Abstract
Noonan syndrome is a common autosomal dominant disorder characterized by short stature, congenital heart disease and facial dysmorphia with an incidence of 1/1000 to 2500 live births. Up to now, several genes have been proven to be involved in the disturbance of the transduction signal through the RAS-MAP Kinase pathway and the manifestation of Noonan syndrome. The first gene described was PTPN11, followed by SOS1, RAF1, KRAS, BRAF, NRAS, MAP2K1, and RIT1, and recently SOS2, LZTR1, and A2ML1, among others. Progressively, the physiopathology and molecular etiology of most signs of Noonan syndrome have been demonstrated, and inheritance patterns as well as genetic counseling have been established. In this review, we summarize the data concerning clinical features frequently observed in Noonan syndrome, and then, we describe the molecular etiology as well as the physiopathology of most Noonan syndrome-causing genes. In the second part of this review, we assess the mutational rate of Noonan syndrome-causing genes reported up to now in most screening studies. This review should give clinicians as well as geneticists a full view of the molecular aspects of Noonan syndrome and the authentic prevalence of the mutational events of its causing-genes. It will also facilitate laying the groundwork for future molecular diagnosis research, and the development of novel treatment strategies.
Keywords: Molecular etiology, MAP kinase signaling pathways, Mutation rate, Noonan syndrome, PTPN11, RAS family
Abbreviations: CDC25, cell division cycle 25; CHD, congenital heart defects; CR, conserved region; CRD, cysteine-rich domain; GAP, GTPase activating protein; GDP, guanosine-DiPhosphate; GEF, guanine exchange factor; GTP, guanosine-TriPhosphate; GH, growth hormone; HCM, hypertrophic cardiomyopathy; IGF-1, insulin-like growth factor I; RBD, RAS binding domain; REM, RAS exchange motif
1. Introduction
Noonan syndrome ([NS1, OMIM 163950]) is a common autosomal dominant disorder characterized by short stature, congenital heart disease, facial dysmorphia and other features such as cryptorchidism, bleeding diathesis, skeletal malformations and mild cognitive delays with variable expressivity. The prevalence of this disorder is estimated to be 1/1000–2500 live births [1], [2], [3]. The varied clinical manifestations observed in Noonan syndrome patients (facial, skeletal cardiac, and hematological, among others) are a result of the involvement of the RAS MAP kinase molecular signaling pathway, as will be shown below.
The previous updates and reviews in the literature have thoroughly discussed Noonan syndrome, often focusing on diagnostic evaluations, clinical guidelines, and management with treatment options, which have amply helped clinicians provide the best care for Noonan syndrome children. The molecular aspects of this disorder have been approached in these reviews progressively with genetic advances. However, considering the fast advances and consecutive progress being made in the physiopathology and genetic studies of Noonan syndrome, it seems important to gather these findings in one paper to help the audience of clinicians and geneticists have a full view of recent advances in the molecular etiology of Noonan syndrome, as well as an authentic prevalence of the mutational rates of its causing-genes. Therefore, this review provides, in the first part, an update on the molecular aspect of the disease, in which we summarize the data concerning clinical features frequently observed, then focus on the molecular etiology, the inheritance pattern and the genetic counseling that should be given to patients. In the second part of this review, we establish and discuss the mutational rate reported up to now in most genes involved in Noonan syndrome.
2. Review
2.1. Clinical features and diagnosis
The diagnosis of Noonan syndrome is based primarily on the clinical features that have been established from the very beginning through several clinical studies that have meticulously defined the criteria and signs of diagnosis [1], [2], [3].
2.1.1. Dysmorphic face
The extreme variability of facial traits in Noonan syndrome from one individual to another makes the assessment of frequency difficult and not very meaningful. Furthermore, the dysmorphic signs could change within the same patient depending on his/her age, to be less perceptible in adulthood than earlier in childhood. The dysmorphology is characterized during the postnatal period by a tall forehead, low-set-posteriorly-rotated ears, a thickened helix, nerve deafness, hypertelorism, ptosis, down slanting palpebral fissures, epicanthal folds, deeply grooved philtrum, a high arched palate and triangular face, with a low posterior hairline and webbed neck. In adulthood, the facial features become more subtle, the eyes are less prominent, with a slightly elongated neck, wrinkled skin and high anterior hairline [3], [4], [5].
2.1.2. Congenital heart defect
The cardiac features are well delineated and are estimated to be present in 50% up to 90% of Noonan syndrome patients [6], [7], [8]. The most common congenital heart defects (CHD) are pulmonic stenosis (50–60%), hypertrophic cardiomyopathy (HCM) (20%) and atrial septal defect (6–10%) [9], [10]. The other CHDs such as ventricular septal defect, atrioventricular canal defect, and aortic coarctation are observed less frequently [6], [8], [11]. Electrocardiographic abnormalities were reported in 87% of patients [3]. Electrocardiograms display wide QRS complexes with a predominant negative pattern in the left precordial leads, a left axis deviation and giant Q waves [9], [10], [11].
2.1.3. Growth/short stature
Weight and height are normal at birth. However, during childhood and at puberty, short stature becomes a prominent common sign of Noonan syndrome. In one series reported by Nora et al, the prevalence of Noonan children with height below the 3rd percentile is approximately 83% [12]. In another study, pubertal growth was found to be delayed by almost two years and the mean height was in the 3rd percentile, with female and male average heights of 151 cm and 161 cm respectively, and the average bone age delayed by two years [8].
2.1.4. Skeletal defects
A chest deformity characterized by superior pectus carinatum and inferior pectus excavatum is observed in up to 95% of Noonan syndrome patients. Half (50%) have cubitus valgus and 30% have a clinobrachydactyly. The other orthopedic features, such as thoracic scoliosis, talipes equinovarus or radiolunar synostosis, are observed less often [3], [7].
2.1.5. Bleeding defect
The coagulation defect is the most common hematologic disorder in Noonan syndrome patients. Approximately 55% of patients have mild to moderate abnormal bleeding, whereas only 3% have major abnormal bleeding [3].
2.1.6. Genito-urinary
Altered spermatogenesis and cryptorchidism are observed in 60–80% of patients [13]. Ten percent of patients have renal abnormalities that often include renal pelvis dilatation [3].
2.1.7. Ophthalmological features
Approximately 55% of Noonan syndrome patients have an abnormal ophthalmological test [3]. The frequent abnormalities are refractive errors (61–70%), strabismus (48–63%), amblyopia (33%) and anterior segment changes (63%) [14], [15].
2.1.8. Other features
Other features have also been reported, including lymphatic abnormalities, hematological malignancy, giant cell lesions and cognitive disability, among others [16], [17], [18], [19].
2.2. Genetic diagnosis and molecular etiology
As shown above, Noonan syndrome is a heterogeneous disorder with various clinical features ranging from facial dysmorphology to short stature and bleeding defects up to congenital heart disease. Those features have been demonstrated by several studies to be the yield of diverse molecular and physiological mechanisms, some of which are well known, while others need to be further elucidated.
The molecular etiology of Noonan syndrome has been established first by the study of two large Noonan syndrome families using linkage analysis [20], [21], [22]. These studies suggested that the concerned locus is located in the 12q24 region. Based on these data, and the data suggesting the involvement of SHP-2 in the signaling pathways that control semilunar valvulogenesis [23], and by means of a positional candidacy approach, Tartaglia et al [24] suggested PTPN11 to be a Noonan syndrome candidate gene, and confirmed this hypothesis by bidirectionally sequencing the fifteen exons of PTPN11, which have shown mutations in 50% of the studied cohort.
