and Sf3b1 MT mice (Figure S3I), with no evidence of lymph node enlargement in Sf3b1 MT mice (n=35).
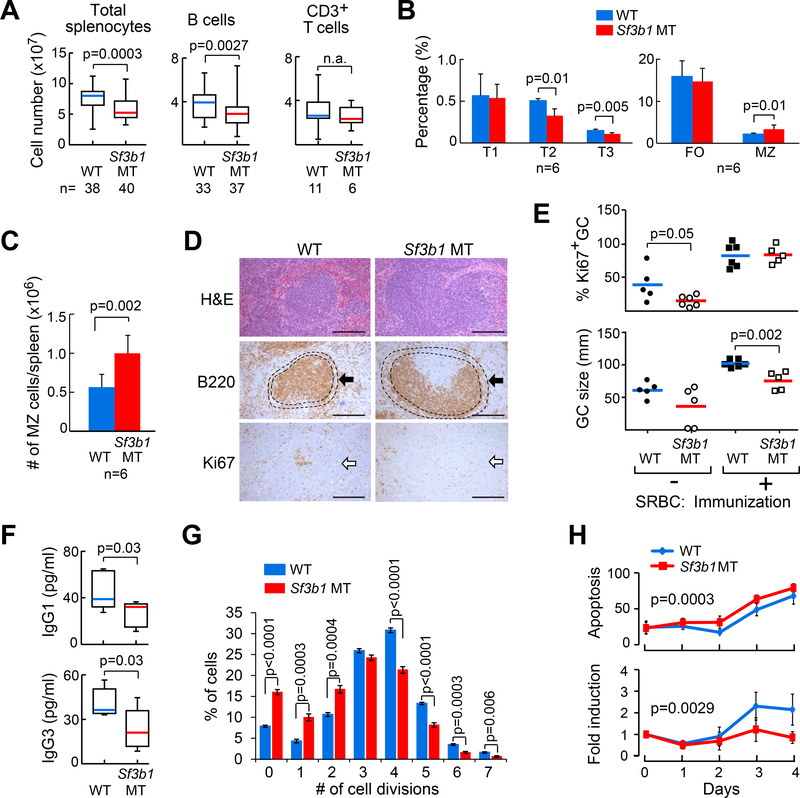
Figure 2. Expression of Sf3b1-K700E affects B cell proliferation in vitro and in vivo.
(A) The number of total splenocytes, splenic B cells and CD3+ T cells in WT and Sf3b1 MT mice are shown. Center lines indicate the means; box limits indicate the 25th and 75th percentiles; whiskers extend to minimum and maximum values. (B) The percentages of different B cell populations in the spleens of WT and Sf3b1 MT mice are shown. T1-T3: transitional B cells, FO: follicular B cells, MZ: marginal zone B cells. (C) The absolute counts of marginal zone B cells per spleen are shown. Average and standard deviation are plotted. For both B and C, data represent Mean±SD of results derived from 6 WT and 6 MT mice, and were analyzed using Student’s t-test. (D) Hematoxylin and Eosin (HE) staining and immunohistochemical staining of spleen sections from WT and MT mice are shown. Arrow indicates marginal zone stained with an α-B220 antibody. Proliferative germinal centers were stained with an α-Ki67 antibody. Scale bar: 250 μm. (E) The percentage and size of germinal centers identified by Ki67 staining in WT and Sf3b1 MT mice are shown. Center lines indicate the means. (F) Abundance of serum IgG3 and IgG1 antibodies in WT (n=9) and Sf3b1 MT (n=7) mice are shown. Center lines indicate the means; box limits indicate the 25th and 75th percentiles; whiskers extend to minimum and maximum values. (G) WT and Sf3b1 MT cells were stimulated with LPS+IL4 for 3 days in vitro and multiple generations were traced using CellTrace violet dye dilution and flow cytometry. Data represent Mean±SD of results derived from 5 WT and 5 MT mice, and were analyzed using ANOVA. (H) Apoptosis and proliferation of cells described in (G) are shown. Apoptosis rates were determined by Annexin V staining and flow cytometry. Data represent Mean±SD of results derived from 6 WT and 7 MT mice. See also Figure S3.
Histological examination of spleen sections revealed consistently enlarged marginal zone B cell regions in Sf3b1 MT mice (Figure 2D). Moreover, both the percentage and size of Ki67+-germinal centers (GC) within the spleens of the Sf3b1 MT mice were reduced compared to WT mice (p=0.002; Figure 2E). With stimulation by immunization with sheep red blood cells (sRBCs), the sizes of the Ki67+ germinal centers were much smaller in Sf3b1 MT mice than in WT mice (Figure 2E, p=0.002), demonstrating an intrinsic defect in B cell proliferation. Consistent with this phenotype, Sf3b1 MT mice demonstrated reduced serum isotype-specific antibody production (all p=0.03, Figure 2F), indicating defects in B cell class switch recombination within the GC in these animals. To determine whether expression of Sf3b1-K700E affects the proliferative capacity of B cells, we stimulated splenic B cells with Lipopolysaccharide (LPS) and IL4 in vitro and examined cell proliferation and apoptosis over 4 days. We found that Sf3b1 MT B cells underwent fewer cell divisions than WT B cells (Figure 2G), and displayed a higher rate of apoptosis (p=0.0003) and proliferation inhibition (p=0.0029, Figure 2H).
Expression of Sf3b1-K700E causes B cell cellular senescence which is overcome by Atm deletion
To investigate whether expression of Sf3b1-K700E directly induces CLL development, we routinely examined peripheral blood of Sf3b1 MT animals for B220+CD5+ CLL cells starting from 6 months of age by flow cytometry. With up to 24-month of observation, the overall survival of the WT (n=35) and Sf3b1 MT (n=40) mice was similar, with no evidence of circulating B220+CD5+ cells in animals (n=40), suggesting that Sf3b1-K700E alone is insufficient to induce CLL.
Since we observed a phenotype of defective cell growth and absence of leukemia development in association with this putative cancer driver alone, we examined whether MT Sf3b1 was linked to cellular senescence, a condition associated with premalignant states (Campisi and d’Adda di Fagagna, 2007). Western blot analysis revealed enhanced levels of senescence markers p16 and p21 in Sf3b1 MT B cells versus WT cells (Figure S3J). Furthermore, elevated levels of the cellular senescence mediators IGFBP6 and IGFBP7 was also detected in the sera of Sf3b1 MT mice (p=0.02 and p=0.04, Figure S3K). Together, these results suggested that Sf3b1-K700E mutation induces cellular senescence and defects in B cells proliferation, but was insufficient to cause CLL, indicating that additional genetic lesions might be required for CLL initiation.
SF3B1 mutations in human CLL cells commonly co-occur with del(11q) (Landau et al., 2015; Nadeu et al., 2016; Wang et al., 2011) (Figure S4), which encompasses ATM. Previous studies had suggested that ATM plays a central role in regulating replication stress and cellular senescence through modulation of mitochondrial function and metabolic processes, and knockdown or inhibition of ATM has been shown to overcome cellular senescence (Aird et al., 2015; Kang et al., 2017). We therefore hypothesized that loss of ATM could lead to the overcoming of the cellular senescence phenotype induced by mutated SF3B1, and thereby synergize with MT SF3B1 for the development of CLL-like disease. We crossed Sf3b1fl/+Atmfl/fl (Zha et al., 2008) and Cd19-Cre+/+ animals to establish mouse lines with only heterozygous Atm deletion (Atm MT, Cd19-Cre+/−Sf3b1+/+Atmfl/+) or with both heterozygous Sf3b1 mutation and Atm deletion (double mutant, or ‘DM’ mice, Cd19-Cre+/−Sf3b1fl/+Atmfl/+). Indeed, we found potent downregulation of senescence markers p16 and p21 in DM mice relative to Sf3b1 MT mice (Figure 3A). Moreover, Atm deletion bypassed the proliferation inhibition induced by Sf3b1 mutation in DM mice (Figure 3B).
