Abstract
Drug abuse is a multifaceted disorder that involves maladaptive decision making. Long-lasting changes in the addicted brain are mediated by a complex circuit of brain reward regions. The prefrontal cortex (PFC) is one region in which chronic drug exposure changes expression and function of upstream transcriptional regulators to alter drug responses and aspects of the addicted phenotype. We reported recently that the transcription factor E2F3a is a critical mediator of cocaine responses in the nucleus accumbens. E2F3a is one of two splice variants of the E2f3 gene; the other is E2F3b. Another recent study predicted E2F3 as an upstream regulator of the transcriptional response to cocaine self-administration (SA) in PFC. Based on previous findings that E2F3a and E2F3b have divergent regulatory roles, we set out to study the putative transcriptional role of these transcripts in PFC in the context of repeated I.P. cocaine exposure. We implemented viral-mediated isoform-specific gene manipulation, RNA-sequencing, advanced bioinformatics analyses, and animal behavior to determine how E2F3a and E2F3b contribute to persistent cocaine-induced transcriptional changes in PFC. We show that E2F3b, but not E2F3a, in PFC is critical for cocaine locomotor and place preference behaviors. Interestingly, RNA-seq of PFC following E2f3b overexpression or I.P. cocaine exposure showed very different effects on expression levels of differentially expressed genes. However, we found that E2F3b drives a similar transcriptomic pattern to that of cocaine SA with overlapping upstream regulators and downstream pathways predicted. These findings reveal a novel transcriptional mechanism in PFC that controls behavioral and molecular responses to cocaine.
Subject terms: Motivation, Epigenetics and behaviour
Introduction
A hallmark of cocaine addiction is the devaluing of other typically rewarding stimuli, such as food, sex, and social interaction, which leads to enhanced drug seeking. These behavioral abnormalities are mediated in part by impairment of the prefrontal cortex (PFC) [1]. The PFC enhances consolidation of reward memory through the overvaluation of specific cues associated with the drug, and sensitization seen in rodents accelerates this consolidation partly through enhanced dopaminergic and glutamatergic signaling [2–4]. Underlying at least some of these functional changes are persistent alterations in gene expression, however, little attention has been paid to transcriptional regulation in PFC as compared with nucleus accumbens (NAc) and other reward regions [5–7].
Recently, we found that repeated cocaine exposure increases nuclear levels of the transcription factor, E2F3a, in NAc and that E2f3a overexpression in this brain region enhances behavioral responses to cocaine via regulation of both gene transcription and alternative splicing [8]. The other E2f3 isoform, E2F3b, was not affected by cocaine in NAc, where it also had no behavioral effect. E2F3a and E2F3b share the same DNA binding, nuclear localization, and transactivation domains, but E2F3b lacks an N-terminus ubiquitin targeting domain. Differing roles for E2F3a and E2F3b are reported depending on the cell or tissue type [9, 10]. Furthermore, a recent study found E2F3 as a predicted upstream regulator via motif analysis in PFC following cocaine self-administration (SA) + withdrawal and cocaine/context re-exposure [11]. Thus, we hypothesized that one or both E2F3 isoforms might mediate some of cocaine’s effects in PFC as well. We investigated both isoforms to uncover possible isoform and regional specificity.
We utilized a combination of animal behavior, viral-mediated gene transfer, RNA-sequencing (RNA-seq), and bioinformatic analyses to address the hypothesis that E2F3a and/or E2F3b contribute to PFC modulation of cocaine-elicited behaviors and that this regulation is driven by transcriptional and/or splicing regulation. We first tested the effect of overexpression or knockdown of either E2F3a or E2F3b on cocaine behaviors. Next, we utilized RNA-seq to compare differential gene expression after either repeated intraperitoneal (I.P.) cocaine exposure or E2F3b overexpression. The RNA-seq results showed that E2F3b overexpression in PFC exerted very different effects on gene expression as I.P. cocaine, but more closely resembled the transcriptomic signature of cocaine self-administration (SA) in this brain region.
Materials and methods
Animals
C57BL/6J male mice (Jackson), 7–8 weeks old and weighing 25–30 g, were habituated to the animal facility 1 week before use and maintained at 22–25 °C on a 12-h light/dark cycle. All animals had access to food and water ad libitum. All experiments were conducted in accordance with the guidelines of Mount Sinai’s IACUC.
Repeated cocaine injection
For RNA-seq, mice were injected I.P. with saline or cocaine (20 mg/kg) once daily for 5 days 48 h after stereotactic surgery to allow the animals to recover per standard procedures [12]. Mice were euthanized 24 h after the final cocaine injection.
Viral-mediated gene transfer
Mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg) and prepared for stereotactic surgery. Thirty-three gauge syringe needles (Hamilton) were used to bilaterally infuse 0.5 µL virus in PFC at a rate of 0.1 µL/min at 1.8 mm anterior, 0.75 mm lateral, and 2.7 mm ventral from Bregma at a 15° angle. We used bicistronic p1005 Herpes simplex virus (HSV) vectors expressing green fluorescent protein (GFP) alone or GFP plus E2F3a, E2F3b, miR targeting E2f3b mRNA, or miR targeting LacZ mRNA (Table 1).
