Abstract
The rapid identification and differentiation of members of the Mycobacterium tuberculosis complex (MTBC) is essential to assess the potential zoonotic risk. Different available molecular methods are time consuming since they depend on cultivation of mycobacteria. High Resolution Melting (HRM) is a low cost, rapid and easy to perform single-tube method not limited to cultured samples. In this study, a HRM assay specifically targeting gyrB was developed to simultaneously identify and differentiate Mycobacterium (M.) tuberculosis, M. microti and M. bovis/M. caprae. To evaluate the performance of this assay, 38 MTBC isolates and 25 directly extracted clinical specimens were analysed. HRM results of all 38 (100%) examined isolates correlated with the results obtained with the commercially available GenoType MTBC test (Hain Lifescience). From the 25 clinical specimens tested, species identification by HRM showed concordant results with the previously used identification methods in 23 samples (92%). The assay demonstrated a good analytical sensitivity, specificity and reproducibility and can be used directly on clinical specimens.
Introduction
Mycobacterium tuberculosis complex (MTBC) consists of the closely related species Mycobacterium (M.) tuberculosis, M. bovis, M. bovis Bacillus Calmette and Guérin (BCG), M. caprae, M. africanum, M. microti, M. pinnipedii, M. canettii and three further species (M. orygis, the dassie bacillus, M. mungi)1. M. tuberculosis and M. africanum are often described as host-specific to humans. According to the recent WHO report from 2017, tuberculosis is still the leading cause of human death from a single infectious agent with 6.3 million new cases and an estimated 1.7 million deaths in 20162. Additionally, several cases of M. tuberculosis infections in animals have been reported3–5. Bovine tuberculosis (bTB) is an important zoonosis most commonly caused by M. bovis and less frequently by M. caprae1. New cases of bTB in 2013 in Switzerland6, resulting in the implementation of a national bTB surveillance program, highlighted the importance of routine species-level identification. This program consists of a systematic microbiological testing of suspicious lymph nodes found linked to meat inspection in slaughterhouses. Fast and reliable identification and differentiation between the species within this MTB complex is important to assess the potential zoonotic risk and is therefore a fundamental procedure for public health surveillance and food safety.
Identification of MTBC is based on different PCR based methods targeting 16S rRNA or IS61107,8. In contrast, differentiation of Mycobacterium species within the MTBC is more laborious. Diverse molecular methods such as the GenoType MTBC test (Hain Lifescience, Nehren, Germany) or restriction fragment length polymorphism9,10 are used although having limitations in the reliance on bacterial cultures to produce a valid amount of bacterial DNA. Mycobacteria especially M. microti requires several months to obtain significant growth underlining a delayed time span to get valid results of MTBC species-level identification. Spoligotyping uses polymorphism on the direct repeat locus for differentiation and typing of MTBC11. A result is obtained in one or two days comprising several working steps such as PCR, membrane preparation, hybridisation, washing steps and detection by chemiluminescence. On the other hand, newer alternatives to conventional spoligotyping such as Luminex technology or Spoligotyping by MALDI-TOF MS requires advanced and expensive equipment12. Numerous single-tube multiplex RT-PCR assays have been previously proposed for MTBC species discrimination, mainly targeting several region of differences (RD)13–17. The complexity of a multiplex RT-PCR reaction can be a problem for intricate veterinary samples often dealing with animal tissues.
High resolution melting (HRM) assay is reported as a rapid and low cost assay to detect single-nucleotide polymorphism (SNP)18. The assay characterizes amplified PCR products according to their dissociation behaviour without requiring additional instrumentation. A fluorescently labelled dye binding to double stranded DNA is combined with amplicons resulting from the PCR reaction. When increasing the temperature, the double stranded DNA dissociates into single strands leading to a decrease in fluorescence intensity. The melting temperature depends on GC content, length and nucleotide sequence. It is an easy to perform and single-tube method leading to a result within approximately two hours. Moreover, HRM is not limited to cultured material but is able to detect DNA in clinical specimens directly extracted from tissue samples. Various HRM assays are already used successfully to identify and differentiate many bacteria species such as Mycoplasma synoviae19, Chlamydiaceae sp.20, Staphylococcus aureus21, Brucella sp.22 and Leptospira sp.23. Among Mycobacteria sp. HRM is a tool often reported to analyse drug resistance in M. tuberculosis24–26. Furthermore, the method is used to differentiate non-tuberculous mycobacteria (NTM) and to distinguish them from MTBC27–29. Wright et al. designed a HRM assay to detect the Region of Deletion 9 (RD9) with the aim of differentiating M. tuberculosis from other members of MTBC in fine-needle aspiration biopsies30. However, the drawback of this study lies in its poor sensitivity of only 51.9%. Other studies combined RD-targeted multiplex RT-PCR assays with HRM14,17. In this study, a novel HRM method based on SNPs located in gyrB was developed to simultaneously identify and differentiate members of MTBC to its species-level.