2.2.1. PTPN11 (protein-tyrosine phosphatase, nonreceptor-type 11)
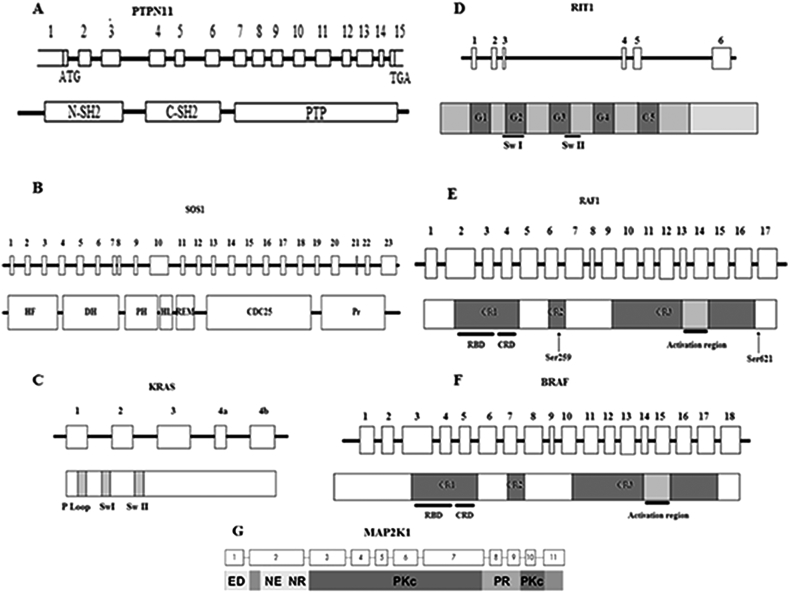
The PTPN11 gene ([OMIM 176876]) (Fig. 1A) is organized into three domains: the N-amino terminal src-homology 2 domain (N-SH2) and the phosphotyrosine phosphatase (PTP) domain, which are the most commonly mutated domains; and the C-amino terminal src-homology 2 domain (C-SH2) and carboxy-terminal tail [25], [26].
Figure 1.
Organization of Noonan syndrome causing genes and domains, A. PTPN11 exons and SHP-2 domains, B. SOS1 exons and domains, C. KRAS coding exons and domains. In most cases, the exon 4a is spliced out. D. RIT1 coding exons and domains. E. RAF1 coding exons and domains with the localization of Ser259 and Ser621 residues that is critical for RAF1 auto-inhibition. F. BRAF coding exons and domains. G. MAP2K1 coding exons and domains. C-SH2: C-amino-terminal src-homology 2, CR: Conserved Region, CRD: Cysteine-Rich Domain, DH: Dbl Homology, HF: Histone-like Fold, HL: Helical Linker, N-SH2: N- amino-terminal src-homology 2, PH: Pleckstrin Homology, Pr: Proline-riche motif, PTP: protein-tyrosine phosphatase, RBD: RAS binding domain, REM: RAS Exchange Motif, Sw: Switch.
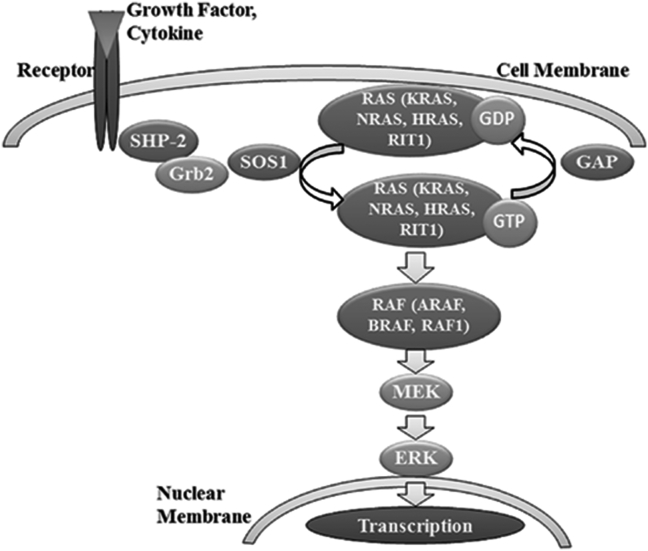
Protein SHP-2 coded by PTPN11 was demonstrated to be involved in several developmental processes such as limb development, semilunar valvulogenesis, hemopoietic cell differentiation and mesodermal patterning [23], [27], [28], [29], and is widely expressed in several tissues such as the heart, muscles, and brain, in which SHP-2 modulates the cellular proliferation, migration or differentiation processes during the developmental stage [30]. SHP-2 is a key element of the signaling molecular RAS-MAP Kinase cascade (Fig. 2). The latter is triggered when cytokines, hormones or growth factors bind to membrane receptors [25], [26], [31]. Therefore, its disturbance may cause alterations of these tissues that lead to the aberrant phenotype shown in Noonan syndrome patients. This fact has been confirmed by De Rocca et al, has confirmed that the SHP-2 mutations in Noonan syndrome were associated with the inhibition of GH-induced IGF-1 via the hyperactivation of RAS/ERK1/2 in a mouse model, which lead to the short stature observed in Noonan syndrome [32].
Figure 2.
Localization of Noonan syndrome causing genes into the RAS-MAP kinase signal transduction pathway.
On the molecular level, SHP-2 is a cytosolic phosphatase protein activated by the binding of a N-SH2 domain to phosphotyrosyl residue, which leads to the conformational change making the catalytic site (PTP) available to the phosphotyrosyl residue. In the absence of substrate, SHP-2 regains its inactive form in which N-SH2 interacts with PTP to hide the catalytic site [33].
Most of the PTPN11 mutations causing Noonan syndrome manifestations cluster in these two domains. The energy-based structured analysis suggests that PTPN11 mutations disrupt the inactive conformation, resulting in the emergence of the catalytic site which triggers the phosphotyrosyl phosphatase activity in the absence of the ligand [24].
Additionally, several screening studies were carried out to define PTPN11 mutations in other populations and to highlight putative novel mutations. PTPN11 mutations were significantly correlated to the manifestation of pulmonary stenosis observed in 45–70% of cases [34], [35].
Seven other genes from the RAS-MAP kinase pathway (Fig. 2) were demonstrated to be mutated and to cause Noonan syndrome.
2.2.2. SOS1 (Son of Sevenless homolog 1)
At present, SOS1 ([OMIM 182530]) is considered the second molecular cause of Noonan syndrome. Roberts et al reported that SOS1 is responsible for approximately 20% of Noonan syndrome patients in the absence of PTPN11 mutations [36].
SOS1 is a guanine exchange factor (GEF) with a major role in the RAS-MAP kinase pathway. It is mapped on the 2p22-p21 region and consists of 23 exons (Fig. 1B) [37], [38] coding for multiple domains containing: histone-like folds domain (HF), Dbl homology domains (DH) and Pleckstrin homology domains (PH) with the following regulatory function; RAS exchanging motif (REM) and Cdc25 domains with catalytic function; helical linker (HL) relating PH and REM, and the PolyProline region [36], [39].
It was reported that Noonan syndrome-causing SOS1 was associated with a high prevalence of ectodermal abnormalities [36], [39], [40], [41].
In the RAS-MAP kinase molecular cascade, RAS protein is inactivated by hydrolyzing GTP to GDP via GTPase Activating Protein (GAP). To be reactivated, RAS protein needs to exchange bound GDP for GTP via SOS1.
It was suggested that the Dbl homology domain inhibits RAS-GEF activity by competing with RAS for binding to the REM-Cdc25 complex designated as an allosteric site, which stabilizes the inactive conformation [42].
Most mutations cluster in these domains and lead to gain-of-function in the RAS-MAP kinase pathway by disrupting the auto-inhibition of SOS1 RAS-GEF activity [36], [39], [43].
Pulmonic stenosis was more frequently observed in patients with SOS1 mutations (83.3%) than in those with PTPN11 mutations [34].