Table 1.
miR target and hairpin sequences
| Target | Target sequence | Top strand | Bottom strand |
|---|---|---|---|
| E2f3b |
CTTACAGCAGC AGGCAAAGCG |
TGCTGCGCTTTGCCTGCTGCT GTAAGGTTTTGGCCACTGACTG ACCTTACAGCCAGGCAAAGCG |
CCTGCGCTTTGCCTGGCTGTAA GGTCAGTCAGTGGCCAAAACCT TACAGCAGCAGGCAAAGCGC |
| LacZ |
GACTACACAAA TCAGCGATTT |
TGCTGAAATCGCTGATTTGTGT AGTCGTTTTGGCCACTGACTGA CGACTACACATCAGCGATTT |
CCTGAAATCGCTGATGTGTAGT CGTCAGTCAGTGGCCAAAACGA CTACACAAATCAGCGATTTC |
In this system, GFP expression is driven by a cytomegalovirus (CMV) promoter, whereas the transgene of interest is driven by the IE4/5 promoter [13]. Viral expression was confirmed during tissue collection using fluorescence microscopy (Leica) to visualize GFP and confirm PFC targeting.
Tissue collection
Tissue was collected after pharmacological or viral manipulation via cervical dislocation followed by dissection of bilateral PFC punches with a 14-gauge needle for viral studies to ensure only virally infected tissue was dissected. Tissue was immediately frozen on dry ice. Single animal samples were used in RNA-seq.
Behavioral assays
Locomotor activity was measured per published protocols [14] with minor modifications. Activity was assessed in the x- and y-planes for horizontal ambulation in a 75 cm−2 chamber using EthoVision XT (Noldus). In overexpression experiments, 48 h after surgery (denoted as Day 0, Fig. 1a) animals were injected I.P. with saline and placed in the locomotor chamber for 15 min, then removed from the chamber, injected with saline again, and returned to the locomotor chamber where activity was measured for 45 min. Three to seven days after surgery (Days 1–5, Fig. 1a) animals were injected with saline and placed in the cage for 15 min, then removed and injected with cocaine (5 mg/kg) and monitored in the locomotor chamber for 45 min. In knockdown experiments, animals underwent the above manipulations starting 4 days after surgery to allow for enough time for E2F3b knockdown prior to beginning behavioral manipulation, using 7.5 mg/kg cocaine (Fig. 1g). Total number of beam breaks were measured and used to assess locomotor activity during the saline and cocaine periods.
Fig. 1.
E2F3b expression in PFC is integral to cocaine-elicited behaviors. a HSV-GFP, HSV-E2F3a, or HSV-E2F3b was infused into PFC. Forty-eight hours post-infusion, mice were tested over 6 days for cocaine (5 mg/kg)-induced locomotor activity. b No effect of virus was observed in mice overexpressing E2F3a when compared with GFP (no main effect of virus, via two-way ANOVA F1, 54 = 2.07, p = 0.1562). c Increase in locomotor response to cocaine across testing days was seen in mice overexpressing E2F3b in PFC (two-way ANOVA, interaction of virus and day F5, 75 = 6.31, p < 0.0001), with significant increases in locomotor response confirmed by post-hoc analysis on days 4 and 5 (Day 4: t14 = 2.869, *p < 0.05; Day 3: t14 = 3.853, **p < 0.01). d HSV-GFP, HSV-E2F3a, or HSV-E2F3a was infused into PFC and 48 h post-infusion, saline or cocaine (5 mg/kg) pairing began. e E2F3a had no effect on cocaine CPP (no main effect by two-way ANOVA (F1, 30 = 0.5579, p = 0.5579). f E2F3b increased preference for cocaine compared with GFP (t8 = 3.113, *p < 0.05). g HSV-miR-LacZ or HSV-miR-E2f3b was infused into PFC. Five days later, at peak knockdown, tissue was collected and RNA isolated. h HSV-miR-E2f3b caused an approximately 70% knockdown of E2f3b but not E2f3a mRNA. i HSV-miR-LacZ or HSV-miR-E2f3b was infused into PFC. Ninety-six hours post-infusion, mice were tested over 6 days for cocaine. j Decrease in locomotor response to cocaine across testing days was seen in mice expressing miR-E2f3b in PFC (two-way ANOVA, interaction of virus and day, F5, 89 = 3.31, p = 0.0087), with significant decrease in locomotor response confirmed by post-hoc analysis on days 3, 4, and 5 (Day 3: t17 = 3.323, **p<0.01; Day 4: t16 = 3.863, **p < 0.01; Day 5: t14 = 3.569, **p < 0.01). k HSV-miR-LacZ or HSV-miR-E2f3b was infused into PFC. At 120 h post-infusion, saline or cocaine (7.5 mg/kg) pairing began. l E2F3b knockdown decreased cocaine preference (interaction of virus and test observed by two-way ANOVA: F1, 7 = 6.4, p = 0.0393; miR-E2f3b Test vs. Pre-test: t6 = 0.337, p > 0.05; miR-LacZ Test vs. Pre-test: t7 = 3.417, *p < 0.05)
Cocaine conditioned place preference (CPP), using an unbiased procedure, was conducted as described [15]. A pre-test was performed 72 h after surgery (Day 4, Fig. 1d) for overexpression experiments, or 120 h (Day 6, Fig. 1i) for knockdown experiments. Mice that showed significant baseline preference for either chamber were excluded from the study (<5% of all mice), and conditioning groups were balanced to adjust for small chamber bias across individual animals and viruses. For 2 consecutive days, animals were injected with saline and confined to one chamber in the morning for 30 min and then injected with cocaine (5 or 7.5 mg/kg, I.P.) and confined to the other chamber in the afternoon for 30 min. On the day of the test, mice were returned to the apparatus and allowed to freely explore both sides of the chamber without treatment for 20 min. Preference was measured as time spent in the cocaine-paired minus time spent in the saline-paired chamber.