Results
HRM Analysis with Cultured Samples
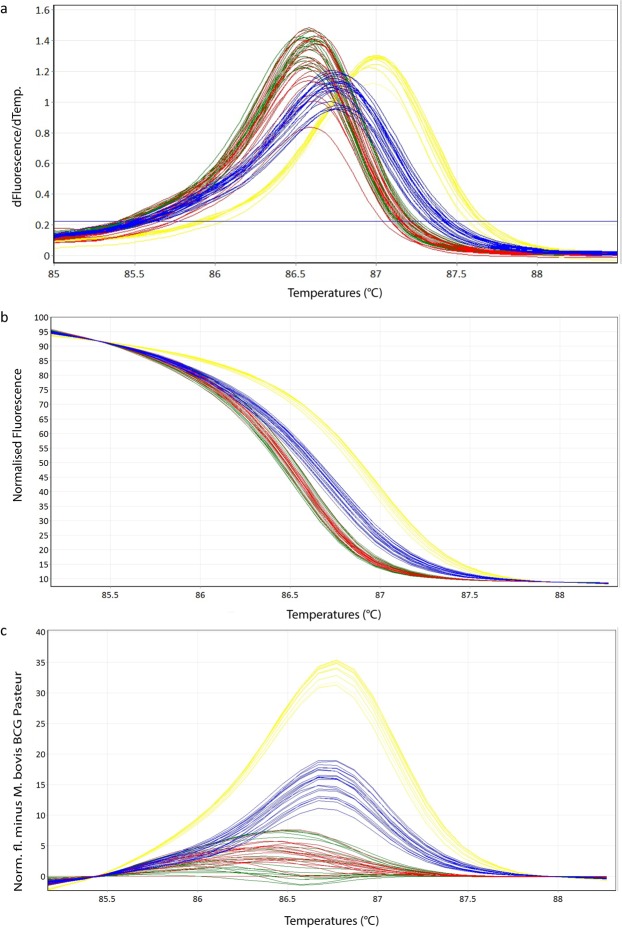
All cultured samples tested were amplified and resulted in a corresponding melting curve. The HRM species-specific melting temperatures Tm (Table 1, Supplementary Table S1) and the corresponding melting curves (Fig. 1a) of the sample-subset used for determination of the intra- and inter-assay reproducibility are shown. Since the Tm ranges are very close to each other, it is difficult to clearly distinguish between members of MTBC. On the other hand, the difference plot (Fig. 1c) as well as the normalized plot (Fig. 1b) allowed a clear species differentiation into the three groups of M. tuberculosis/M. africanum, M. microti and M. bovis/M. caprae. The intra-assay coefficiences of variation (CVs) and the inter-assay CVs showed values ranging between 0.01–0.02% and 0.02–0.03%, respectively (Table 1, Supplementary Table S1). Species identification results of all 38 (100%) tested cultured samples correlated with the GenoType MTBC test (Hain Lifescience) results.
Table 1.
Melting temperatures (mean and standard deviation) of the intra- and inter-assay of a randomly chosen subset of cultured samples for different MTBC species with its corresponding coefficients of variation (CV) in % are listed.
| Run 1 | Run 2 | Run 3 | Inter-Assay | |||||
|---|---|---|---|---|---|---|---|---|
| CV% | Tm values | CV% | Tm values | CV% | Tm values | CV% | Tm values | |
| M. tuberculosis H37Rv | 86.93 | 86.98 | 86.95 | |||||
| M. bovis BCG Pasteur ATCC 35734 | 86.60 | 86.58 | 86.58 | |||||
| M. microti ATCC 19422 | 86.75 | 86.72 | 86.73 | |||||
| M. tuberculosis (n = 3) | 0.02 | 86.99 ± 0.04 | 0.01 | 86.98 ± 0.03 | 0.02 | 87.00 ± 0.03 | 0.02 | 86.99 ± 0.04 |
| M. caprae (n = 6) | 0.02 | 86.57 ± 0.04 | 0.02 | 86.58 ± 0.05 | 0.01 | 86.62 ± 0.04 | 0.03 | 86.59 ± 0.06 |
| M. bovis (n = 6) | 0.02 | 86.58 ± 0.05 | 0.02 | 86.60 ± 0.05 | 0.02 | 86.64 ± 0.04 | 0.03 | 86.60 ± 0.07 |
| M. microti (n = 7) | 0.02 | 86.74 ± 0.07 | 0.02 | 86.73 ± 0.05 | 0.01 | 86.75 ± 0.05 | 0.03 | 86.74 ± 0.07 |
Figure 1.