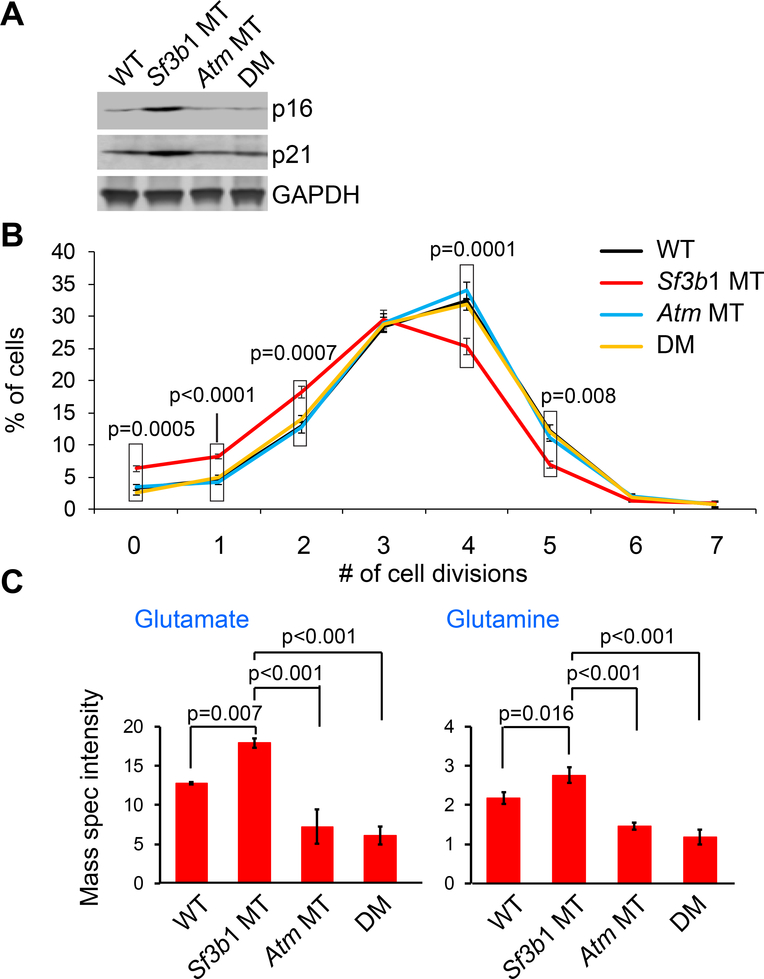
Figure 3. Atm deletion overcomes Sf3b1 mutation induced-cellular senescence.
(A) Western blot of senescence markers p16 and p21 in B cells from the different groups of mice are shown. (B) The percentage of B cells undergoing different numbers of cell divisions upon stimulation with LPS+IL4 were shown. Data represent Mean±SD of three replicates. The p values indicate the difference between Sf3b1 MT and other groups in each division point, using ANOVA. (C) Abundance of glutamate and glutamine in splenic B cells from the different mice groups are shown. Data represent Mean±SD of mass spectrometry intensities of three replicates and were analyzed using a one-way ANOVA with the Scheffe correction for multiplicity of testing. See also Figures S4.
The accumulation of glutamine and glutamate is a hallmark of cellular senescence in mouse models (Liu et al., 2008). Conversely, the overcoming cellular senescence by Atm deletion was previously noted to involve enhanced glutamine and glutamate consumption (Aird et al., 2015), which provides important nitrogen and carbon sources for an array of growth-promoting pathways. Consistent with these studies, we found marked accumulation of both glutamate and glutamine in Sf3b1 MT B cells compared to WT B cells (p=0.007 and p=0.016, respectively; Figure 3C), indicating reduced glutamine and glutamate consumption. In contrast, Atm deletion resulted in reduced glutamate and glutamine levels in DM cells (both p<0.001). These results suggest that Atm deletion restores a metabolic profile and overcomes cellular senescence.
Development of CLL with combined expression of Sf3b1-K700E and Atm deletion
To investigate whether Sf3b1 mutation in combination with Atm deletion could lead to CLL-like disease, we established 2 additional cohorts of mice (i.e. 31 Atm MT and 33 DM mice) and monitored their peripheral blood for the appearance of B220+CD5+ cells for up to 24 months of age. Starting from 18 months of age, we detected the appearance of B220+CD5+ CLL-like cells within peripheral blood mononuclear cells (PBMCs), splenocytes and bone marrow mononuclear cells (BMCs) in 3 of 33 (9%) DM mice (‘DM-CLL’ mice) (Figure 4A-B). In these mice, the percentage of circulating B220+CD5+ cells rose to 30–50% by 24 months of age.
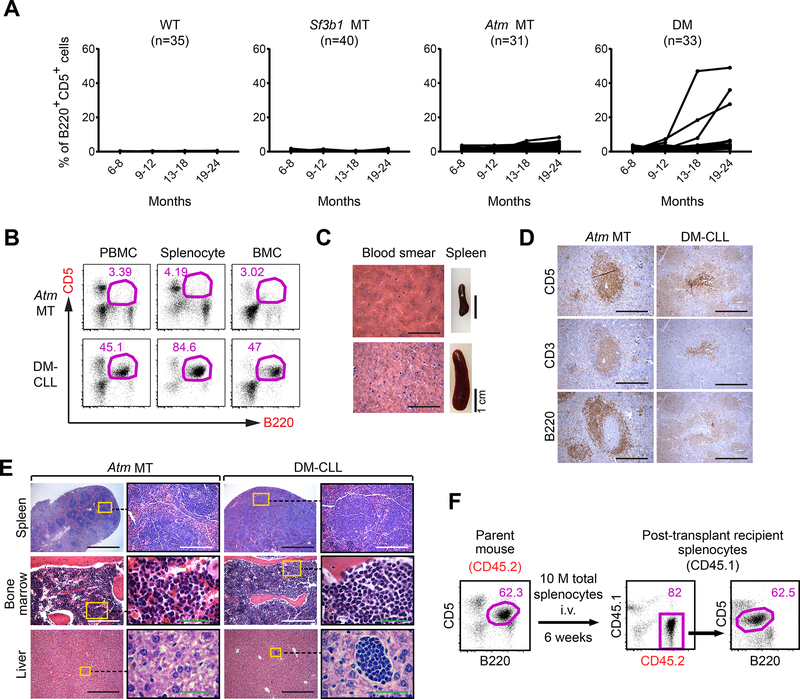
Figure 4. Combined Sf3b1 mutation and Atm deletion cause CLL.
(A) The percentage of CLL-like cells (B220+CD5+) within the lymphocyte population in the different groups of mice over time are shown. (B) Flow cytometry data identified B220+CD5+ cells within peripheral blood mononuclear cells (PBMCs), splenocytes and bone marrow mononuclear cells (BMCs) of Atm MT and DM-CLL mice. (C) H&E staining of blood smears and spleen of Atm MT and DM-CLL mice are shown. Scale bar for blood smear images:1 mm. (D) Immunohistochemical staining show CD5, CD3 and B220 signals on the consective sections of spleens from Atm MT and DM-CLL mice. Scale bar: 250 μm. (E) H&E staining shows leukemia cells infiltration in the spleen, bone marrow and liver tissues in the Atm MT and DM-CLL mice. Black scale bar: 1 mm; White scale bar: 250 μm; Green scale bar: 50 μm. (F) 10 million total splenocytes and bone marrow cells from a DM-CLL mouse were engrafted into either immunodeficient (NSG) or immunocompetant (CD45.1) mice 6 weeks after the engraftment. mice were sacrificed and B220+CD5+ in the spleen were detected by flow cytometry. See also Figure S5 and Table S2.
We performed a detailed histopathologic analysis on 2 of 3 DM-CLL animals. H&E staining of peripheral blood smears of the DM-CLL mice confirmed the presence of cells with morphology highly similar to leukemia, and their spleens were also enlarged (Figure 4C). Immunohistochemistry analysis demonstrated the presence of B220+CD5+ cells on spleen sections of DM-CLL mice (Figure 4D), while H&E staining showed infiltration of CLL-like cells in multiple tissues including spleen, bone marrow and liver (Figure 4E). We confirmed that the CLL-like cells were clonal based on Igκ expression and immunoglobulin heavy chain gene variable region (IGHV) mutational status (Table S2, all CLL-like cells were IGHV-unmutated).