The RAS family (KRAS, HRAS, NRAS and RIT1) is a small GTPase protein family responsible of signal propagation through the RAS-MAP kinase cascade. It acts as a molecular switch cycling between an inactive GDP bound conformation and an active GTP bound conformation (Fig. 2) [44], [45].
2.2.3. KRAS (kirsten rat sarcoma viral oncogene homolog)
The KRAS ([OMIM 190070]) gene is mapped to the 12p12 region, and consists of 6 exons coding for the P loop, and switch I and switch II domains (Fig. 1C) [46]. In response to a transduction signal generated from membrane receptor binding, KRAS regains its active bound GTP conformation via the guanine nucleotide exchange factor, and entails an activation cascade through the downstream effectors of RAS-MAP kinase (such as RAF1). KRAS has intrinsic GTPase activity, which, supported by RAS-GAPs (RAS-GTPase activating protein), hydrolyzes bound GTP to GDP to inactive itself [44], [45].
Lee et al and Schubbert et al have reported that KRAS mutations cause approximately 5% of Noonan syndrome cases in the absence of PTPN11 mutations [34], [47]. These mutations lead to a gain in the function effect through the RAS-MAP kinase pathway by altering the intrinsic GTPase activity of KRAS and creating insensitivity to RAS-GAPs [46], [47].
Noonan syndrome patients with KRAS mutations were reported to have severe phenotypes with severe mental retardation [46], [47], [48].
2.2.4. NRAS (neuroblastoma RAS viral oncogene homolog)
The NRAS ([OMIM 164790]) gene mapped on 1p13.2 comprises 6 coding exons [49]. Cirstea et al have reported that NRAS mutations are involved in less than 1% of Noonan syndrome cases [50].
2.2.5. RIT1(Ric-like protein without Caax motif 1)
Interestingly, in 2013, Aoki et al reported that mutations in RIT1 ([OMIM 609591]) cause Noonan syndrome [51]. RIT1 is a member of the RAS subfamily of small GTPases (Fig. 2) [52], and consists of 6 exons and is located in the 1q22 region (Fig. 1D).
Aoki et al identified nine missense mutations in seventeen individuals from 180 patients (9%) with Noonan syndrome or related disorders and without mutations in known Noonan syndrome-causing genes, and observed that the frequency of RIT mutations in the Noonan syndrome cohort was seemingly similar to the frequency of RAF1 mutations. This finding was subsequently confirmed by Bertola et al, in 2014, who found the same prevalence (9%), and Gos et al, in 2014, who found a lower mutation rate (3.8%). Those mutation clusters in the G1, Switch I, and more frequently in Switch II domains, were proven to entail a significant activation of the RAS MAPK pathway by hyper-activating transcription factor ELK1. [51], [53], [54].
Noonan syndrome patients with RIT1 mutations were characterized by a high incidence of congenital heart disease (94%), especially hypertrophic cardiomyopathy (71%) and pulmonic stenosis (65%). The frequency of hypertrophic cardiomyopathy among RIT1 mutation-positive patients is similar to that of RAF1 mutation-positive subjects with Noonan syndrome, which leads to the conclusion that RIT1 and RAF1 interact with each other and have the same effect on cardiac development [51], [55], [56].
The RAF family (ARAF, BRAF and RAF1) has an activation role upstream of the MEK-ERK cascade into the RAS-MAPK pathway (Fig. 2).
2.2.6. RAF1 (v-raf-1 murine leukemia viral oncogene homolog 1)
RAF1 ([OMIM 164760]) mutations cause 3–17% of Noonan syndrome cases. RAF1 consists of 17 exons coding for multidomain protein that act as a serine–threonine kinase (Fig. 1E) [55], [57], [58]. This protein comprises three conserved regions (CR): CR1 that contains the RAS binding domain (RBD) and cysteine-rich domain (CRD), which are both suggested to be involved in the negative regulation of RAF1 by direct physical interaction [59]; CR2, which is an important region where many RAF1-activating mutations and clusters lie. Eighty percent (80%) RAF1 mutations in Noonan syndrome have HCM. It contains Ser259 residue, which is critical to RAF1 auto-inhibition [55]; and the third conserved region (CR3) in the C-terminal region of RAF1 that is responsible for catalytic activity [55], [57]. The RAF1 inactive conformation is maintained via 14-3-3 protein dimers that bind to phosphorylated Ser259 and Ser621 in such a way that the catalytic site remains hidden [60]. Mutations in these conserved residues or in its flanking residues (such as Arg256 and Pro261, among others) prevent the 14-3-3 binding leading to an auto-inhibition fail and the activation of the RAS-MAP kinase cascade [57].
Noonan syndrome-causing RAF1 mutations were strongly correlated to hypertrophic cardiomyopathy. The latter was observed in 95% of Noonan syndrome patients, and seems to have an allele specificity that is associated with Ser259 and Ser612 mutations (P < .0001) [57].
In 2015, De Iriarte Rodrıguez et al proved that reduced levels of RAF1 in mice may lead to hearing impairments [61].
2.2.7. BRAF (V-Raf murine sarcoma viral oncogene homolog B1)
BRAF ([OMIM 164757]), a member of the RAF family, was proven to be involved in Noonan syndrome pathogenesis by enhancing ERK activation [55], [62]. However, the frequency of these mutations reported in a large cohort screening of Noonan syndrome patients were lower than those observed in RAF1, approximately 1.7–1.9%. These mutations cluster predominantly in conserved regions of CR1 and CR3 (Fig. 1F), but modestly increase kinase activity compared to RAF1 [34], [63].
2.2.8. MAP2K1 (mitogen activated protein kinase 1)
MAP2K1 ([OMIM 176872]) is mapped to the 15q22 region and comprises 11 exons encoding the MEK protein, which is a dual-specificity kinase with a major protein kinase domain that activates the extracellular-signal-regulated (Erk) mitogen-activated protein (MAP) kinases (Fig. 1G) [64].
Previous studies confirmed that MEK plays a crucial role in the embryonic developmental process, especially in cell migration and placental development [64]. In 2007, Nava et al identified MAP2K1 mutations in 4.2% of Noonan syndrome patients who are negative for PTPN11 and SO1 mutations [65]. This finding proved that mutations in the MEK protein kinase domain increase MEK basal kinase activity. This hyperactivity entails an increased phosphorylation of the downstream targets (such as ERK1 and ERK2) [66].
Recently, with the progress of molecular screening tools, whole genome sequencing (WGS) and whole exome sequencing (WES) have become increasingly used in the screening of developmental diseases. As a result, several novel Noonan syndrome-causing genes were highlighted in the last two years.
2.2.9. SOS2 (Son of Sevenless homolog 2)
In 2015, Yamamoto et al reported that SOS2 ([OMIM 601247]), the homolog of SOS1, is responsible of 4% of Noonan syndrome cases with no mutation in the genes previously associated with Noonan syndrome. SOS1 and SOS2 are 70% homologous. The reported mutations cluster in the DH domain, which plays a pivotal role in the stabilization of inactive conformation. Similar to SOS1, these mutations were associated with ectodermal defects [67].
2.2.10. LZTR1 (leucine-zipper-like transcription regulator 1)
In the same study, Yamamoto et al identified pathogenic mutations in a candidate gene not associated with the RAS/MAPK pathway, the LZTR1 gene ([OMIM 600574]). The prevalence of these mutations in the same studied cohort was 8% [67].
The LZTR1 gene is located in 22q11.21 and consists of 21 exons and encodes a protein member of the BTB-kelch superfamily. It was suggested that this gene may have a crucial role in the control of fundamental cellular processes (such as the cell cycle and the regulation of chromatin conformation) [68], [69].
However, it is worth mentioning that LZTR1 is already known to be associated with the Schwannomatosis, a form of neurofibromatosis [70].