Representative high resolution melting graphs corresponding to one high resolution melting analysis of a subset of cultured samples (n = 22). Curves of tested samples previously identified as M. tuberculosis are shown in yellow, M. microti in blue, M. bovis/M. bovis BCG in red and M. caprae in green. (a) Melting curves; (b) Normalized plot; (c) Difference plot.
HRM Analysis with Clinical Specimens
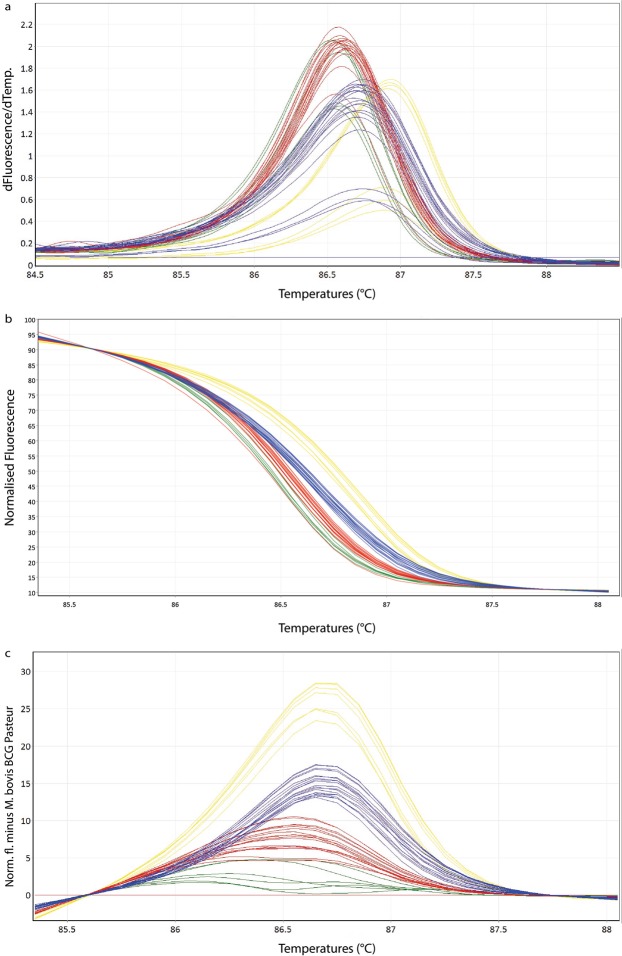
25 clinical specimens were tested in the HRM assay and resulted in three main groups consistent with the expected MTBC species. The obtained normalized and difference plots of the tested subset of clinical specimens showed a clear discrimination (Fig. 2b,c). The intra-assay CVs (Table 2, Supplementary Table S2) were between 0.01–0.02% for M. tuberculosis and M. caprae, 0.01% for M. bovis and 0.05% for M. microti. The inter-assay CVs (Table 2, Supplementary Table S2) were higher than in the cultured samples with values between 0.12–0.15%. From those 25 clinical specimens 23 (92%) showed concordant identification results. Two samples revealed lower Tm values at approximately 84 °C, compared to the other sample results. The corresponding culture of one of those two clinical specimens (sample 17–2287) showed a correct species identification by HRM. The second clinical specimen (sample 17-1063) is not successfully cultured yet and further investigations are still ongoing.
Figure 2.
Representative high resolution melting graphs corresponding to one high resolution melting analysis of a subset of clinical specimens (n = 19). Curves of tested samples previously identified as M. tuberculosis are shown in yellow, M. microti in blue, M. bovis/M. bovis BCG in red and M. caprae in green. (a) Melting curves; (b) Normalized plot; (c) Difference plot.
Table 2.