We furthermore examined the transplantability of the DM-CLL cells in both immunocompetent (CD45.1) and immunodeficient (NSG) mice. Transplanted CLL were detectable in peripheral blood of all recipient animals (n=30 each) 4–6 weeks following engraftment (Figure 4F and Figure S5A), with leukemic cell infiltration in multiple tissues (Figure S5B). Examination of multiple lineage-specific markers during serial transplantation revealed preservation of the phenotype of the engrafted CLL-like cells compared to parental cells in immunodeficient recipient mice. Within the immunocompetent recipients, however, slight subclonal changes of the engrafted cells were observed (i.e. enhanced expression of Igκ), indicating the effects of immune-based selective pressure on the leukemia cells within these mice (Figure S5A). Consistent with human CLL, the engrafted DM-CLL cells were sensitive to the BCR signaling inhibitor ibrutinib, suggesting preserved signaling circuitry in these animals (Figure S5C). Our transplanted model thus provides opportunities not only for further dissection of disease mechanisms but also for the testing of therapeutic targets.
Sf3b1-K700E and heterozygous Atm deletion are associated with increased DNA damage
To understand the molecular mechanisms underlying leukemogenesis in relationship to Sf3b1 and Atm mutations, we performed RNA-seq analysis of splenic B cells derived from WT, Sf3b1 MT, Atm MT, and DM mice with (DM-CLL) or without CLL-like changes. All samples were collected from aged mice (18 months), with 2–6 samples per group. As shown in Table S3, we observed that the aged Sf3b1 MT mice accumulated a larger number of mis-spliced events other than alternative 3’ss usage, including markedly reduced intron retention events, compared to the young (3-month old) animals (Figure 1D, Figure 5A). This profile was highly concordant with that observed in human CLL samples (Figure S1A). In contrast, over 70% of the mis-spliced events associated with heterozygous deletion of Atm involved increased intron retention (Figure 5A). A combined effect of the two mutations on splicing was observed, as DM and DM-CLL cells showed a hybrid of the mis-spliced profiles of the two alterations (Figure 5A and Figure S6A).
Figure 5. Sf3b1 mutation and Atm deletion are associated with increased DNA damage.
(A) The number of alternative splicing events significantly included or excluded in MT groups versus WT are shown. (B) Top Gene Ontology categories significantly enriched for mis-spliced transcripts in different MT versus WT groups are shown. (C) Representative data of γH2AX immunofluoresent staining in splenic B cells obtained from WT and different MT mice. Scale bars, 5 μm. (D) γH2AX signal intensities in WT and different MT mice. 70–91 cells were measured for each group. Center lines show the medians, box limits indicate the 25th and 75th percentiles and whiskers extend to minimum and maximum values. (E) Chromosomal copy number variants are detected using whole-genome sequencing. The copy number ratio to WT mice are shown. Red arrows indicate chromosome amplification and deletions. (F) Heatmap showing the normalized gene expression of 146 human genes (homologs of mouse Chr 15 and 17 genes) significantly upregulated (p<0.05) in 11 human CLL with SF3B1 mutations versus 8 normal B cells. Genes involved in MAPK/ERK signaling are labeled red. See also Figure S6 and Tables S3–S5.
Gene Ontology (GO) analysis of the mis-spliced genes revealed high enrichment for the DNA damage response pathway in all groups (all p<4.0×10−6, Figure 5B), consistent with the notion that deregulation of DNA damage repair pathways is a common mechanism for malignant transformation (Di Micco et al., 2006). Using RT-PCR, we validated splicing defects of several DNA damage response genes in DM-CLL cells (Figure S6B). As a defective DNA damage response could lead to accumulation of mutations and genomic instability, we evaluated the mouse lines for formation of DNA double-strand breaks (DSBs) in splenic B cells by immunofluorescence staining of H2AX phosphorylation (γH2AX) (Figure 5C). Consistent with previous findings in human CLLs with SF3B1 mutations (Te Raa et al., 2015; Wang et al., 2016), significantly higher levels of γH2AX signal were observed in Sf3b1 MT mice compared to WT mice (Figure 5C-D), and even higher levels of γH2AX signals were detected in DM and DM-CLL cells compared to WT cells (Figure 5C-D), indicative of a synergistic effect of Sf3b1 mutation and Atm deletion in inducing DNA damage.
The observed increased DNA damage and long latency of appearance of leukemic cells led us to query whether DM-CLL cells carried additional genetic alterations compared to non-CLL DM cells. We therefore performed whole-genome sequencing (WGS) analysis of splenic B cell DNA isolated from the various mouse lines (n=2 per line). By analysis of single nucleotide variants (SNVs), we identified 169 and 159 clonal mutations in the two DM-CLLs (Figure S6C, Table S4), with only 0–2 nonsilent mutations in genes unlikely to be drivers of CLL. By analysis of copy number variants (CNVs), however, we observed amplification of chromosomes (Chr) 15 and 17 in both DM-CLLs, and a deletion on Chr 5 in only DM-CLL2 (Figure 5E). We further performed WGS analysis of splenocytes transplanted from DM-CLL2 cells into NGS recipient mice. The engrafted cells were found to preserve 79% (125/159) of the SNVs (Figure S6D) and all CNVs of the original CLL-like cells (Figure S6E), indicating the overall fidelity of the transplanted model to the original DM-CLL cells.
RNA-seq analysis revealed that the amplifications were associated with overexpression of 835 of 987 (85%) Chr15 and Chr17 genes detected in DM-CLL cells versus DM cells (Figure S6F). Of note, 146 genes that are the human homologs of the mouse Chr15 and Chr17 genes were overexpressed in human CLLs with SF3B1 mutations compared to normal B cells (p<0.05, Figure 5F and Table S5). Gene Ontology analysis revealed that these upregulated genes in human CLLs were most significantly enriched in the MAPK/ERK signaling pathway (p=0.0027, Figure 5F-red genes), which has been previously implicated in driving CLL development (Landau et al., 2015). Altogether, these results established enhanced genome instability leading to chromosomal amplification as a potential mechanism underlying the generation of leukemia.
Integrated transcriptome and proteome analysis of murine CLL-like cells implicate dysregulated BCR signaling
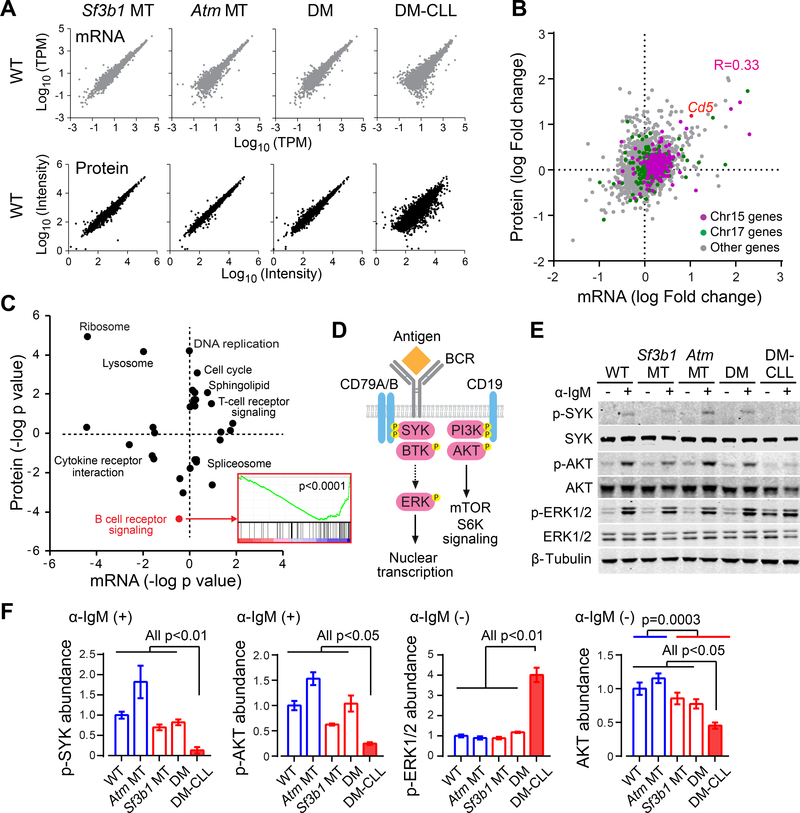
In parallel with the RNA-seq analysis, we performed a quantitative proteomic analysis of splenic B cells from different mouse lines (n=2 each), to identify mRNA- and protein-level changes that putatively contribute to CLL development. Globally, both mRNA (n=9,174) and protein (n=7,096) expression in the Sf3b1 MT, Atm MT and DM mice were highly concordant with that of WT mice, while more extensive changes were observed in DM-CLL mice (Figure 6A, Figure S6G and Table S6). Focusing on the comparison between DM and DM-CLL cells, we observed a moderate correlation between the fold changes in mRNA and protein expression (Figure 6B, Pearson correlation coefficient R=0.33). As an example, both CD5 mRNA and protein levels were markedly elevated in DM-CLL cells (Figure 6B). Consistent with the copy number changes, Chr15 and Chr17 genes showed consistently enhanced mRNA and protein expression in DM-CLL cells versus DM cells, suggesting a strong effect of chromosome amplification on gene expression (Figure 6B).