RNA isolation, RNA-seq, and library preparation
Tissue was collected 24 h after the final cocaine or saline injection and RNA-seq was performed on samples from individual animals in the afternoon—similar timing used by Walker et al. [11]. RNA was isolated with TriZol reagent (Invitrogen) and purified with RNAeasy micro kits (Qiagen). Library preparation was performed as described with minor modifications [16]. RNA purity and integrity were assessed using an Agilent 2100 Bioanalyzer with the RNA 6000 Nano assay (Agilent, Santa Clara, CA). All samples had a RIN value above 8. Libraries were prepared using the TruSeq mRNA Stranded Sample Prep Kit v2 protocol (Illumina, San Diego, CA). Briefly, complementary DNA was synthesized from polyA(+) fragmented RNA, followed by end-repair and ligation with sequencing adaptors. The libraries were size selected and purified using AMPure XP beads (Beckman Coulter, Brea, CA). Each library was assigned an individual adaptor to allow for pooling. Library size and size selection were confirmed on a Bioanalyzer using DNA 1000 kit. Equal molar concentrations of each library were pooled into two samples, and each pool was sequenced over two lanes. Libraries were sequenced on the Illumina HiSeq 2500 System using V4 chemistry with 125 base pair single-end reads at Beckman Coulter Genomics (now GeneWiz) with the goal of achieving ~60 million reads/sample (average: 76.32 M; lowest: 60.89 M). Average mapping rate was 85.07% (lowest: 77.61%), with an average of 65.03 M mapped reads (lowest: 50.70 M). Ribosomal and mitochondrial RNA rates averaged 0.020 and 3.85%, respectively (highest rRNA: 0.022%; highest mtRNA: 4.90%).
Bioinformatics data analysis
All bioinformatics analyses were carried out utilizing established methods with some modifications [16, 17].
Differential expression analysis
Sequencing reads were aligned to the mouse mm10 reference transcriptome using Tophat2 [18]. Read count normalization and gene expression estimation were performed by HTSeq. Samples were filtered for protein-coding and long non-coding RNAs, raw counts were summed across all samples, and the bottom 25% were removed to eliminate very low-expressed genes. Subsequently, pair-wise differential expression comparisons were performed using Voom Limma [19]. A nominal significance threshold of p < 0.05 and fold change > 1.3 was used. Full list of transcriptomic changes is in Supplementary Table S1.
Fisher’s exact tests
Enrichment between gene lists was analyzed using the GeneOverlap R Package, which utilizes Fisher’s exact tests (FETs) to determine whether enrichment is significant (p < 0.05), and reports an odds ratio (OR) to denote the level of enrichment [20].
Rank rank hypergeometric overlap
To evaluate the similarity of differential expression between I.P. cocaine, E2F3b overexpression, and cocaine SA, rank rank hypergeometric overlap (RRHO) was conducted on the full differential expression lists with no significance thresholds applied [21]. Gene lists were ranked by the –log10(p-value) multiplied by the sign of the fold change. We used a script modified from the original that visualizes both positive and negative correlations simultaneously, and also illustrate each quadrant separately with size based on the length of gene lists [22].
Cell-type enrichment
Analysis of cell-type enrichment was performed using web-tools [23]. We used the Benjamini–Hochberg corrected p-values from FETs performed on gene lists from our datasets compared with the curated cell-type gene lists.
Upstream regulator analysis
Upstream regulators were determined by using HOMER (v.4.10, [25]) and Ingenuity Pathway Analysis (IPA; QIAGEN Inc., v. 44691306, https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). HOMER was performed on separate lists of DEGs to reveal enriched consensus sequences. IPA was performed on all three lists of differentially expressed genes (DEGs) to determine positively and negatively associated upstream regulators. A cutoff of p < 0.01 and |z-score| > 2 was used. Full list is in Supplementary Table S2.
Pathway enrichment analysis
Overrepresentation of pathways was determined using Protein Analysis Through Evolutionary Relationships (PANTHER, pantherdb.org, version 11). PANTHER is an online resource used to determine functional enrichment of gene ontology annotations, which is updated monthly, in large-scale biological datasets [26, 27]. Full list is in Supplementary Table S3.
Statistics
All analyses were performed in Prism (GraphPad). One-way or two-way analysis of variances (ANOVAs) were used for all multiple comparisons, followed by Bonferroni post-hoc tests where appropriate.
Results
Overexpression of E2f3b, but not E2f3a, in PFC enhances behavioral responses to cocaine
Mice underwent stereotactic surgery to infuse either HSV-Gfp, -E2f3a, or -E2f3b into PFC to assess isoform-specific effects on cocaine-induced locomotor activity using a relatively low drug dose (5 mg/kg; Fig. 1a). E2f3a overexpression had no effect across all days (no main effect of virus, via two-way ANOVA; F1, 54 = 2.07, p = 0.1562), whereas overexpression of E2f3b greatly increased sensitivity to cocaine (interaction of virus and day F5, 75 = 3.31, p < 0.0001; Fig. 1b, c). Post-hoc analysis showed that E2f3b overexpressing animals exhibited a significant increase in locomotor activity compared with Gfp controls on days 4 and 5 (Day 4: t14 = 2.869, p < 0.05; Day 5: t14 = 3.853, p < 0.01; Fig. 1c). Neither E2f3a nor E2f3b overexpression altered saline-induced locomotor activity (Figure S1A & B).
We next studied the effect of E2F3 isoform overexpression on motivational effects of cocaine using CPP. As with locomotor behavior, using a low dose of cocaine (5 mg/kg), E2f3a overexpression had no effect on the animals’ preference for cocaine (no main effect of virus, two-way ANOVA; F1, 30 = 0.5579, p = 0.5579; Fig. 1d). In contrast, enhanced preference for the cocaine-paired side of the chamber after training was seen in the E2F3b group (E2F3b Test vs. Pre-test: t8 = 3.113, p < 0.05, Fig. 1e, f). Together, these data show that E2f3b overexpression in PFC is sufficient to enhance cocaine-elicited behaviors, with no effect seen for E2f3a. These observations are opposite to what was recently reported in NAc, where E2f3a overexpression increased behavioral responses to cocaine, with E2f3b overexpression having no effect [8].