Melting temperatures (mean and standard deviation) of the intra- and inter-assay of a randomly chosen subset of clinical specimens for different MTBC species with its corresponding coefficients of variation (CV) in % are listed.
| Run 1 | Run 2 | Run 3 | Inter-Assay | |||||
|---|---|---|---|---|---|---|---|---|
| CV% | Tm values | CV% | Tm values | CV% | Tm values | CV% | Tm values | |
| M. tuberculosis H37Rv | 86.80 | 86.93 | 86.75 | |||||
| M. bovis BCG Pasteur ATCC 35734 | 86.38 | 86.55 | 86.37 | |||||
| M. microti ATCC 19422 | 86.52 | 86.70 | 86.48 | |||||
| M. tuberculosis (n = 2) | 0.01 | 86.73 ± 0.01 | 0.02 | 86.91 ± 0.03 | 0.02 | 86.70 ± 0.03 | 0.13 | 86.80 ± 0.13 |
| M. caprae (n = 5) | 0.01 | 86.27 ± 0.17 | 0.02 | 86.45 ± 0.15 | 0.02 | 86.34 ± 0.12 | 0.12 | 86.35 ± 0.25 |
| M. bovis (n = 6) | 0.01 | 86.41 ± 0.06 | 0.01 | 86.59 ± 0.04 | 0.01 | 86.36 ± 0.04 | 0.15 | 86.48 ± 0.16 |
| M. microti (n = 7) | 0.03 | 86.59 ± 0.04 | 0.02 | 86.73 ± 0.05 | 0.02 | 86.48 ± 0.05 | 0.14 | 86.60 ± 0.17 |
Analytical Specificity
In the exclusivity run the tested 41 NTMs and Nocardia paucivorans, Escherichia coli and Streptococcus suis either showed no melting curves or melting curves with completely different Tm values compared to those obtained from samples of the MTBC. Therefore, the assay showed an analytical specificity of 100%.
Limit of Detection
The limit of detection (LOD) for the lowest dilution of which the acceptance criteria were fulfilled was 10 genome equivalents (GE) for M. tuberculosis, M. bovis/M. caprae and M. microti (Table 3).
Table 3.
Limit of detection of the real-time PCR step within the HRM assay.
| MTBC Member | Genome equivalents | Ct | SD |
|---|---|---|---|
| M. tuberculosis H37Rv | 1'000'000 | 16.42 | 0.10 |
| 100'000 | 19.89 | 0.02 | |
| 10'000 | 23.76 | 0.07 | |
| 1000 | 27.48 | 0.08 | |
| 100 | 31.21 | 0.32 | |
| 10 | 35.04 | 0.27 | |
| 1 | — | — | |
| M. bovis BCG pasteur ATCC 35734 | 1'000'000 | 15.13 | 0.20 |
| 100'000 | 18.61 | 0.16 | |
| 10'000 | 22.05 | 0.16 | |
| 1000 | 25.75 | 0.17 | |
| 100 | 29.38 | 0.20 | |
| 10 | 33.15 | 0.50 | |
| 1 | 35.51 | 0.12 | |
| M. microti ATCC 19422 | 1'000'000 | 15.73 | 0.06 |
| 100'000 | 19.03 | 0.05 | |
| 10'000 | 22.98 | 0.07 | |
| 1000 | 26.28 | 0.13 | |
| 100 | 30.18 | 0.17 | |
| 10 | 33.92 | 0.14 | |
| 1 | 37.82 | 0.73 |
Determination of Ct values and its standard deviation (SD) of 3 replicates for a dilution series ranging from 1 to 1'000'000 genome equivalents using the three reference strains M. tuberculosis H37Rv, M. bovis Pasteur ATCC 35734 and M. microti ATCC 19422.
Efficiency
The efficiencies of the RT-PCR were 87% for M. microti, 94% for M. bovis and 85% for H37Rv (Supplementary Fig. S1).
Discussion
This study reports the development of a HRM assay to identify and distinguish the main members of the MTBC complex of clinical specimens and cultured samples in approximately two hours. M. microti, M. tuberculosis/M. africanum and M. bovis/M. caprae can be clearly and reliably distinguished from each other by unique difference plots. The Tm alone is not sufficient to discriminate the main species because of partially overlapping Tm values (Tables 1 and 2). However, after appropriate transformation of the melting curves into normalized and difference plots by applying algorithms of the Rotor-Gene Q Software 2.3.1 (Qiagen Hilden, Germany), the members of the MTBC can be clearly distinguished into three groups (Figs 1c and 2c). Using this strategy, the species-specific melting profiles showed an unambiguous picture.