Figure 6. Integrative transcriptome and proteomics analyses of CLL cells.
(A) Correlations in mRNA and protein levels between different MT and WT cells are shown. Expression values were log transformed. Abundance of mRNA was measured by TPM (Transcripts Per Kilobase Million) and protein abundance was measured by Tandem mass tag (TMT) signal intensity. (B) Correlation between changes in mRNA expression and changes in protein expression between DM-CLL and DM are shown. DM-CLL/DM fold changes were log transformed, with positive values indicating upregulation and negative values indicating downregulation in DM-CLL versus DM. (C) GSEA analysis of KEGG pathways enriched for differentially expressed genes/proteins between DM-CLL and DM. KEGG pathways were plotted by the log transformed enrichment p values, with upregulated or downregulated pathways indicated by positive or negative values, respectively. GSEA enrichment plot for BCR signaling genes is shown. (D) A schematic shows a simplified overview of the BCR signaling pathway. (E) Western blot of BCR signaling components in non-CLL or CLL cells either untreated or treated with 20 μg/ml α-IgM for 5 min. (F) Quantification of the western blot results. Abundances of p-SYK and p-AKT upon α-IgM stimuli, as well as of p-ERK1/2 and total AKT without α-IgM stimuli, are shown. Data represent Mean±SD of three independent experiments. See also Figure S6.
Gene Set Enrichment Analysis (GSEA) of differentially expressed genes/proteins associated with DM-CLL versus DM B cells identified proteins involved in B cell receptor (BCR) signaling as the most dramatically downregulated in DM-CLL cells (p<0.0001, Figure 6C). BCR signaling is highly active in CLL cells, but variation in the activity of this pathway has been identified (Burger and Wiestner, 2018; Kipps et al., 2017). Our proteome observations led us to investigate the basal and induced BCR signaling activity in the different mice lines. As shown in Figure 6D-E, upon α-IgM stimuli, splenic B cells from non-CLL mice exhibited potent increases in the phosphorylation (p-) of SYK, AKT and ERK1/2 without affecting the overall protein levels, indicating robust ligand-dependent BCR signaling transduction in these cells. In striking contrast, however, DM-CLL cells were defective in initiating SYK and AKT phosphorylation upon stimulation (all p<0.05, Figure 6E-F). In addition, different from non-CLL cells, DM-CLL cells showed constitutive phosphorylation of ERK1/2 even without α-IgM stimuli (all p<0.01, Figure 6E-F), a phenotype previously described in self-reactive anergic B cells and subset of human CLLs (Blix et al., 2012; Duhren-von Minden et al., 2012; Herve et al., 2005; Marklin et al., 2017; Muzio et al., 2008; Yarkoni et al., 2010). Furthermore, we noted that DM-CLL cells had lower basal expression of AKT protein than the non-CLL mouse lines (Figure 6E-F, all p<0.05). Indeed, B cells from mice with Sf3b1 mutation (Sf3b1 MT, DM and DM-CLL) had lower AKT expression than those without Sf3b1 mutation (WT and Atm MT) (Figure 6F, p=0.0003).
Distinct patterns of BCR signaling in CLL cells with SF3B1 mutation
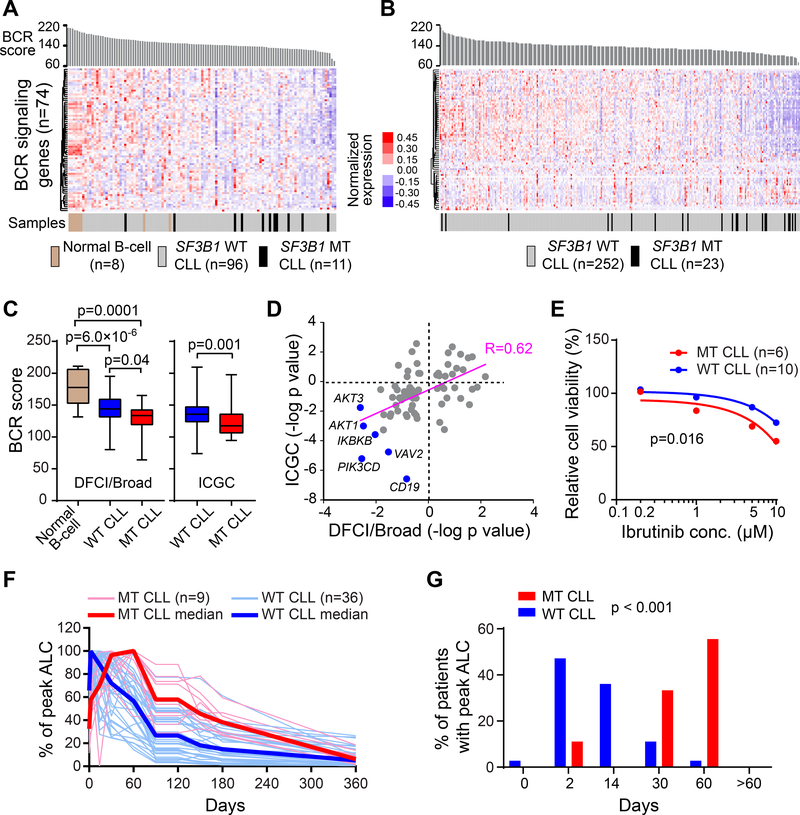
To further investigate these findings in human CLLs, we analyzed expression of BCR signaling genes (n=74, consisting of core components and modulators of BCR signaling, curated from MSigDB gene sets) using RNA-seq data of two independent human CLL cohorts. The DFCI/Broad cohort (Landau et al., 2014) contains 107 CLL cases (96 SF3B1 WT and 11 SF3B1 MT, all previously untreated) and 8 normal CD19+ B cell samples (Figure 7A and Table S7), while the ICGC cohort (Ferreira et al., 2014) contains 275 CLL cases (252 SF3B1 WT and 23 SF3B1 MT CLLs) (Figure 7B and Table S7). As shown in Figure 7A-C, ranking samples by ‘BCR score’ (mean expression of all 74 BCR signaling genes), normal B cells generally had higher BCR scores than CLL cells, while SF3B1 MT CLLs showed even lower expression of BCR signaling genes than SF3B1 WT CLLs, supporting a role of SF3B1 mutation in altering BCR signaling gene expression. Similar results were observed when we limited the analysis to 48 core components (Table S7) of BCR signaling pathway in both cohorts (Figure S7A). Of note, the two cohorts were highly correlated for changes in BCR signaling gene expression (Figure 7D, Pearson correlation coefficient R=0.62). In line with the observations in mouse DM-CLL cells (Figure 6E-F), AKT1 and AKT3 were among the consistently downregulated BCR signaling genes in human SF3B1 MT CLLs versus WT CLLs, together with their upstream regulators CD19 and PIK3CD (Figure 7D).
Figure 7. Distinct patterns of BCR signaling in CLL cells with SF3B1 mutation.