Knockdown of E2f3b in PFC reduces behavioral responses to cocaine
It was important to determine if E2F3b expression is also necessary for cocaine’s behavioral responses. Knockdown experiments were performed using a miRNA targeting E2f3b mRNA. This virus shows selective knockdown of E2f3b mRNA (Fig. 1g, h). Using a higher dose of cocaine (7.5 mg/kg) to induce a strong locomotor response in control animals (Fig. 1i), reduced locomotor effects of cocaine were observed upon E2f3b knockdown across the course of the experiment (interaction of virus and day by two-way ANOVA; F5, 89 = 3.31, p = 0.0087). On days 3–5, E2f3b knockdown significantly decreased locomotor activity by Bonferroni post-hoc analysis (Day 3: t17 = 3.323, p < 0.01; Day 4: t16 = 3.863, p < 0.01; Day 3: t14 = 3.569, p < 0.01; Fig. 1j). E2f3b knockdown had no effect on saline-induced locomotor activity (Figure S1C).
Similarly, knocking down E2f3b in PFC within a CPP paradigm using a higher dose of cocaine (Fig. 1k) revealed an interaction of virus and test (F1, 7 = 6.4, p = 0.0393; 7.5 mg/kg). No preference was formed in the miR-E2f3b group, whereas the miR-LacZ control group formed a preference on Test day (miR-E2f3b Test vs. Pre-test t6 = 0.3370, p > 0.05; miR-LacZ Test vs. Pre-test: t7 = 3.417, p < 0.05; Fig. 1l). Thus, E2F3b knockdown in PFC blocked the animals from forming a preference for the cocaine-paired chamber, showing that E2F3b expression in PFC is required for the onset of cocaine-induced behavioral responses. In contrast to observations in NAc, where chronic cocaine increases expression of E2F3a but not E2F3b, we observed no effect of the drug on either E2f3 isoform by quantitative PCR or western blotting (data not shown).
E2f3b overexpression in PFC induces a transcriptomic response distinct from investigator-administered cocaine
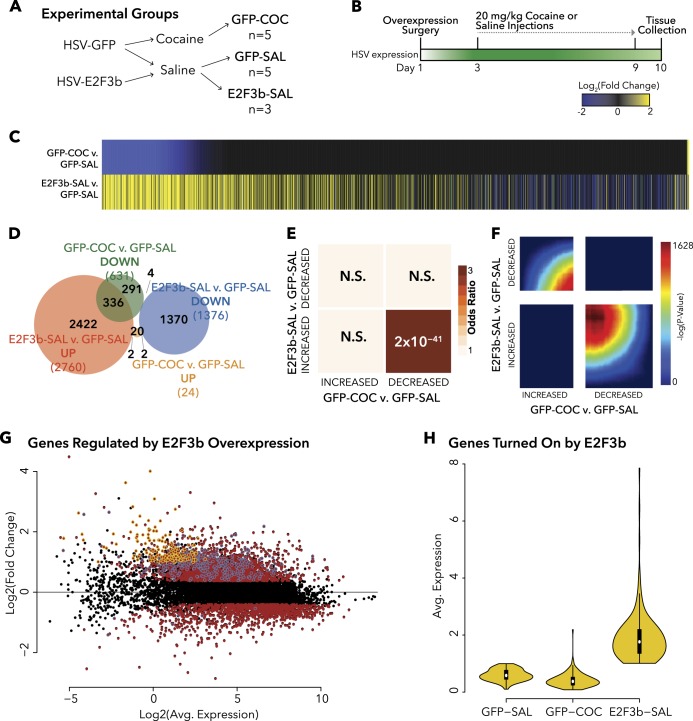
After establishing that E2F3b regulates cocaine-elicited behavioral responses, RNA-seq was utilized to compare cocaine’s transcriptome-wide changes in PFC with those elicited by increased E2f3b expression in this brain region to identify the potential transcriptional mechanisms underlying the behavioral effects (experimental groups and timeline outlined in Fig. 2a, b). E2f3b overexpression elicited a much larger transcriptional response than cocaine exposure. We identified 4119 DEGs in the E2F3b-SAL (saline) vs. GFP-SAL comparison and only 654 DEGs in the GFP-COC (cocaine) vs. GFP-SAL comparison (Table S1). Furthermore, the majority of the E2F3b-SAL vs. GFP-SAL DEGs were upregulated, whereas the majority of the GFP-COC vs. GFP-SAL DEGs were downregulated (Fig. 2c). These two DEG lists have very little overlap, with the most overlap occurring between upregulated E2F3b-SAL vs GFP-SAL and downregulated GFP-COC vs. GFP-SAL DEGs (Fig. 2d). To test this directly, we performed a FET on these two DEG lists. This analysis compares the entire list of up- or downregulated DEGs across both conditions. Thus, we report a single p-value and OR for each comparison. This analysis revealed significant overlap of genes upregulated by E2f3b overexpression but downregulated by cocaine (OR = 4.67; p = 3 × 10−73), whereas no other comparison showed significant overlap (Fig. 2e).
Fig. 2.