2.2.11. A2ML1 (α-2-macroglobulin (A2M)-like-1)
Vissers et al carried out exome sequencing in one Noonan syndrome case-parent trio and found a de novo mutation affecting a highly conserved residue of A2ML1 ([OMIM 610627]), encouraging them to screen an additional cohort of 155 Noonan syndrome patients. This study showed the involvement of A2ML1 mutations in approximately 1% of Noonan syndrome subjects negative for the other NS major genes [71]. A2ML1 is a member of the α-macroglobulin superfamily, localized in 12p13 region with 35 coding exons. It acts as a protease inhibitor upstream of the MAPK pathway [72]. Mutations in A2ML1 are said to cause developmental defects, but the ways in which mutation affects the MAP kinase pathway needs to be further elucidated [71].
Other genes carrying rare variants in a Noonan syndrome population were also highlighted recently, in particular, RASA2, MAP3K8 and SPRY [73].
2.2.12. Other causes
Some studies have suggested other causes of some Noonan syndrome manifestations that are not related to a particular gene. For instance, wide-spaced nipples, cryptorchidism and some traits of facial dysmorphia such as hypertelorism, down slanting palpebral fissures, ptosis and low-set posteriorly rotated ears are suggested to be the result of tissue migration disruption or organ displacement caused by lymphedema at the intrauterine stage. Webbing of the neck and prominence of the trapezius may result from cystic hygroma at early intrauterine stages [74].
Recently, it was suggested that the impairment of testicular function responsible for delayed puberty or infertility is due to Sertoli and Leydig cell dysfunction rather than cryptorchidism [75].
The coagulation defect in Noonan syndrome was explained by the fact that the genes causing Noonan syndrome interact with the regulation process of the genes involved in the coagulation pathway [76].
2.3. Assessment of the mutation rate
In the second part of this review, we gathered and discussed mutational screening data reported in previous studies, to produce valid frequencies and authentic conclusions about mutational rates in Noonan syndrome ([NS, OMIM 163950]).
2.3.1. Search strategy
The data gathered were extracted from original articles published up to now (December 2015) in the “PubMed”, “ScienceDirect” and “Wiley Online Library” databases using the following key words:
-
-
“Noonan” in ([Abstract]).
-
-
“Noonan” in ([Title/Abstract]) AND “PTPN11” in ([Title]),
-
-
“Noonan” in ([Title/Abstract]) AND “SOS1” in ([Title]), and the same goes for the other genes (RAF1, KRAS, BRAF, NRAS and RIT1).
2.3.2. Inclusion criteria
From all of the articles gathered, we considered only studies comprising genetic screening of patient's series. We also considered clinical or endocrinological Noonan syndrome studies based initially on genetic screening. The genes considered in this part of the paper should have been screened by at least 3 studies meeting the inclusion criteria.
On the other hand, to determine the exact frequency of Noonan syndrome mutations, we excluded all data obtained from the screening of cohorts initially presenting with a particular condition (i.e., the mutational screening of Noonan syndrome patients with cardiac hypertrophy). The data gathered from the 82 included studies are summarized in Table 1 [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108].
Table 1.
Summary of the prevalence of Noonan syndrome-causing genes through previous studies.
| Genes | N° of mutated subjects/total of subjects (% of mutations) | % of gene in genetic etiology | First year of gene study | N° of studies | References |
|---|---|---|---|---|---|
| PTPN11 | 814/1917 (42.5%) | 52.6 | 2001 | 29 | [19], [24], [34], [35], [58], [77], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105] |
| SOS1 | 242/1472 (16.4%) | 20.3 | 2007 | 16 | [19], [34], [36], [39], [43], [58], [78], [79], [97], [98], [102], [103], [104], [105], [106], [107] |
| RIT1 | 31/383 (8%) | 10 | 2013 | 4 | [51], [53], [54], [73] |
| RAF1 | 76/961 (8%) | 10 | 2007 | 11 | [34], [55], [56], [57], [58], [78], [79], [98], [103], [105], [107] |
| KRAS | 31/1087 (2.8%) | 3.4 | 2006 | 11 | [46], [47], [48], [58], [65], [79], [89], [98], [102], [103], [105], [108] |
| BRAF | 19/815 (2.3%) | 2.8 | 2009 | 7 | [34], [63], [78], [98], [103], [105], [108] |
| NRAS | 10/1213 (0.8%) | 1 | 2010 | 4 | [50], [80], [81], [82] |
2.3.3. Mutational rate of Noonan syndrome-causing genes
Through the analyzed data (Table 1), we found that PTPN11 is the most studied gene in Noonan syndrome populations (29 studies vs. 16 studies or less for other genes). This may be because PTPN11 was the first gene of the RAS MAPK pathway to be highlighted in 2001, whereas the second gene involved (KRAS) was found 5 years later, followed by SHP-2. Indeed, the second gene, KRAS, was known beforehand to be involved in malignancy disorders through somatic mutations, while its germline mutations were associated for the first time with Noonan syndrome in 2006.
Then, in 2007, three genes were highlighted, (SOS1, RAF1 and MAP2K1), after which the other RAS/MAP kinase genes, BRAF, NRAS and RIT1, were proven to be involved in Noonan syndrome in more recent, in 2009, 2010 and 2013, respectively.
It should be noted that the chronology of the gene identification does not consistently occur in the “more involved” to “less involved” direction. Indeed, RIT1, which was discovered in 2013, was proven to be involved in 8% of cases. This frequency is much higher than the frequency of some genes (BRAF, KRAS and NRAS) that were found first, while their incidences do not exceed 3% (2.3%, 2.8% and 0.8%, respectively).
The first most common genetic cause of Noonan syndrome is still PTPN11 mutations that are observed in 42.5% of Noonan syndrome patients. Those mutations constitute 52.6% of all mutations detected up to now (Table 1).
The second most-involved gene is SOS1, which is altered in 16.4% of patients. On the other hand, we found that RIT1 and RAF1 have the same prevalence (8%) and are the third most-involved genes.
Taken together, PTPN11, SOS1, RAF1 and RIT1 cover 93% of reported mutations (Table 1). Therefore, these genes should be systematically considered in the genetic diagnosis of Noonan syndrome.
Among the RAS subfamily members of the RAS/MAP kinase pathway reported in Noonan syndrome, RIT1 seems to be the most involved gene compared with KRAS and NRAS, which have the lowest frequency among all reported cases (2.8% and 0.8%, respectively); however, among RAF family members, RAF1 is the most involved gene compared to BRAF (8% versus 2.3%, respectively). Among these two families, neither HRAS nor ARAF were reported to cause Noonan syndrome. These divergences should attract more attention from researchers.
It is worth mentioning that there were no mutations in the SHOC2, RRAS and CBL genes in Noonan syndrome patients. These genes were reported to be involved in Noonan-like syndrome (NLS) only.
2.4. Inheritance and genetic counseling
Noonan syndrome is a common disorder that could occur as a sporadic condition or can be inherited in an autosomal dominant pattern. In the latter case, the affected parent who transmits the disorder is predominantly the mother. This may be explained by the adverse impact of Noonan syndrome on male fertility [7], [13], [108]. However, rare autosomal recessive cases of Noonan syndrome with consanguineous parents have also been reported [109].
When there is molecular evidence in one Noonan syndrome child, the clinical and molecular evaluation of parents becomes primordial in the determination of recurrence risks. In Noonan syndrome-causing genes with heterozygous mutations, the recurrence risk in one genetically affected parent is 50% per each pregnancy, while in an unaffected parent with Noonan syndrome child, the recurrence risk in subsequent pregnancies is very low, approximately 1–5% owing to putative germline mosaicism.
The Noonan syndrome patient and their family should be made aware of the consequences of some Noonan syndrome features that may arise or complicate with adulthood, such as fertility problems in cryptorchidism cases and cardiac and hematological complications, which need rigorous follow-up [110].