(A and B) Heatmap of the expression of BCR signaling pathway genes in DFCI/Broad cohort (n=115) (A) and ICGC cohort (n=275) (B). Samples were ranked by BCR scores, which indicate the mean TPM of 74 BCR signaling genes within individual samples. (C) Statistics of the BCR scores in different groups are shown. Center lines indicate the means; box limits indicate the 25th and 75th percentiles; whiskers extend to minimum and maximum values. (D) Correlation between changes in the expression of BCR signaling pathway genes in the two CLL patient cohorts. Positive log-transformed p values indicate upregulation and negative values indicate downregulation in SF3B1 MT compared to WT CLLs. (E) 6 SF3B1 MT and 10 WT human CLLs treated with different doses of ibrutinib for 48 hr in vitro, and the relative cell viability compared to the DMSO control group were calculated. The mean percentage viability of all SF3B1 WT or MT CLL samples upon ibrutinib treatment is shown. (F) Changes in absolute lymphocyte count (ALC) overtime in 36 cases and 9 cases with WT and MT SF3B1, respectively, CLL treated with ibrutinib. ALC was measured by 2, 14 days and 1, 2, 3, 4, 5, 6 and 12 months on ibrutinib and was presented as percentage of the peak ALC for individual cases (thin red lines, MT CLLs; thin blue lines, WT CLLs). Red line - Median values of all MT CLLs; blue line – median value of all WT CLLs. (G) The percentage of cases reaching peak circulating ALC at each time point. All cases reached their peak by 60 days; p value was calculated using the Kruskal-Wallis test. See also Figure S7 and Table S7
Targeted inhibitors of BCR signaling have transformed the therapeutic landscape of CLL (Byrd et al., 2013). Given the intrinsic differences in BCR signaling gene expression between SF3B1 WT and MT CLLs, we asked if these two patient groups respond differently to BCR signaling inhibition. To this end, we exposed SF3B1 WT CLL patient samples (n=10) and SF3B1 MT CLL patient samples (n=6, MT allele frequency 33–55%) to different concentrations of ibrutinib in vitro. Cell viability relative to DMSO control was calculated for each group (Figure S7B). Consistent with downregulation of BCR signaling genes in SF3B1 MT CLLs and hence a shorter ‘distance’ to B cell death when exposed to abrogation of BCR signaling (Muschen, 2018), we detected SF3B1 MT CLL samples to be more sensitive to ibrutinib treatment than SF3B1 WT CLLs in vitro (p=0.016, Figure 7E, Figure S7B). We further examined data from a genomically characterized subset of CLL cases (45 of 86 [52%] cases; 36 SF3B1 WT, 9 SF3B1 MT) enrolled on a clinical trial of ibrutinib monotherapy (NCT01500733) (Herman et al., 2014; Landau et al., 2017). Treatment with ibrutinib is known to commonly lead to early transient lymphocytosis (Herman et al., 2014; Wiestner, 2016; Woyach et al., 2014). We observed a delayed onset of peak absolute lymphocyte count (ALC) in cases with SF3B1 MT (p<0.001, Figure 7F-G), suggesting altered kinetics of disease response. The observed suppressed BCR signaling gene expression in SF3B1 MT CLLs, together with heightened in vitro sensitivity to the BCR signaling inhibitor and altered in vivo drug response kinetics, indicate an altered circuitry of this pathway in the presence of SF3B1 mutation. Together, these data revealed a role of SF3B1 mutation in regulating BCR signaling and ibrutinib response kinetics, highlighting our animal model as powerful tool for cancer research and drug discovery.
DISCUSSION
Here we report that the combined B cell restricted expression of heterozygous Sf3b1 mutation and Atm deletion can generate clonal expansions of mature B cells consistent with clinical pathologic features of CLL. Our results are consistent with a growing literature supporting the idea that cooperative events are needed for leukemogenesis in order to overcome key checkpoints in the clonal expansion of malignant cells. Our finding that SF3B1 mutation is associated with cellular senescence provides mechanistic understanding of how this lesion by itself may not lead to leukemia, and yet provides numerous adaptive changes in gene expression that are impactful on essential cellular pathways. Thus, our results point to the idea that SF3B1 mutation creates a reservoir of cells with numerous alterations ‘in-waiting’ that do not proliferate until the occurrence of some additional event that overcomes a cancer generating checkpoint. These data further support the idea that SF3B1 mutation is not a driver in the classical sense (i.e. absence of generation of leukemia by itself) but functions to efficiently execute numerous changes that are pro-oncogenic. In our study, that cooperating event was Atm deletion. We established that this alteration alone could override cellular senescence and could also alter the RNA splice isoform landscape of the cell. Indeed, previous studies have suggested that overcoming oncogene-induced senescence through regulation of the DNA damage repair pathway (e.g., ATM, CHK2, and TP53) is a common mechanism for malignant transformation (Bartkova et al., 2006). Altogether, our mouse model supports the notion that combination of Sf3b1 mutation and Atm deletion increases genomic instability, selecting for alterations that confer heightened cellular fitness e.g. proliferative effect (MAPK/ERK pathway) and overcome separate cancer checkpoints.
Our integrated transcriptome and proteomic analyses highlighted the numerous altered pathways associated with onset of malignancy, including the BCR signaling pathway. Prior studies have noted human CLL to have a more anergic BCR signaling signature compared to normal B cells, a circuitry that was exactly reflected in our DM-CLL mouse lines, and accentuated in the mouse lines with SF3B1 mutation. Translating forward from the mouse characterizations, we noted that the same deregulated circuitry in SF3B1-mutated animals was also evident in two independent human cohorts and was reflected by altered sensitivity of human CLL cells with SF3B1 mutation to BCR-targeting inhibitors in vitro and in vivo. These findings may not be surprising, given that alternative splicing of CD79b has been implicated in altered BCR expression in CLL (Alfarano et al., 1999). Indeed, analysis of our mouse lines revealed altered-splicing of transcripts from 12 genes within the BCR signaling pathway associated with Sf3b1 mutation or Atm deletion alone or combination, including Cd22, Chuk, Cr2, Ikbkb, Inpp5d, Malt1, Pik3cd, Pik3r1, Pik3r2, Pik3r5, Syk and Vav2. Our results support a ‘goldilocks’ model of BCR signaling and B cell survival, which states that B cell survival is dependent on the tuning of BCR signaling such that it is neither overly strong nor overly weak (Muschen, 2018). Within this context, cells with inherent decreased BCR signaling would be anticipated to be more sensitive to a BTK inhibitor like ibrutinib, as these cells are already closer to the minimum threshold of BCR signaling needed for cell survival. In keeping with this line of reasoning, the ‘distance’ to cell death afforded by abrogation of BCR signaling via a targeted BTK inhibitor is shorter when the target cells already have dampened BCR signaling, as we detected in the CLL samples with mutated SF3B1, and thereby would be more sensitive the drug. Targeting this so-called autoimmunity checkpoint has been suggested to be an attractive approach to eradicate B cell malignancies (Muschen, 2018).
Our study thus opens the possibility of using human genetic data to model subvariants of CLL, from which we anticipate opportunities for discovery of mechanistic biology and for studying the impact of diverse therapeutics. This approach is increasingly important in the age of precision medicine, which will require detailed mappings of disease sub-classifications with therapeutic vulnerabilities.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lili Wang (lilwang@coh.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animals were housed at Dana-Farber Cancer Institute (DFCI). All animal procedures were completed in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at DFCI. Sf3b1-K700E floxed mice (C57BL/6J × 129 hybrids) were generated as previously described (Obeng et al., 2016). Atm floxed mice (C57BL/6J × 129 hybrids) (Zha et al., 2008) were provided by Dr. Fred Alt (Children’s Hospital, Boston). The Sf3b1-K700E floxed mice and Atm floxed were bred with C57BL/6 mice for more than 7 generations before crossed with homozygous Cd19-Cre mice (Cd19-Cre+/+) to yield both WT (Cd19-Cre+/−Sf3b1+/+) and heterozygous Sf3b1 MT mice (Sf3b1 MT, Cd19-Cre+/−Sf3b1fl/+). Similarly, to obtain heterozygous expression of Sf3b1 mutation and Atm deletion in B cells, we crossed Sf3b1-K700E floxed mice with Atm floxed mice to generate Sf3b1fl/+Atmfl/fl mice, which were then crossed with CD19Cre (Cd19-Cre+/+) mice to obtain Cd19-Cre+/−Sf3b1fl/+Atmfl/+ (Atm MT) and Cd19-Cre+/−Sf3b1fl/+Atmfl/+ (Sf3b1 and Atm double mutant, DM) mice.