E2f3b overexpression in PFC drives a disparate transcriptomic profile from repeated I.P. cocaine. a, b Schematic diagrams detailing experimental design and groups. Animals underwent stereotactic surgery to infuse HSV-GFP or HSV-E2F3b into PFC. After 48 h, cocaine (20 mg/kg) or saline I.P. injections commenced, and tissue was collected 24 h after the final injection for RNA-seq. Analysis was performed as outlined in Materials and methods. c Union heat map shows fold change of all genes significantly differentially expressed (FC > 1.3, p < 0.05) for GFP-COC vs. GFP-SAL (top panel) or E2F3b-SAL vs. GFP-SAL (bottom panel), rank ordered by fold change in the GFP-COC vs. GFP-SAL comparison. d Venn diagram depicting the number of DEGs in each group and the amount of overlap. e Matrix summarizes enrichment for genes across the two comparisons using Fisher’s exact test, corrected for multiple comparisons. Darker colors indicate increasing odds Rratio (OR). -Value is noted in each quadrant. Significant overlap was found between DEGs upregulated in E2F3b-SAL vs. GFP-SAL and downregulated in GFP-COC vs. GFP-SAL (OR = 4.67; p = 3 × 10−73). f RRHO maps compare threshold-free differential expression between the two gene lists. Each pixel represents the overlap between the transcriptome of the two manipulations (I.P. cocaine or E2f3b overexpression) with the significance of overlap [–log10(p-value) of a hypergeometric test] color coded, with warmer colors indicating increasing significance. The results reveal significant overlap in genes increased in E2F3b-SAL vs. GFP-SAL and decreased in GFP-COC vs. GFP-SAL (lower right-hand corner) and in genes decreased in E2F3b-SAL vs. GFP-SAL and increased in GFP-COC vs. GFP-SAL (upper left-hand corner). g MA-plot depicting all genes in E2F3b-SAL vs. GFP-SAL. Red = significantly regulated (FC > 1.3, p < 0.05). Blue = uniquely regulated by E2F3b. Yellow = uniquely turned on by E2F3b (Avg. RPKM < 1 in GFP-SAL; Avg. RPKM > 1 and FC > 2 in E2F3b-SAL). h Avg. Expression (RPKM) of genes turned on by E2F3b are shown here in each comparison
We further investigated this pattern using a genome-wide, threshold-free analysis—RRHO (Fig. 2e), which compares two ranked lists transcript-by-transcript (our lists are threshold-free and ranked by –log2(p-value) from differential expression analysis with a sign applied based on direction of fold change). Thus, the heat map shows a spectrum of –log(p-values) indicating the degree to which similarly ranked groups of transcripts overlap. We observed a highly significant overlap in the lower right quadrant, consistent with the FET analysis, representing genes that are upregulated in PFC by E2F3b, but downregulated by cocaine (max –log(p-value) = 1628). There was also a strong signal in the upper left quadrant signifying significant overlap between genes downregulated by E2F3b and upregulated by cocaine (Fig. 2f), an observation not significant by FET analysis (Fig. 2e). The lack of FET signal is likely due to the small number of upregulated DEGs meeting significance threshold in the GFP-COC vs. GFP-SAL condition.
As mentioned, we observed a ~6-fold greater number of genes whose expression was affected in PFC by E2f3b overexpression than by cocaine. Of these genes, there were 3775 uniquely regulated by E2F3b (blue, Fig. 2g), with 157 of them uniquely turned on (yellow, Fig. 2g). These genes were defined as E2F3b-induced genes if they had an average <1 reads per kilobase million (RPKM) in GFP-SAL, were not significantly increased in GFP-COC, and had an average >1 RPKM in E2F3b-SAL, and were also significantly upregulated compared with GFP-SAL (Fig. 2h).
E2f3b overexpression more similarly models the transcriptomic profile of SA cocaine
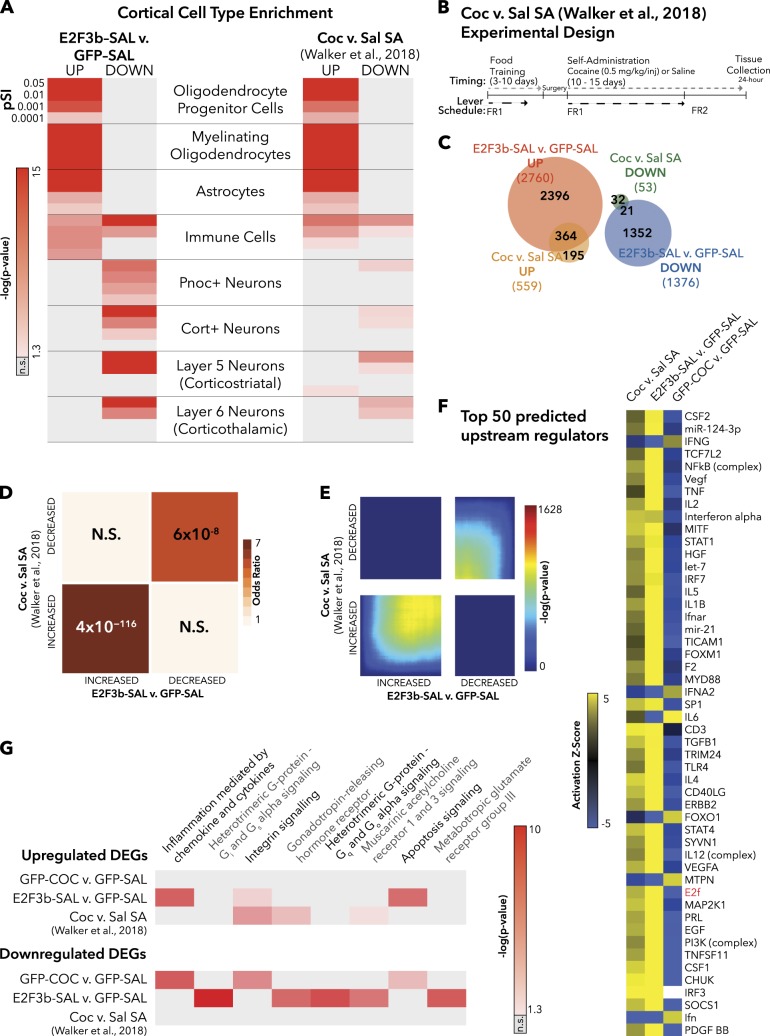
To better understand the genes regulated by E2F3b in PFC, we ran cell-type analysis using data from [23], which includes multiple sub-types of cortical cells. We found that E2F3b-regulated DEGs were enriched for oligodendrocytes and oligodendrocyte progenitor cell (OPC) markers (Fig. 3a). This observation is particularly striking because HSV vectors infect neurons only [24]. It was recently shown that OPCs and oligodendrocyte differentiation and maturation in PFC are critical in regulating SA of heroin [28]. Thus, we chose to compare our E2f3b overexpression dataset with the cocaine SA dataset from the Walker et al. [11] study that most closely mimicked our experimental design—animals that underwent 2 weeks of cocaine SA compared with saline SA that were euthanized 24 h after the last session (Fig. 3b). Cell-type analysis revealed a nearly identical pattern as seen for E2f3b overexpression (Fig. 3a), and we observed an overlap in the DEGs changing in the same direction in both conditions (Fig. 3c). FET showed a highly significant overlap of DEGs upregulated both by E2f3b overexpression and by cocaine SA (OR = 7.82; p = 4 × 10−116), as well as DEGs downregulated in both conditions (OR = 5.61; p = 6 × 10−8; Fig. 3d). RRHO analysis was consistent, showing significant overlap in genes that are changed in the same direction (Fig. 3e).