The de-novo PTPN11 mutations are known to be more recurrent in Noonan syndrome. Tartaglia et al suggested that de-novo mutations have a predominantly paternal origin. They also demonstrated that the paternal average age in sporadic Noonan cases was significantly higher than that of the general population [111].
3. Conclusion
Noonan syndrome is a common multigenic disease. Its clinical side has been profoundly discussed by several researchers and diagnostic guides as support has been developed. This review completes the efforts of researchers focusing on the genetic etiology side of this syndrome and explains the physiopathological intervention of different genes in the manifestation of Noonan syndrome traits. In the second part, this paper provides pediatricians and geneticists with the mutational prevalence of genes involved in Noonan syndrome, which was deduced from the analysis of most studies carried out so far.
The result of this prevalence analysis leads to interesting conclusions, in particular, the high mutation rates observed in PTPN11, SOS1, RAF1 and RIT1, which cover 93% of reported mutations, suggesting that those genes should be systematically considered in the genetic diagnosis of Noonan syndrome. Secondly, we draw attention to the significant differences observed between mutational rates in genes of the same family (genes of the same level of the cascade). The explanation of this divergence and the other points discussed in this review may lead to new insights in the genetic research field of Noonan syndrome.
Conflict of interest
The authors have no conflict of interest to report.
Footnotes
Peer review under responsibility of King Faisal Specialist Hospital & Research Centre (General Organization), Saudi Arabia.
References
- 1.Duncan W.J., Fowler R.S., Farkas L.G., Ross R.B., Wright A.W., Bloom K.R. A comprehensive scoring system for evaluating Noonan syndrome. Am J Med Genet. 1981;10:37–50. doi: 10.1002/ajmg.1320100106. [DOI] [PubMed] [Google Scholar]
- 2.Mendez H.M., Opitz J.M. Noonan syndrome: a review. Am J Med Genet. 1985;21:493–506. doi: 10.1002/ajmg.1320210312. [DOI] [PubMed] [Google Scholar]
- 3.Sharland M., Burch M., McKenna W.M., Paton M.A. A clinical study of Noonan syndrome. Arch Dis Child. 1992;67:178–183. doi: 10.1136/adc.67.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allanson J.E. Noonan syndrome. In: Cassidy S.B., Allanson J.E., editors. Management of genetic syndromes. Wiley-Blackwell; 2010. pp. 569–586. [Google Scholar]
- 5.Allanson J.E., Hall J.G., Hughes H.E., Preus M., Witt R.D. Noonan syndrome: the changing phenotype. Am J Med Genet. 1985;21:507–514. doi: 10.1002/ajmg.1320210313. [DOI] [PubMed] [Google Scholar]
- 6.Patton M.A. Noonan syndrome: a review. Growth Genet Horm. 1994;33(10):1–3. [Google Scholar]
- 7.Allanson J.E. Noonan syndrome. J Med Genet. 1987;24:9–13. doi: 10.1136/jmg.24.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw A.C., Kalidas K., Crosby A.H., Jeffery S., Patton M.A. The natural history of Noonan syndrome: a long-term follow-up study. Arch Dis Child. 2007;92:128–132. doi: 10.1136/adc.2006.104547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pernot C., Marcon F., Worms A.M., Cloez J.L., Gilgenkrantz S., Marios L. La dysplasie cardio-vasculaire du syndrome de Noonan. Arch Mal Coeur. 1987;80:434–443. [PubMed] [Google Scholar]
- 10.Lin A.E. Noonan syndrome. J Med Genet. 1988;25:64–65. doi: 10.1136/jmg.25.1.64-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Cascos A. The Noonan syndrome. Eur Heart J. 1983;4:223–229. doi: 10.1093/oxfordjournals.eurheartj.a061452. [DOI] [PubMed] [Google Scholar]
- 12.Nora J.J., Nora A.H., Sinha A.K., Spangler R.D., Lubs H.A. The Ullrich-Noonan syndrome (Turner phenotype) Am J Dis Child. 1974;127:48–55. doi: 10.1001/archpedi.1974.02110200050007. [DOI] [PubMed] [Google Scholar]
- 13.Marcus K.A., Sweep C.G.J., van der Burgt I., Noordam C. Impaired Sertoli cell function in males diagnosed with Noonan syndrome. J Pediatr Endocrinol Metab. 2008;21:1079–1084. doi: 10.1515/jpem.2008.21.11.1079. [DOI] [PubMed] [Google Scholar]
- 14.Lee N.B., Kelly L., Sharland M. Ocular manifestations of Noonan syndrome. Eye. 1992;6:328–334. doi: 10.1038/eye.1992.66. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds D.J., Rubin S.E., Fox J., Kodsi S.R. Ocular manifestations of Noonan syndrome in the pediatric patient. J AAPOS. 2004;8:282–283. doi: 10.1016/j.jaapos.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 16.White S.W. Lymphedema in Noonan's syndrome. Int J Dermatol. 1984;23:656–657. doi: 10.1111/j.1365-4362.1984.tb01226.x. [DOI] [PubMed] [Google Scholar]
- 17.Bader-Meunier B., Tchernia G., Mielot F., Fontaine J.L., Thomas C., Lyonnet S. Occurrence of myeloproliferative disorder in patients with Noonan syndrome. J Pediatr. 1997;130:885–889. doi: 10.1016/s0022-3476(97)70273-7. [DOI] [PubMed] [Google Scholar]
- 18.Cohen M.M., Gorlin R.J. Noonan-like/multiple giant cell lesion syndrome. Am J Med Genet. 1991;40:159–166. doi: 10.1002/ajmg.1320400208. [DOI] [PubMed] [Google Scholar]
- 19.Pierpont E.I., Pierpont M.E., Mendelsohn N.J., Roberts A.E., Tworog-Dube E., Seidenberg M.S. Genotype differences in cognitive functioning in Noonan syndrome. Genes Brain Behav. 2009;8:275–282. doi: 10.1111/j.1601-183X.2008.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamieson C.R., van der Burgt I., Brady A.F., van Reen M., Elsawi M.M., Hol F. Mapping a gene for Noonan syndrome to the long arm of chromosome 12. Nat Gene. 1994;8:357–360. doi: 10.1038/ng1294-357. [DOI] [PubMed] [Google Scholar]
- 21.Brady A.F., Jamieson C.R., van der Burgt I., Crosby A., van Reen M., Kremer H. Further delineation of the critical region for Noonan syndrome on the long arm of chromosome 12. Eur J Hum Genet. 1997;5:336–337. [PubMed] [Google Scholar]
- 22.Legius E., Schollen E., Matthijs G., Fryns J.P. Fine mapping of Noonan/cardio-facio cutaneous syndrome in a large family. Eur J Hum Genet. 1998;6:32–37. doi: 10.1038/sj.ejhg.5200150. [DOI] [PubMed] [Google Scholar]
- 23.Chen B., Bronson R.T., Klaman L.D., Hampton T.G., Wang J.F., Green P.J. Mice mutants for Egfr and Shp2 have defective cardiac semilunar valvulogenesis. Nat Genet. 2000;24:296–299. doi: 10.1038/73528. [DOI] [PubMed] [Google Scholar]
- 24.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 25.Feng G.S. Shp-2 tyrosine phosphatase: signaling one cell or many. Exp Cell Res. 1999;253:47–54. doi: 10.1006/excr.1999.4668. [DOI] [PubMed] [Google Scholar]
- 26.Stein-Gerlach M., Wallasch C., Ullrich A. SHP-2, SH2-containing protein tyrosine phosphatase-2. Int J Biochem Cell Biol. 1998;30:559–566. doi: 10.1016/s1357-2725(98)00002-8. [DOI] [PubMed] [Google Scholar]