Primary Human CLL Samples
Studies were approved by the Institutional Review Boards of DFCI and conducted in accordance to the Declaration of Helsinki protocol. All primary CLL samples were obtained from CLL Research Consortium. CLL cases provided samples after their informed consent and primary human de-identified CLL samples derived from peripheral blood mononuclear cells were used.
METHOD DETAILS
Disease blood monitoring in mouse cohorts
100 μl of blood was collected from mice via submandibular bleeds into EDTA-containing tubes to prevent clotting. Erythrocyte lysis was carried out by incubating the blood in 1 ml of ACK buffer for 5 min and then washing with PBS supplemented with 2% FCS and 2 mM EDTA. The cells were then stained with a cocktail of antibodies against CD5, B220, CD11b, CD3 and IgKappa for 15 min at 4°C. Cells were then washed and analyzed using flow cytometry.
Cell subpopulation analysis
To analyze specific cell surface markers, erythrocyte lysed single cell suspensions prepared from mouse spleen, bone marrow, and peritoneal cavity were washed with PBS supplemented with 2% FCS and 2 mM EDTA and then incubated with antibodies for 15 min at 4°C. The marginal B cell population: CD93, CD23, B220, CD21, IgD and IgM (PE-Cy7). Hardy fraction in the bone marrow cells: IgM, ly-51, B220, CD43, IgD and CD24. Peritoneal cavity cells: CD5, B220 and CD11b.
Culture and analysis of splenic B cells
Mice were euthanized in a CO2 chamber and the harvested spleens were mechanically dissociated between frosted slides to form a single cell suspension. Erythrocyte lysis was carried out using ACK buffer and B cells were immunomagnetically selected from the resulting splenocytes. Negatively enriched splenic B cells were used to perform all experiments, and this enrichment method usually yields more than 90% CD19+ B cells. Splenic B cells were cultured at a density of 0.5 × 106 – 0.8 × 106 cells/ml in RPMI media supplemented with 10% FCS, 1% penicillin/streptomycin, 50 μg/ml lipopolysaccharides from Salmonella enterica, 10 ng/ml murine Il-4 and 0.0572 mM beta-mercaptoethanol. Cells were cultured at 37 ˚C in a humidified incubator with 5% CO2 for 24 – 96 hr before they were collected for analysis. Mice were alternatively stimulated with immunization with fresh sheep red blood cells (sRBCs). SRBCs were washed in sterile PBS twice and re-suspended in sterile PBS to concentration of 200 × 106 cells/ 100μl. Mice were injected via intravenous tail vein injection with 200 μl of the cell suspension. After 10 days, mice were euthanized for analysis. Proliferation was measured using CellTrace Violet Cell Proliferation Kit and flow cytometry, per manufacturer’s instruction. Dye intensity was determined every 24 hr following labelling. Cell apoptosis was measured by flow cytometric evaluation of Annexin V-PE and 7-AAD staining.
IHC staining, quantification of Ki67 positive GC
All spleens were fixed in 10% buffered neutral formalin overnight followed by 70% ethanol until the tissues were processed. Spleens were then paraffin embedded and sectioned into 20μm for IHC staining. Cell proliferation was estimated using Ki67 specific antibodies and a horseradish peroxidase (HRP)-conjugated secondary antibody to reveal the diaminobenzidine (DAB) staining. Slides were scanned into Aperio eSlide Manager and observed at a magnification of 10x. The diameter of each Ki67 positive germinal center (indicated by brown staining) was then measured. B220, CD5, and CD5 staining were performed on the section with standard procedure.
Pyrosequencing
Quantitative targeted sequencing to detect a region of genetic change within cDNA was performed, as previously described (Landau et al., 2014). In brief, biotinylated amplicons generated from PCR of transcript surrounding SF3B1 mutation were generated. Immobilized biotinylated single stranded DNA fragments were isolated per manufacturer’s protocol and sequencing undertaken using an automated pyrosequencing instrument (PSQ96; Qiagen, Valencia, CA), followed by quantitative analysis using Pyrosequencing software (Qiagen). Forward (CAGAGCACTGATGGTCCGAA) and reverse (Bio CAAGCGAGGCACACTGGTAT) primers are used to amplify the region across the K700E mutation site from the cDNA. The probe sequence is CTGATGGTCCGAACTT.
Genome sequencing and analysis
350 ng genomic DNA was used for library preparation. DNA libraries were constructed using the KAPA Library Prep Kits (KAPA Biosystems) using palindromic forked adapters with unique 8 base index sequences embedded within the adapter (IDT). Following sample preparation, libraries were quantified using quantitative PCR with probes specific to the ends of the adapters. Samples were then combined with HiSeq X Cluster Amp Mix 1,2 and 3 into single wells on a strip tube using the Hamilton Starlet Liquid Handling system. Cluster amplification of the templates was performed according to the manufacturer’s protocol (Illumina) using the Illumina cBot. Flowcells were sequenced using the Illumina HiSeqX platform (Illumina). Whole genome sequencing data analyses were performed as described previously (Berger et al., 2011). Briefly, sequencing output was post-processed with the Picard and GATK toolkits (Broad Institute), which includes marking duplicated reads and recalibrating base qualities, and all BAM files were aligned to Genome Reference Consortium Mouse Build 38, or mm10. From the aligned BAM files, both somatic and germline alterations were identified using a set of tools developed at the Broad Institute (www.broadinstitute.org/cancer/cga). Somatic single nucleotide variations (sSNVs) were detected using MuTect1 (Cibulskis et al., 2013) (version 1.1.6). Variants were annotated using Variant Effect Predictor (McLaren et al., 2016). Variants called from Mutect1 were further filtered by removing any calls that had less than 8 reads on the tumor. Furthermore, variants that belonged to lower case masked regions, as specified by the mm10 reference sequence, were discarded. The (updated) copy number analysis was conducted using the coverage-only copy number variant pipeline from GATK 4 (Broad Institute). The purity of the samples was calculated from the heterozygous site split and the copy number calls. From the rejected Mutect1 calls, sites were considered as heterozygous sites if their allelic fraction fell between 0.48–0.53 in the normal sample site. Only sites with 30 or more reads in the tumor, and between 20 and 50 reads in the normal were considered for analysis. To track the clonal trajectories between the parental DM-CLLs and engrafted CLL cells, we first measured the variant allele fraction (VAF) of all sSNVs/sIndels identified in the available WES data per mouse line. VAFs were transformed to cancer cell fractions (CCFs) using ABSOLUTE, followed by n-dimensional clustering. This procedure allowed us to infer discrete clusters of somatic mutations that define the clonal cluster and each subclone and to calculate the CCF of each subclone at a time point.
RNA-seq analysis
RNA-seq analyses for splicing and differential gene expression were performed as previously described (Wang et al., 2016). Alternative splicing analysis was performed using JuncBASE (Brooks et al., 2011). TPM (Transcripts Per Kilobase Million) value was used to measure gene expression and genes with poor expression (mean TPM<1) were filtered. The DFCI/Broad cohort was described in (Landau et al., 2014) and the ICGC cohort was described in (Ferreira et al., 2014).
Quantitative Proteomics
Tandem mass tag (TMT) 10-plex labeling was performed and peptides were fractionated via BPRP HPLC. An 1100 pump (Agilent, Santa Clara, CA) equipped with a degasser and a photodiode array (PDA) detector (ThermoFisher Scientific, Waltham, MA) was used. Peptides were subjected to a linear gradient from 3 to 25% acetonitrile in 0.125% formic acid using an Agilent 300 Extend-C18 column (Agilent, Santa Clara, CA), and were fractionated into a total of 96 fractions. Mass spectrometry was performed using an Orbitrap Fusion mass spectrometer (ThermoFisher Scientific, Waltham, MA) coupled to a Proxeon EASY-nLC 1000 liquid chromatography (LC) pump (ThermoFisher Scientific, Waltham, MA). Peptides were detected (MS1) and quantified (MS3) in the Orbitrap. MS2 spectra were searched using the SEQUEST algorithm against a Uniprot composite database derived from the mouse proteome containing its reversed complement and known contaminants. Peptide spectral matches were filtered to a 1% false discovery rate (FDR) using the target-decoy strategy combined with linear discriminant analysis. The detected proteins were filtered to a <1% FDR and were quantified only from peptides with a summed SN threshold of >=200 and isolation specificity of 0.5. Statistical analysis of the proteome was performed based on the normalized intensities of the TMT-reporter ions. Proteins that exhibited more than 2-fold difference between sample types were considered differentially expressed. Quantitative metabolomics profiling was performed by the Proteomics/Metabolomics Core of Beth Israel Deaconess Medical Center using a selected reaction monitoring (SRM) method as previously described (Yuan et al., 2012).