Fig. 3.
E2f3b overexpression in PFC and cocaine SA induce similar transcriptomic profiles. a Cell-type enrichment of E2F3b-SAL and chronic cocaine SA (Coc vs. Sal SA) [11]. pSI = Specificity Index thresholds of varying stringency (e.g., pSI < 0.01 identifies a larger number of enriched transcripts, pSI < 0.0001 is a shorter list of transcripts more specific to the cell type). p-Value is indicated for Fisher’s exact test (FET) with Benjamini–Hochberg correction. b Experimental design for cocaine SA group from Walker et al. [11] study (modified with permission). c Venn diagram depicting the number of DEGs in each group and the amount of overlap. d Matrix summarizes enrichment for genes across the two comparisons using FET, corrected for multiple comparisons. Significant overlap was found between DEGs upregulated in both E2F3b-SAL vs. GFP-SAL and C24 vs. S24 (OR = 7.82, p = 4 × 10−116), as well DEGs downregulated in both conditions (OR = 5.61, p = 6 × 10−8). e RRHO maps compare threshold-free differential expression between the two gene lists. This analysis shows a significant overlap in genes increased in E2F3b-SAL vs. GFP-SAL and Coc vs. Sal SA (lower left-hand corner) and in genes decreased in both conditions (upper left-hand corner). f The top 50 upstream regulators predicted by IPA are depicted in a heat map. Yellow indicates a positive activation z-score and blue indicates a negative activation z-score. g The pathways predicted by PANTHER with a cutoff of an average of p < 0.05 across all groups. Gray = not significant [37]
Further, HOMER analysis on E2F3b-regulated genes revealed transcription factor motifs overlapping with those found in PFC for two patterns of gene expression related to cocaine SA behavior in a recent paper, including SMAD3, EGR1, and ATF1 [11], as well as the SOX family, which was shown to directly affect SA behavior [28] (Supplemental Table S2).
E2F3b and cocaine SA have similar upstream regulators and downstream effects
Based on these observations, and the fact that E2F3 was itself a predicted upstream regulator of transcriptional responses in PFC to cocaine SA [11], we predicted that these conditions may have common upstream regulators. We explored upstream regulators using IPA (Qiagen) for E2F3b-SAL vs. GFP-SAL and chronic cocaine SA; we also included GFP-COC vs. GFP-SAL to investigate how the opposing transcriptional profile of investigator-delivered drug might be regulated. We only included upstream regulators with an activation Z-score > 2 or < −2 and p-value < 0.01 in at least one condition; the top 50 molecules are displayed in Fig. 3f (full list in Supplementary Table S2). Overall, E2f3b overexpression and cocaine SA have strikingly similar predicted upstream regulators acting in the same direction (Fig. 3f). Conversely, although the same upstream regulators were predicted for the investigator-administrated cocaine, they were nearly all predicted as having an opposite direction of association as the other two conditions. As expected, the E2F family was predicted as a positively associated upstream regulator in E2F3b-SAL vs. GFP-SAL and, in concurrence with the data showing transcriptomic overlap, in the chronic cocaine SA group. Interestingly, E2F was negatively associated with the GFP-COC vs. GFP-SAL condition, further highlighting the divergence of these groups. Note that many of the predicted upstream regulators have been implicated in cocaine responses in PFC. Specifically, nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB), miRNA-124, and vascular endothelial growth factor (VEGF) have been shown to be regulated by chronic cocaine [29–31]. Although the majority of predicted upstream regulators had positive activation scores in both E2F3b-SAL vs. GFP-SAL and chronic cocaine SA, of the top 50 predicted regulators there were five with negative activation scores in both conditions: three associated with interferon (Ifn family, IFNG, and IFNA2), as well as FOXO1 and MTPN.
Using PANTHER (v.11), we investigated the pathways overrepresented in each condition. We separated the analysis into upregulated and downregulated DEGs and included pathways with an average p-value <0.05 across all groups (Fig. 3g). Similar to what we found with the upstream regulators, E2f3b overexpression and cocaine SA result in enrichment of overlapping pathways, whereas investigator-administrated cocaine has an opposing pattern of enrichment. It is important to note that cocaine SA downregulated and GFP-COC vs. GFP-SAL upregulated DEGs were so few that none of the listed pathways rose to the level of significance.