- 27.Saxton T.M., Ciruna B.G., Holmyard D., Kulkarni S., Harpal K., Rossant J. The SH2 tyrosine phosphatase shp2 is required for mammalian limb development. Nat Genet. 2000;24:420–423. doi: 10.1038/74279. [DOI] [PubMed] [Google Scholar]
- 28.Qu C.K., Yu W.M., Azzarelli B., Cooper S., Broxmeyer H.E., Feng G.S. Biased suppression of hematopoiesis and multiple developmental defects in chimeric mice containing Shp-2 mutant cells. Mol Cell Biol. 1998;18:6075–6082. doi: 10.1128/mcb.18.10.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang T.L., Freeman R.M., Jr., O'Reilly A.M., Neel B.G., Sokol S.Y. The SH2-containing protein-tyrosine phosphatase SH-PTP2 is required upstream of MAP kinase for early Xenopus development. Cell. 1995;80:473–483. doi: 10.1016/0092-8674(95)90498-0. [DOI] [PubMed] [Google Scholar]
- 30.Neel B.G., Gu H., Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 31.Tamir I., Dal Porto J.M., Cambier J.C. Cytoplasmic protein tyrosine phosphatase SHP-1 and SHP-2: regulators of B cell signal transduction. Curr Opin Immunol. 2000;12:307–315. doi: 10.1016/s0952-7915(00)00092-3. [DOI] [PubMed] [Google Scholar]
- 32.De Rocca S.N.A., Edouard T., Tréguer K., Tajan M., Araki T., Dance M. Noonan syndrome-causing SHP2 mutants inhibit insulin-like growth factor 1 release via growth hormone-induced ERK hyperactivation, which contributes to short stature. Proc Natl Acad Sci U S A. 2012;109:4257–4262. doi: 10.1073/pnas.1119803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hof P., Pluskey S., Dhe-Paganon S., Eck M.J., Shoelson S.E. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 34.Lee B.H., Kim J.M., Jin H.Y., Kim G.H., Choi J.H., Yoo H.W. Spectrum of mutations in Noonan syndrome and their correlation with phenotypes. J Pediatr. 2011;159:1029–1035. doi: 10.1016/j.jpeds.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 35.Tartaglia M., Kalidas K., Shaw A., Song X., Musat D.L., van der Burgt I. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70:1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts A.E., Araki T., Swanson K.D., Montgomery K.T., Schiripo T.A., Joshi V.A. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 37.Hart T.C., Zhang Y., Gorry M.C., Hart P.S., Cooper M., Marazita M.L. A mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1. Am J Hum Genet. 2002;70:943–954. doi: 10.1086/339689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb G.C., Jenkins N.A., Largaespada D.A., Copeland N.G., Fernandez C.S., Bowtell D.D. Mammalian homologues of the Drosophila Son of sevenless gene map to murine chromosomes 17 and 12 and to human chromosomes 2 and 14. Genomics. 1993;18:14–19. doi: 10.1006/geno.1993.1421. [DOI] [PubMed] [Google Scholar]
- 39.Lepri F., De Luca A., Stella L., Rossi C., Baldassarre G., Pantaleoni F. SOS1 mutations in Noonan syndrome: molecular spectrum, structural insights on pathogenic effects, and genotype-phenotype correlations. Hum Mut. 2011;32:760–772. doi: 10.1002/humu.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zenker M., Horn D., Wieczorek D., Allanson J., Pauli S., van der Burgt I. SOS1 is the second most common Noonan gene but plays no major role in cardio-facio-cutaneous syndrome. J Med Genet. 2007;44:651–656. doi: 10.1136/jmg.2007.051276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nimnual A., Bar-Sagi D. The two hats of SOS. Sci STKE. 2002;145:36. doi: 10.1126/stke.2002.145.pe36. [DOI] [PubMed] [Google Scholar]
- 42.Sondermann H., Soisson S.M., Boykevisch S., Yang S.S., Bar-Sagi D., Kuriyan J. Structural analysis of autoinhibition in the Ras activator Son of Sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Tartaglia M., Pennacchio L.A., Zhao C., Yadav K.K., Fodale V., Sarkozy A. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 44.Donovan S., Shannon K.M., Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- 45.Vetter I.R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 46.Carta C., Pantaleoni F., Bocchinfuso G., Stella L., Vasta I., Sarkozy A. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubbert S., Zenker M., Rowe S.L., Böll S., Klein C., Bollag G. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 48.Zenker M., Lehmann K., Schulz A.L., Barth H., Hansmann D., Koenig R. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2007;44:131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell E.L., Jones D., White G.R., Varley J.M., Santibanez Koref M.F. Determination of the gene order of the three loci CD2, NGFB, and NRAS at human chromosome band 1p13 and refinement of their localisation at the subband level by fluorescence in situ hybridization. Cytogenet Cell Genet. 1995;70:183–185. doi: 10.1159/000134028. [DOI] [PubMed] [Google Scholar]
- 50.Cirstea I.C., Kutsche K., Dvorsky R., Gremer L., Carta C., Horn D. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat Genet. 2010;42:27–29. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aoki Y., Niihori T., Banjo T., Okamoto N., Mizuno S., Kurosawa K. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am J Hum Genet. 2013;93:173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hynds D.L., Spencer M.L., Andres D.A., Snow D.M. Rit promotes MEK-independent neurite branching in human neuroblastoma cells. J Cell Sci. 2003;116:1925–1935. doi: 10.1242/jcs.00401. [DOI] [PubMed] [Google Scholar]
- 53.Bertola D.R., Yamamoto G.L., Almeida T.F., Buscarilli M., Jorge A.A.L., Malaquias A.C. Further evidence of the importance of RIT1 in noonan syndrome. Am J Med Genet Part A. 2014;9999:1–6. doi: 10.1002/ajmg.a.36722. [DOI] [PubMed] [Google Scholar]
- 54.Gos M., Fahiminiya S., Poznański J., Klapecki J., Obersztyn E., Piotrowicz M. Contribution of RIT1 mutations to the pathogenesis of Noonan syndrome: four new cases and further evidence of heterogeneity. Am J Med Genet Part A. 2014;9999:1–7. doi: 10.1002/ajmg.a.36646. [DOI] [PubMed] [Google Scholar]
- 55.Razzaque M.A., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi T., Aoki Y., Niihori T., Cavé H., Verloes A., Okamoto N. Molecular and clinical analysis of RAF1 in Noonan syndrome and related disorders: dephosphorylation of serine 259 as the essential mechanism for mutant activation. Hum Mutat. 2010;31:284–294. doi: 10.1002/humu.21187. [DOI] [PubMed] [Google Scholar]
- 57.Pandit B., Sarkozy A., Pennacchio L.A., Carta C., Oishi K., Martinelli S. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 58.Ko J.M., Kim J.M., Kim G.H., Yoo H.W. PTPN11, SOS1, KRAS, and RAF1 gene analysis, and genotype-phenotype correlation in Korean patients with Noonan syndrome. J Hum Genet. 2008;53:999–1006. doi: 10.1007/s10038-008-0343-6. [DOI] [PubMed] [Google Scholar]