BCR stimulation
For analysis of BCR signaling transduction, purified splenic B cells or CLL cells (10 × 106) were stimulated in 500 μl RPMI 1640 medium (Life Technologies, Woburn, MA) in the presence of 20 μg/ml goat α-mouse IgM for 5 min at 37°C. Cells were harvested, washed twice with cold phosphate-buffered saline (PBS), and lysed for 30 min on ice with RIPA buffer (Boston Bioproducts, Boston, MA) supplemented with a protease inhibitor mix (1 tablet in 10 ml RIPA buffer, Roche, San Francisco, CA). Cell debris was removed by centrifugation at 12,000×g for 5 min, and cell extracts were fractionated by SDS-PAGE followed by western blot analysis of kinase activation (SYK, AKT and ERK1/2). Antibodies used in the study include: α-SYK, α-p-SYK (Y352), α-AKT (40D4), α-p-AKT S473 (D9E), α-ERK1/2 (137F5) and α-p-ERK1/2 (D13.14.4E).
Immunofluorescence
For immunofluorescence staining, purified splenic B cells or CLL cells were fixed in 4% paraformaldehyde in PBS for 15 min, washed once with PBS, and permeabilized in 0.5% Triton X-100/PBS for 15 min at room temperature. After washed once with PBS, cells were blocked in blocking buffer (1% bovine serum albumin in PBS) for 30 min. Immunostaining was performed using α-SF3B1 or α-SF3A1–3 antibody at 4 °C overnight, or α-γH2AX antibody for 1 h at room temperature. After 3× 10 min wash with PBS, cells were stained with secondary antibody conjugated to FITC at room temperature for 1h. Images were captured with a Nikon TE2000U inverted microscope.
Human CLLs treated with ibrutinib in vitro
CLL primary patient samples were obtained from The CLL Research Consortium (CRC). CLL samples were resuspended in RPMI1640 medium supplemented with 10% FBS and cultured in 96-well tissue culture plates (50,000 cells/100 μL). Ibrutinib (Selleckchem, Houston, TX) was diluted serially in medium and was added to corresponding wells at final concentrations ranging from 0 to 10 μM. After incubation for 48 hr at 37 °C with 5% CO2, cell viability was measured by CellTiter-Glo® Luminescent Assay and normalized by cells with no drug treatment.
Mouse CLL transplantation and ibrutinib treatment
The transplantation studies were performed on 8–12 weeks old immunocompetent CD45.1 C57BL/6 mice (pre-conditioned with 400 rads split dose irradiation) or immunodeficient recipient NSG mice using viably cryopreserved splenocytes from DM-CLL animals. Ten million DM-CLL cells/recipient were resuspended in 100 microliters PBS and injected intravenously into CD45.1 C57BL/6 or NSG. In ibrutinib single agent treatment studies, CLL burden in the peripheral blood of the transplanted recipients (NSGs) was analyzed by flow cytometry and the treatment was initiated two weeks after transplantation when 20–40% CLL cells were detectable. Vehicle control (1% HP-β-CD in water) or 25 mg/kg ibrutinib was administered daily by oral gavage to NSG mice for 6 days, then animals were observed for survival; criteria for euthanasia included hunched posture, difficulties breathing or moving, visible hepatosplenomegaly, weight loss equal to 15% body weight. Disease burden in the peripheral blood, spleen and bone marrow were evaluated by flow cytometry and spleen and liver sections were examined by IHC at euthanasia.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Statistical analysis was performed using GraphPad Prism 6. p values were calculated with a two-sample Student t-test (after confirming homogeneity of variance) or Mann Whitney test. The impact of ibrutinib dosing on cell viability was modeled as a longitudinal mixed effects model which included patient sample as a random factor as well as fixed factors for SF3B1 mutation status and dose; this approach was also used for the studies of apoptosis and cell proliferation. Modeling was performed in R using package ‘nlme’. The Kruskal-Wallis test was used to assess the shift in time of peak lymphocytosis after initiation of ibrutinib treatment.
DATA AND SOFTWARE AVAILABILITY
Accession Codes
Gene Expression Omnibus: The RNA-seq data reported in this paper are deposited into the GEO database (GSE122668) and whole genome sequencing data are deposited into SRA database (PRJNA509881).
Supplementary Material
Table S1, related to Figure 1. Altered splicing events associated with Sf3b1 mutation in mouse B cells.
Table S2, related to Figure 4. IGHV junction characterization in DM-CLL1 and DM-CLL2.
Table S3, related to Figure 5. Mis-splicing events associated with each MT group versus WT in old mice.
Table S4, related to Figure 5. Clonal SNVs identified in DM-CLL1 and DM-CLL2 cells.
Table S5, related to Figure 5. Human homologs of mice Chr15 and Chr17 genes significantly upregulated in SF3B1 MT CLL versus normal B cells.
Table S6, related to Figure 6. Differentially expressed genes and proteins between different mouse lines.
Table S7, related to Figure 7. Expression of BCR signaling genes in normal B cells and CLL cases with or without SF3B1 mutation.
SIGNIFICANCE.
Although SF3B1 is frequently mutated in CLL, its role in the pathogenesis of CLL remains elusive. Here we generated a murine line conditionally expresses heterozygous Sf3b1-K700E mutation in B cells. Expression of this mutant splicing factor disrupted B cell growth and function, and led to cellular senescence. Combining Sf3b1 mutation with Atm deletion in B cells overcame cellular senescence and resulted in lowly penetrant CLL in aged mice, with clonally expanded B220+CD5+ cells present in blood, bone marrow and spleen, recapitulating features of human CLL. Dissection of this murine CLL model revealed the involvement of mutant SF3B1 in BCR signaling dysregulation, leading us to discover altered response of human CLL cells with SF3B1 mutations to ibrutinib treatment.
ACKNOWLEDGEMENTS
The authors thank Drs. Markus Műschen and Hassan Jumaa for their critical insights, and Ms. Sarah Hergert for excellent technical support. This work was in part supported by NCI 1R01 CA21673-01 (L.W. and C.J.W.), NCI 1R01 CA155010-01A1 (C.J.W.), NCI 1U10 CA180861-01 (C.J.W.), NHLBI 1R01 HL103532-01 (C.J.W.), NHLBI 1R01 HL116452-01 (C.J.W.), the Lymphoma Research Foundation (C.J.W.), a Leukemia Lymphoma Translational Research Program Award (C.J.W.); NIH R01 DK087992 (M.D.F.); NIH P01 CA81534 (T.J.K.), NIH Cancer Center Core Grant 5P30 CA006516 (D.N.); NIH R35 GM122524 (R.R.); NHLBI ZIA HL002346-13 (A.W.); US Public Health Service grants R01 CA63113 (J.A.D.), R01 CA173023 (J.A.D.), P01 CA203655 (J.A.D.); the Leukemia and Lymphoma Society fellowship (E.T.H.) and National Science Foundation Graduate Research Fellowship DGE1144152 (J.F.).
Footnotes
DECLARATION OF INTERESTS
C.J.W. is a co-founder of Neon therapeutics. C.J.W., D.N. and G.G. receive research funding from Pharmacyclics. B.L.E. has been a consultant for H3 Biomedicine and received research funding from Celgene. G.G receives research funds from IBM. G.G. is an inventor on patent applications related to MuTect, ABSOLUTE and other bioinformatics methods. J.A.D. has received honoraria for participation in advisory board from Merck. J.A.D. has received research funding from Constellation Pharmaceuticals. J.S. is a current employee of Moderna Therapeutics. All other authors do not have any relevant conflict of interest.