Discussion
This study demonstrates that E2F3b is both necessary and sufficient to regulate the PFC contribution to cocaine-elicited behaviors and, in conjunction with previous findings [8], demonstrates regional specificity of E2F3 isoforms on cocaine responses. HSV-mediated overexpression of E2f3b, but not E2f3a, in PFC enhances cocaine-elicited locomotor and place preference behaviors. These behaviors are blunted upon reduction of E2f3b expression in this brain region via HSV-mediated knockdown. These observations are very different from what we reported in NAc, where E2F3a is both necessary and sufficient for enhanced behavioral responses to cocaine, with E2F3b having no effect. However, unlike the observed effects of E2f3a overexpression in NAc, which recapitulated a large portion of cocaine-induced transcriptional regulation and alternative splicing, E2f3b overexpression in PFC does not recapitulate the effects of investigator-administered cocaine. In fact, for a subset of genes, E2f3b overexpression induced an opposing transcriptomic profile to that of I.P. cocaine injections. Upon further investigation, we found that, by contrast, SA cocaine induced a similar pattern of gene expression to that of E2f3b overexpression in PFC. The DEGs following both cocaine SA and E2f3b overexpression showed nearly identical cell-type enrichment patterns as well as similar upstream regulator predictions and affected biological pathways.
We were initially surprised by the opposing effect of E2F3b and I.P. cocaine on the PFC transcriptome. Cell-type enrichment analysis revealed that E2F3b preferentially upregulated genes specific to glia, with the most significant enrichment in OPCs and myelinating oligodendrocytes. As mentioned, these cells in PFC were recently implicated in the regulation of heroin SA in rats [28]. This led us to analyze the overlap of E2F3b-induced expression with cocaine SA from a previous study. Although E2F3b appears to regulate mostly distinct transcriptional pathways from I.P. cocaine, we know that some similar behaviors (enhanced locomotion and place conditioning) are seen with I.P. and SA cocaine. Thus, E2F3b’s induction of a transcriptional profile similar to that of SA cocaine would likely still enhance motivation and locomotor behaviors. In the Martin et al. [28] study, the authors showed that Sox10, a gene involved in oligodendrocyte differentiation and maturation, was downregulated in PFC following heroin SA, and that Sox10 overexpression decreased motivation to SA heroin. In our analyses, HOMER predicted Sox10 as an upstream regulator of E2F3b and cocaine SA upregulated DEGs (Supplementary Table S2). This suggests a similar role for Sox10 in PFC in cocaine SA as seen in the heroin SA study, and that E2F3b may be involved in regulating this process.
Since our HSV vectors infect neurons solely (Neve et al. 2005), our data suggest that E2F3b activity in PFC neurons exerts a dramatic influence on surrounding oligodendrocytes and OPCs. We did not detect an effect of E2F3b overexpression or knockdown on the health or morphology of PFC neurons, however, future studies are required to understand this interesting observation and the underlying mechanisms involved.
One of the pathways enriched in DEGs downregulated by I.P. cocaine and DEGs upregulated by cocaine SA or E2F3b is integrin signaling. Studies of integrins in cocaine addiction have focused mainly on NAc, where they have been shown to play an important role in drug-seeking and drug-induced reinstatement via regulation of glutamatergic synaptic plasticity [32]. Interestingly, Wiggins et al. showed that, when rats were I.P. injected with cocaine vs. undergoing cocaine SA, NAc protein levels of beta-integrins in the postsynaptic density were altered in opposing directions [32, 33]. While this work was carried out in NAc, a large proportion of the glutamatergic input onto NAc neurons is from PFC. Thus, regulation of integrins in NAc in these studies is likely mediating stability of synapses between these two regions. Furthermore, we know that oligodendrocytes express integrins and that beta1 integrins regulate oligodendrocyte-mediated myelin sheath outgrowth in the CNS [34]. Specifically, oligodendrocyte integrins interact with laminins on astrocytes to promote oligodendrocyte survival in adult mouse brain in turn helping to maintain myelin homeostasis [35]. Our sequencing data show that RNA expression of the beta1-integrin subunit and two laminins (Lama2, Lama5) are upregulated by cocaine SA and E2F3b and downregulated by I.P. cocaine in PFC (Supplementary Table S1; [11]). These results highlight the possibility that cocaine SA strengthens the connection between PFC and downstream regions, such as NAc, and that the integrin-mediated component in PFC is mediated by E2F3b.
Although the focus here is E2F3b and similarities and differences it shows to cocaine SA- and I.P. cocaine-induced transcriptional changes, respectively, our findings make the point that I.P. cocaine and SA cocaine induce disparate transcriptional profiles in PFC (Supplementary Figure S1). This is not unsurprising given the PFC’s role in decision-making and executive function, rather than the more behaviorally conserved reward role of NAc, for example. In a similar way, I.P. cocaine vs. SA cocaine exerted very similar effects on another transcription factor, ∆FosB, in NAc, but different effects in PFC [36]. We performed the RNA-seq experiments outlined in this study to define the transcriptional component of E2F3b manipulation on I.P. cocaine-driven behaviors. We found unexpectedly that the transcriptional responses were vastly different. In contrast, we found that E2F3b overexpression better emulated the transcriptional response seen after cocaine SA. As SA behavior is ethologically more relevant to human drug use, results of the present study underscore the importance of utilizing this drug administration procedure particularly when studying PFC.
Results of the present study identify a novel transcriptional mechanism—focused on E2F3b—that controls behavioral and molecular responses to cocaine in PFC. Given that this brain region is crucial for the impaired decision making and drug craving that characterizes cocaine addiction, our findings raise the possibility that E2F3b and some of its target genes could potentially be therapeutic targets for cocaine use disorders.