- 59.Cutler R.E., Stephens R.M., Saracino M.R., Morrison D.K. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci U S A. 1998;95(16):9209–9214. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer A., Baljuls A., Reinders J., Nekhoroshkova E., Sibilski C., Metz R. Regulation of RAF activity by 14-3-3 proteins: RAF kinases associate functionally with both homo- and heterodimeric forms of 14-3-3 proteins. J Biol Chem. 2009;284(5):3183–3194. doi: 10.1074/jbc.M804795200. [DOI] [PubMed] [Google Scholar]
- 61.de Iriarte Rodriguez R., Magarinos M., Pfeiffer V., Rapp U.R. Varela-Nieto I.C-Raf deficiency leads to hearing loss and increased noise susceptibility. Cell Mol Life Sci. 2015;72(20):3983–3998. doi: 10.1007/s00018-015-1919-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wan P.T., Garnett M.J., Roe S.M., Lee S., Niculescu-Duvaz D., Good V.M. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 63.Sarkozy A., Carta C., Moretti S., Zampino G., Digilio M.C., Pantaleoni F. Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum Mutat. 2009;30:695–702. doi: 10.1002/humu.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giroux S., Tremblay M., Bernard D., Cardin-Girard J.F., Aubry S., Larouche L. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr Biol. 1999;9(7):369–376. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 65.Nava C., Hanna N., Michot C., Pereira S., Pouvreau N., Niihori T. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: genotypephenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delaney A.M., Printen J.A., Chen H., Fauman E.B., Dudley D.T. Identification of a novel mitogen-activated protein kinase kinase activation domain recognized by the inhibitor PD 184352. Mol Cell Biol. 2002;22:7593–7602. doi: 10.1128/MCB.22.21.7593-7602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto G.L., Aguena M., Gos M., Hung C., Pilch J., Fahiminiya S. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J Med Genet. 2015;52(6):413–421. doi: 10.1136/jmedgenet-2015-103018. [DOI] [PubMed] [Google Scholar]
- 68.Perez-Torrado R., Yamada D., Defossez P.A. Born to bind the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 69.Nacak T.G., Leptien K., Fellner D., Augustin H.G., Kroll J. The BTB-kelch protein LZTR-1 is a novel Golgi protein that is degraded upon induction of apoptosis. J Biol Chem. 2006;281:5065–5071. doi: 10.1074/jbc.M509073200. [DOI] [PubMed] [Google Scholar]
- 70.Piotrowski A., Xie J., Liu Y.F., Poplawski A.B., Gomes A.R., Madanecki P. Germline loss-of-function mutations in LZTR1 predispose to an inherited disorder of multiple schwannomas. Nat Genet. 2014;46:182–187. doi: 10.1038/ng.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vissers L.E., Bonetti M., Paardekooper O.J., Nillesen W.M., Frints S.G., de Ligt J. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur J Hum Genet. 2015;23(3):317–324. doi: 10.1038/ejhg.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galliano M.F., Toulza E., Gallinaro H., Jonca N., Ishida-Yamamoto A., Serre G. A novel protease inhibitor of the alpha2-macroglobulin family expressed in the human epidermis. J Biol Chem. 2006;281:5780–5789. doi: 10.1074/jbc.M508017200. [DOI] [PubMed] [Google Scholar]
- 73.Chen P.C., Yin J., Yu H.W., Yuan T., Fernandez M., Yung C.K. Next-generation sequencing identifies rare variants associated with Noonan syndrome. Proc Natl Acad Sci U S A. 2014;111:11473–11478. doi: 10.1073/pnas.1324128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Witt D.R., Hoyme H.E., Zonana J., Manchester D.K., Fryns J.P., Stevenson J.G. Clues to pathogenesis and prenatal diagnosis and review of the literature. Am J Med Genet. 1987;27(4):841–856. doi: 10.1002/ajmg.1320270412. [DOI] [PubMed] [Google Scholar]
- 75.Ankarberg-Lindgren C., Westphal O., Dahlgren J. Testicular size development and reproductive hormones in boys and adult males with Noonan syndrome: a longitudinal study. Eur J Endocrinol. 2011;165:137–144. doi: 10.1530/EJE-11-0092. [DOI] [PubMed] [Google Scholar]
- 76.Bertola D.R., Carneiro J.D., D'Amico E.A., Kim C.A., Albano L.M., Sugayama S.M. Hematological findings in Noonan syndrome. Rev Hosp Clin Fac Med Sao Paulo. 2003;58(1):5–8. doi: 10.1590/s0041-87812003000100002. [DOI] [PubMed] [Google Scholar]
- 77.Sznajer Y., Keren B., Baumann C., Pereira S., Alberti C., Elion J. The spectrum of cardiac anomalies in noonan syndrome as a result of mutations in the PTPN11 gene. Pediatrics. 2007;119(6):1325–1331. doi: 10.1542/peds.2006-0211. [DOI] [PubMed] [Google Scholar]
- 78.Denayer E., Devriendt K., de Ravel T., Van Buggenhout G., Smeets E., Francois I. Tumor spectrum in children with Noonan syndrome and SOS1 or RAF1 mutations. Genes Chromosomes Cancer. 2010;49(3):242–252. doi: 10.1002/gcc.20735. [DOI] [PubMed] [Google Scholar]
- 79.Croonen E.A., Nillesen W., Schrander C., Jongmans M., Scheffer H., Noordam C. Comparing mutation-positive with mutation-negative Dutch patients. Mol Syndromol. 2013;4:227–234. doi: 10.1159/000350686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Runtuwene V., van Eekelen M., Overvoorde J., Rehmann H., Yntema H.G., Nillesen W.M. Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis Model Mech. 2011;4:393–399. doi: 10.1242/dmm.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Denayer E., Peeters H., Sevenants L., Derbent M., Fryns J.P., Legius E. NRAS mutations in Noonan syndrome. Mol Syndromol. 2012;3:34–38. doi: 10.1159/000338467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kraoua L., Journel H., Bonnet P., Amiel J., Pouvreau N., Baumann C. Constitutional NRAS mutations are rare among patients with Noonan syndrome or juvenile myelomonocytic leukemia. Am J Med Genet Part A. 2012;158A:2407–2411. doi: 10.1002/ajmg.a.35513. [DOI] [PubMed] [Google Scholar]
- 83.Kosaki K., Suzuki T., Muroya K., Hasegawa T., Sato S., Matsuo N. PTPN11 (proteintyrosine phosphatase, nonreceptor-type 11) mutations in seven Japanese patients with Noonan syndrome. J Clin Endocrinol Metab. 2002;87:3529–3533. doi: 10.1210/jcem.87.8.8694. [DOI] [PubMed] [Google Scholar]
- 84.Musante L., Kehl H.G., Majewski F., Meinecke P., Schweiger S., Gillessen-Kaesbach G. Spectrum of mutations in PTPN11 and genotype-phenotype correlation in 96 patients with Noonan syndrome and five patients with cardio-facio-cutaneous syndrome. Eur J Hum Genet. 2003;11:201–206. doi: 10.1038/sj.ejhg.5200935. [DOI] [PubMed] [Google Scholar]
- 85.Sarkozy A., Conti E., Seripa D., Digilio M.C., Grifone N., Tandoi C. Correlation between PTPN11 gene mutations and congenital heart defects in Noonan and LEOPARD syndromes. J Med Genet. 2003;40:704–708. doi: 10.1136/jmg.40.9.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zenker M., Buheitel G., Rauch R., Koenig R., Bosse K., Kress W. Genotype-phenotype correlations in Noonan syndrome. J Pediatr. 2004;144:368–374. doi: 10.1016/j.jpeds.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 87.Yoshida R., Hasegawa T., Hasegawa Y., Nagai T., Kinoshita E., Tanaka Y. Protein-tyrosine phosphatase, nonreceptor type 11 mutation analysis and clinical assessment in 45 patients with Noonan syndrome. J Clin Endocrinol Metab. 2004;89:3359–3364. doi: 10.1210/jc.2003-032091. [DOI] [PubMed] [Google Scholar]