REFERENCE
- Aird KM, Worth AJ, Snyder NW, Lee JV, Sivanand S, Liu Q, Blair IA, Wellen KE, and Zhang R (2015). ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell reports 11, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarano A, Indraccolo S, Circosta P, Minuzzo S, Vallario A, Zamarchi R, Fregonese A, Calderazzo F, Faldella A, Aragno M, et al. (1999). An alternatively spliced form of CD79b gene may account for altered B-cell receptor expression in B-chronic lymphocytic leukemia. Blood 93, 2327–2335. [PubMed] [Google Scholar]
- Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, et al. (2016). Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nature communications 7, 10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. (2006). Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637. [DOI] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. (2011). The genomic complexity of primary human prostate cancer. Nature 470, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blix ES, Irish JM, Husebekk A, Delabie J, Forfang L, Tierens AM, Myklebust JH, and Kolstad A (2012). Phospho-specific flow cytometry identifies aberrant signaling in indolent B-cell lymphoma. BMC cancer 12, 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, and Graveley BR (2011). Conservation of an RNA regulatory map between Drosophila and mammals. Genome research 21, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JA, and Wiestner A (2018). Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nature reviews Cancer 18, 148–167. [DOI] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, et al. (2013). Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. The New England journal of medicine 369, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, and d’Adda di Fagagna F (2007). Cellular senescence: when bad things happen to good cells. Nature reviews Molecular cell biology 8, 729–740. [DOI] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, and Getz G (2013). Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvelo A, Hallegger M, Smith CW, and Eyras E (2010). Genome-wide association between branch point properties and alternative splicing. PLoS computational biology 6, e1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, et al. (2015). Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3? Splice Site Selection through Use of a Different Branch Point. Cell reports 13, 1033–1045. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. (2006). Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642. [DOI] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, and Lichter P (2000). Genomic aberrations and survival in chronic lymphocytic leukemia. The New England journal of medicine 343, 1910–1916. [DOI] [PubMed] [Google Scholar]
- Duhren-von Minden M, Ubelhart R, Schneider D, Wossning T, Bach MP, Buchner M, Hofmann D, Surova E, Follo M, Kohler F, et al. (2012). Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 489, 309–312. [DOI] [PubMed] [Google Scholar]
- Ferreira PG, Jares P, Rico D, Gomez-Lopez G, Martinez-Trillos A, Villamor N, Ecker S, Gonzalez-Perez A, Knowles DG, Monlong J, et al. (2014). Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome research 24, 212–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman SE, Niemann CU, Farooqui M, Jones J, Mustafa RZ, Lipsky A, Saba N, Martyr S, Soto S, Valdez J, et al. (2014). Ibrutinib-induced lymphocytosis in patients with chronic lymphocytic leukemia: correlative analyses from a phase II study. Leukemia 28, 2188–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT, Chiorazzi N, and Meffre E (2005). Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. The Journal of clinical investigation 115, 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. (2014). Age-related clonal hematopoiesis associated with adverse outcomes. The New England journal of medicine 371, 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HT, Park JT, Choi K, Kim Y, Choi HJC, Jung CW, Lee YS, and Park SC (2017). Chemical screening identifies ATM as a target for alleviating senescence. Nature chemical biology 13, 616–623. [DOI] [PubMed] [Google Scholar]
- Kipps TJ, Stevenson FK, Wu CJ, Croce CM, Packham G, Wierda WG, O’Brien S, Gribben J, and Rai K (2017). Chronic lymphocytic leukaemia. Nature reviews Disease primers 3, 17008. [DOI] [PubMed] [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, and Dalla-Favera R (2010). The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer cell 17, 28–40. [DOI] [PubMed] [Google Scholar]
- Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, et al. (2014). Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer cell 26, 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Sun C, Rosebrock D, Herman SEM, Fein J, Sivina M, Underbayev C, Liu D, Hoellenriegel J, Ravichandran S, et al. (2017). The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nature communications 8, 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Bottcher S, et al. (2015). Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He J, Ji S, Wang Q, Pu H, Jiang T, Meng L, Yang X, and Ji J (2008). Comparative studies of early liver dysfunction in senescence-accelerated mouse using mitochondrial proteomics approaches. Molecular & cellular proteomics : MCP 7, 1737–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklin M, Heitmann JS, Fuchs AR, Truckenmuller FM, Gutknecht M, Bugl S, Saur SJ, Lazarus J, Kohlhofer U, Quintanilla-Martinez L, et al. (2017). NFAT2 is a critical regulator of the anergic phenotype in chronic lymphocytic leukaemia. Nature communications 8, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, and Cunningham F (2016). The Ensembl Variant Effect Predictor. Genome biology 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschen M (2018). Autoimmunity checkpoints as therapeutic targets in B cell malignancies. Nature reviews Cancer 18, 103–116. [DOI] [PubMed] [Google Scholar]
- Muzio M, Apollonio B, Scielzo C, Frenquelli M, Vandoni I, Boussiotis V, Caligaris-Cappio F, and Ghia P (2008). Constitutive activation of distinct BCR-signaling pathways in a subset of CLL patients: a molecular signature of anergy. Blood 112, 188–195. [DOI] [PubMed] [Google Scholar]
- Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, Jares P, Navarro A, Martin-Garcia D, Bea S, et al. (2016). Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood 127, 2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, Schneider RK, Lord AM, Wang L, Gambe RG, et al. (2016). Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer cell 30, 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha J, Secreto C, Rabe K, Ayres-Silva J, Tschumper R, Dyke DV, Slager S, Fonseca R, Shanafelt T, Kay N, and Braggio E (2014). Monoclonal B-cell lymphocytosis is characterized by mutations in CLL putative driver genes and clonal heterogeneity many years before disease progression. Leukemia 28, 2395–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, Munar M, Rubio-Perez C, Jares P, Aymerich M, et al. (2015). Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. [DOI] [PubMed] [Google Scholar]
- Shay T, and Kang J (2013). Immunological Genome Project and systems immunology. Trends in immunology 34, 602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Raa GD, Derks IA, Navrkalova V, Skowronska A, Moerland PD, van Laar J, Oldreive C, Monsuur H, Trbusek M, Malcikova J, et al. (2015). The impact of SF3B1 mutations in CLL on the DNA-damage response. Leukemia 29, 1133–1142. [DOI] [PubMed] [Google Scholar]
- Wang L, Brooks AN, Fan J, Wan Y, Gambe R, Li S, Hergert S, Yin S, Freeman SS, Levin JZ, et al. (2016). Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer cell 30, 750–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. (2011). SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. The New England journal of medicine 365, 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiestner A (2016). Choosing frontline chemoimmunotherapy for CLL. The Lancet Oncology 17, 852–854. [DOI] [PubMed] [Google Scholar]
- Woyach JA, Smucker K, Smith LL, Lozanski A, Zhong Y, Ruppert AS, Lucas D, Williams K, Zhao W, Rassenti L, et al. (2014). Prolonged lymphocytosis during ibrutinib therapy is associated with distinct molecular characteristics and does not indicate a suboptimal response to therapy. Blood 123, 1810–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. (2014). Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine 20, 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni Y, Getahun A, and Cambier JC (2010). Molecular underpinning of B-cell anergy. Immunological reviews 237, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, and Asara JM (2012). A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nature protocols 7, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Sekiguchi J, Brush JW, Bassing CH, and Alt FW (2008). Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proceedings of the National Academy of Sciences of the United States of America 105, 9302–9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 1. Altered splicing events associated with Sf3b1 mutation in mouse B cells.
Table S2, related to Figure 4. IGHV junction characterization in DM-CLL1 and DM-CLL2.
Table S3, related to Figure 5. Mis-splicing events associated with each MT group versus WT in old mice.
Table S4, related to Figure 5. Clonal SNVs identified in DM-CLL1 and DM-CLL2 cells.
Table S5, related to Figure 5. Human homologs of mice Chr15 and Chr17 genes significantly upregulated in SF3B1 MT CLL versus normal B cells.
Table S6, related to Figure 6. Differentially expressed genes and proteins between different mouse lines.
Table S7, related to Figure 7. Expression of BCR signaling genes in normal B cells and CLL cases with or without SF3B1 mutation.
Data Availability Statement
Accession Codes
Gene Expression Omnibus: The RNA-seq data reported in this paper are deposited into the GEO database (GSE122668) and whole genome sequencing data are deposited into SRA database (PRJNA509881).