Funding and disclosure
Funding was provided by NIDA (R01DA07359). The authors declare no competing interests.
Supplementary information
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41386-018-0296-1).
References
- 1.Montague PR, Hyman SE, Cohen JD. Computational roles for dopamine in behavioural control. Nature. 2004;431:760–7. doi: 10.1038/nature03015. [DOI] [PubMed] [Google Scholar]
- 2.Everitt BJ, Dickinson A, Robbins TW. The neuropsychological basis of addictive behaviour. Brain Res Brain Res Rev. 2001;36:129–38. doi: 10.1016/S0165-0173(01)00088-1. [DOI] [PubMed] [Google Scholar]
- 3.Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat Neurosci. 2005;8:1481–9. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- 4.Berke JD, Hyman SE. Addiction, dopamine, and the molecular mechanisms of memory. Neuron. 2000;25:515–32. doi: 10.1016/S0896-6273(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 5.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–98. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 6.Kalivas PW, Volkow N, Seamans J. Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron. 2005;45:647–50. doi: 10.1016/j.neuron.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–37. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cates HM, Heller EA, Lardner CK, Purushothaman I, Peña CJ, Walker DM, et al. Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol Psychiatry. 2018;84:167–79. doi: 10.1016/j.biopsych.2017.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Julian LM, Vandenbosch R, Pakenham CA, Andrusiak MG, Nguyen AP, McClellan KA, et al. Opposing regulation of Sox2 by cell-cycle effectors E2f3a and E2f3b in neural stem cells. Cell Stem Cell. 2013;12:440–52. doi: 10.1016/j.stem.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, et al. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–32. doi: 10.1128/MCB.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker DM, Cates HM, Loh Y-HE, Purushothaman I, Ramakrishnan A, Cahill KM, et al. Cocaine self-administration alters transcriptome-wide responses in the brain’s reward circuitry. Biol Psychiatry. 2018;84:867–80. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 12.Cates HM, Thibault M, Pfau M, Heller E, Eagle A, Gajewski P, et al. Threonine 149 phosphorylation enhances ΔFosB transcriptional activity to control psychomotor responses to cocaine. J Neurosci. 2014;34:11461–9. doi: 10.1523/JNEUROSCI.1611-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–90. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–29. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 16.Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang J, et al. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron. 2016;90:969–83. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L. GeneOverlap - R package for testing and visualizing gene overlaps. 2013. 10.18129/B9.bioc.GeneOverlap.
- 21.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cahill KM, Huo Z, Tseng GC, Logan RW, Seney ML. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci Rep. 2018;8:9588. doi: 10.1038/s41598-018-27903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–30. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neve RL, Neve KA, Nestler EJ, Carlezon WA. Use of herpes virus amplicon vectors to study brain disorders. Biotechniques. 2005;39:381–91. [DOI] [PubMed]
- 25.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–9. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin JA, Caccamise A, Werner CT, Viswanathan R, Polanco JJ, Stewart AF, et al. A novel role for oligodendrocyte precursor cells (OPCs) and Sox10 in mediating cellular and behavioral responses to heroin. Neuropsychopharmacology. 2018;43:1385–94. doi: 10.1038/npp.2017.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannotti G, Caffino L, Calabrese F, Racagni G, Riva MA, Fumagalli F. Prolonged abstinence from developmental cocaine exposure dysregulates BDNF and its signaling network in the medial prefrontal cortex of adult rats. Int J Neuropsychopharmacol. 2014;17:625–34. doi: 10.1017/S1461145713001454. [DOI] [PubMed] [Google Scholar]
- 30.Orso R, Creutzberg KC, Centeno-Silva A, Carapeços MS, Levandowski ML, Wearick-Silva LE, et al. NFκB1 and NFκB2 gene expression in the prefrontal cortex and hippocampus of early life stressed mice exposed to cocaine-induced conditioned place preference during adolescence. Neurosci Lett. 2017;658:27–31. doi: 10.1016/j.neulet.2017.08.032. [DOI] [PubMed] [Google Scholar]
- 31.Yin W, Clare K, Zhang Q, Volkow ND, Du C. Chronic cocaine induces HIF-VEGF pathway activation along with angiogenesis in the brain. PLoS One. 2017;12:e0175499. doi: 10.1371/journal.pone.0175499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiggins A, Smith RJ, Shen HW, Kalivas PW. Integrins modulate relapse to cocaine-seeking. J Neurosci. 2011;31:16177–84. doi: 10.1523/JNEUROSCI.3816-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiggins AT, Pacchioni AM, Kalivas PW. Integrin expression is altered after acute and chronic cocaine. Neurosci Lett. 2009;450:321–3. doi: 10.1016/j.neulet.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barros CS, Nguyen T, Spencer KSR, Nishiyama A, Colognato H, Müller U. Beta1 integrins are required for normal CNS myelination and promote AKT-dependent myelin outgrowth. Development. 2009;136:2717–24. doi: 10.1242/dev.038679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domingues HS, Portugal CC, Socodato R, Relvas JB. Oligodendrocyte, astrocyte, and microglia crosstalk in myelin development, damage, and repair. Front Cell Dev Biol. 2016;4:71. doi: 10.3389/fcell.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winstanley CA, LaPlant Q, Theobald DE, Green TA, Bachtell RK, Perrotti LI, DiLeone RJ, Russo SJ, Garth WJ, Self DW, Nestler EJ. DeltaFosB induction in orbitofrontal cortex mediates tolerance to cocaine-induced cognitive dysfunction. J Neurosci. 2007;27:10497–507. [DOI] [PMC free article] [PubMed]
- 37.CSEA tool. Dougherty Lab: from autism to astrocytes. 2017. http://genetics.wustl.edu/jdlab/csea-tool-2/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.