- 88.Niihori T., Aoki Y., Ohashi H., Kurosawa K., Kondoh T., Ishikiriyama S. Functional analysis of PTPN11/SHP-2 mutants identified in Noonan syndrome and childhood leukemia. J Hum Genet. 2005;50(4):192–202. doi: 10.1007/s10038-005-0239-7. [DOI] [PubMed] [Google Scholar]
- 89.Ferreira L.V., Souza S.A., Arnhold I.J., Mendonca B.B., Jorge A.A. PTPN11 (protein tyrosine phosphatase, nonreceptor type 11) mutations and response to growth hormone therapy in children with Noonan syndrome. J Clin Endocrinol Metab. 2005;90:5156–5160. doi: 10.1210/jc.2004-2559. [DOI] [PubMed] [Google Scholar]
- 90.Binder G., Neuer K., Ranke M.B., Wittekindt N.E. PTPN11 mutations are associated with mild growth hormone resistance in individuals with Noonan syndrome. J Clin Endocrinol Metab. 2005;90:5377–5381. doi: 10.1210/jc.2005-0995. [DOI] [PubMed] [Google Scholar]
- 91.Jongmans M., Sistermans E.A., Rikken A., Nillesen W.M., Tamminga R., Patton M. Genotypic and phenotypic characterization of Noonan syndrome: new data and review of the literature. Am J Med Genet Part A. 2005;134A:165–170. doi: 10.1002/ajmg.a.30598. [DOI] [PubMed] [Google Scholar]
- 92.Bertola D.R., Pereira A.C., Albano L.M.J., De Oliveira P.S.L., Kim C.A., Krieger J.E. PTPN11 gene analysis in 74 Brazilian patients with Noonan syndrome or Noonan-like phenotype. Genet Test. 2006;10:186–191. doi: 10.1089/gte.2006.10.186. [DOI] [PubMed] [Google Scholar]
- 93.Lee S.T., Ki C.S., Lee H.J. Mutation analysis of the genes involved in the Ras-mitogen-activated protein kinase (MAPK) pathway in Korean patients with Noonan syndrome. Clin Genet. 2007;72:150–155. doi: 10.1111/j.1399-0004.2007.00839.x. [DOI] [PubMed] [Google Scholar]
- 94.Hung C.S., Lin J.L., Lee Y.J., Lin S.P., Chao M.C., Lo F.S. Mutational analysis of PTPN11 gene in Taiwanese children with Noonan syndrome. J Formos Med Assoc. 2007;106(2):169–172. doi: 10.1016/S0929-6646(09)60235-7. [DOI] [PubMed] [Google Scholar]
- 95.Ferreira L.V., Souza S.A., Montenegro L.R., Arnhold I.J., Pasqualini T., Heinrich J.J. Variabilidade do fenótipo de pacientes com síndrome de Noonan com e sem mutações no gene PTPN11. Arq Bras Endocrinol Metab. 2007;51(3):450–456. doi: 10.1590/s0004-27302007000300014. [DOI] [PubMed] [Google Scholar]
- 96.Croonen E.A., van der Burgt I., Kapusta L., Draaisma J.M.T. Electrocardiography in Noonan syndrome PTPN11 gene mutation-phenotype characterization. Am J Med Genet Part A. 2008;146A:350–353. doi: 10.1002/ajmg.a.32140. [DOI] [PubMed] [Google Scholar]
- 97.Ferrero G.B., Baldassarre G., Delmonaco A.G., Biamino E., Banaudi E., Carta C. Clinical and molecular characterization of 40 patients with Noonan syndrome. Eur J Med Genet. 2008;51:566–572. doi: 10.1016/j.ejmg.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 98.Pierpont E.I., Pierpont M.E., Mendelsohn N.J., Roberts A.E., Tworog-Dube E., Rauen K.A. Effects of germline mutations in the Ras/MAPK signaling pathway on adaptive behavior: cardiofaciocutaneous syndrome and Noonan syndrome. Am J Med Genet Part A. 2010;152A:591–600. doi: 10.1002/ajmg.a.33268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brasil A.S., Pereira A.C., Wanderley L.T., Kim C.A., Malaquias A.C., Jorge A.A. PTPN11 and KRAS gene analysis in patients with Noonan and Noonan-like syndromes. Gent Test Mol Biomark. 2010;14(3):425–432. doi: 10.1089/gtmb.2009.0192. [DOI] [PubMed] [Google Scholar]
- 100.Derbent M., Oncel Y., Tokel K., Varan B., Haberal A., Yazıcı A.C. Clinical and hematologic findings in Noonan syndrome patients with PTPN11 gene mutations. Am J Med Genet Part A. 2010;152A:2768–2774. doi: 10.1002/ajmg.a.33713. [DOI] [PubMed] [Google Scholar]
- 101.Papadopoulou A., Issakidis M., Gole E., Kosma K., Fryssira H., Fretzayas A. Phenotypic spectrum of 80 Greek patients referred as Noonan syndrome and PTPN11 mutation analysis: the value of initial clinical assessment. Eur J Pediatr. 2012;171:51–58. doi: 10.1007/s00431-011-1487-5. [DOI] [PubMed] [Google Scholar]
- 102.Choi J.H., Lee B.H., Jung C.W., Kim Y.M., Jin H.Y., Kim J.M. Response to growth hormone therapy in children with noonan syndrome: correlation with or without PTPN11 gene mutation. Horm Res Paediatr. 2012;77:388–393. doi: 10.1159/000339677. [DOI] [PubMed] [Google Scholar]
- 103.Razzaque M.A., Komoike Y., Nishizawa T., Inai K., Furutani M., Higashinakagawa Characterization of a novel KRAS mutation identified in Noonan syndrome. Am J Med Genet Part A. 2012;158A:524–532. doi: 10.1002/ajmg.a.34419. [DOI] [PubMed] [Google Scholar]
- 104.Şimşek-Kiper P.Ö., Alanay Y., Gülhan B., Lissewski C., Türkyilmaz D., Alehan D. Clinical and molecular analysis of RASopathies in a group of Turkish patients. Clin Genet. 2013;83(2):181–186. doi: 10.1111/j.1399-0004.2012.01875.x. [DOI] [PubMed] [Google Scholar]
- 105.Prendiville T.W., Gauvreau K., Tworog-Dube E., Patkin L., Kucherlapati R.S., Roberts A.E. Cardiovascular disease in Noonan syndrome. Arch Dis Child. 2014;99(7):629–634. doi: 10.1136/archdischild-2013-305047. [DOI] [PubMed] [Google Scholar]
- 106.Narumi Y., Aoki Y., Niihori T., Sakurai M., Cavé H., Verloes A. Clinical manifestations in patients with SOS1 mutations range from Noonan syndrome to CFC syndrome. J Hum Genet. 2008;53:834–841. doi: 10.1007/s10038-008-0320-0. [DOI] [PubMed] [Google Scholar]
- 107.Longoni M., Moncini S., Cisternino M., Morella I.M., Ferraiuolo S., Russo S. Noonan syndrome associated with both a new Jnk-activating familial SOS1 and a de novo RAF1 mutations. Am J Med Genet Part A. 2010;152A:2176–2184. doi: 10.1002/ajmg.a.33564. [DOI] [PubMed] [Google Scholar]
- 108.Elsawi M.M., Pryor J.P., Klufio G., Barnes C., Patton M.A. Genital-tract function in men with Noonan syndrome. J Med Genet. 1994;31:468–470. doi: 10.1136/jmg.31.6.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van der Burgt I., Brunner H. Genetic heterogeneity in Noonan syndrome: evidence for an autosomal recessive form. Am J Med Genet. 2000;94:46–51. doi: 10.1002/1096-8628(20000904)94:1<46::aid-ajmg10>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 110.van der Burgt I. Noonan syndrome. Orphanet J Rare Dis. 2007;14:2–4. doi: 10.1186/1750-1172-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tartaglia M., Cordeddu V., Chang H., Shaw A., Kalidas K., Crosby A. Paternal germline origin and sex-ratio distortion in transmission of PTPN11 mutations in Noonan syndrome. Am J Hum Genet. 2004;75:492–497. doi: 10.1086/423493. [DOI] [PMC free article] [PubMed] [Google Scholar]