To date, MTBC species identification is often based on methods requiring cultured samples10 or based on time-consuming procedures11. Halse et al. reported the development of a multiplex RT-PCR method for clinical specimens, however, this assay is more expensive due to the need of five different probes16. Furthermore, the complexity of such a multiplex reaction can be challenging when analysing tissue samples comprising various substances containing large amounts of co-extracted host DNA and ingredients, which can lead to inhibition of the PCR reaction31. Other studies evaluated mainly cultured isolates13–15,17. The main advantage of our developed HRM assay compared to previous studies is the implementation of a relatively cheap and straightforward singleplex method for directly extracted clinical specimens.
The current HRM assay identified MTBC positive cultured samples in complete agreement with results of the GenoType MTBC test (Hain Lifescience). The clinical specimens showed a concordance of 92%. The remaining 8% (n = 2) showed unspecific melting curves not allowing to assign the samples to MTBC using the developed method. Both samples derived from alpaca tissues, either from spleen or from a mix of different tissues including lymph node, lung, heart, liver and cervical vertebra. All 23 clinical samples lead to an unambiguous and correct result derived from lymph nodes, lung or liver tissues (Supplementary Table S3). It is likely that the content of these particular alpaca tissues interfered with the melting procedure. Further investigations to clarify this finding are continuing.
The intra- and inter-assay CVs showed very low values demonstrating a very good reproducibility of the method. The analytical specificity displayed a perfect value of 100% indicating a MTBC specific assay. Furthermore, the assay demonstrated a good PCR efficiency of more than 85% and a good sensitivity with a LOD of 10 GE.
One limitation of the assay is its inability to distinguish between M. bovis, M. bovis BCG and M. caprae with this particular primer set. Moreover, M. africanum cannot be separated from M. tuberculosis. In order to design a HRM assay having a high resolution detecting SNPs, the PCR amplicon should optimally not exceed 150 bp since longer amplicons would have a negative impact on the resolution of the assay. SNPs distantly located within a gene, as in the case of gyrB, are impossible to analyse by HRM using just one primer pair. Our primary goal was to clearly distinguish M. microti from other members of the MTBC complex since its proper identification in directly extracted clinical samples are advantageous considering its long cultivation time. Therefore, our developed HRM assay was restricted to the detection of 2 out of 5 possible SNPs within the gyrB gene10 resulting in a clear and rapid identification and differentiation of the three main MTBC species most relevant to veterinarians. In order to overcome the described limitation, there is the possibility to extend the HRM assay with a second primer pair, resulting in a two-reaction HRM paradigm, targeting a region to further discriminate M. bovis from M. caprae and M. africanum from M. tuberculosis. In addition, another drawback of the developed assay seems to lie in the failure of detection of samples deriving from certain alpacas (two out of four tested alpacas), especially samples containing spleen or bone.
Conclusions
The developed HRM assay enables the simultaneous identification and differentiation of MTBC between the three clinically most relevant groups namely M. tuberculosis/M. africanum, M. microti and M. bovis/M. caprae from tissue samples as well as from cultured material. Therefore, the use of this powerful assay may save several months of cultivation time to differentiate between species of MTBC. It is an easy to perform, cheap, sensitive and specific assay leading to a result in less than two hours. Since tuberculosis is one of the top 10 causes of death worldwide, it is expected that a cost-effective and easy to set-up assay could be implemented in laboratories with moderate resources as a high-throughput screening and confirmatory tool for MTBC infections. This would significantly contribute to develop efficient public health and veterinary surveillance strategies worldwide.
Methods
Ethics statement
This study was carried out in accordance with the recommendations of Swiss federal regulations (TSV 916.401 and VSFK 817.190). Analysis of animal specimens was carried out within an official context of monitoring bovine tuberculosis and NTM infections, meaning that no animals were killed for the purposes of this research project and ethical approval was not necessary.
Reference strains and samples
62 MTBC positive samples originating from 40 different animals and tissues (Table 4, Supplementary Table S3) were used for assay development. One additional isolate of a wild boar was kindly provided by Lucía de Juan Ferré and Beatriz Romero Martinez from the European Union Reference Laboratory for Bovine Tuberculosis in Madrid. Finally, a total of 63 samples comprising 38 isolates and 25 clinical specimens were tested. Reference strains M. microti ATCC 19422, M. bovis BCG Pasteur ATCC 35734 and M. tuberculosis H37Rv were included as positive controls in each run. To determine the specificity of the optimized HRM assay a set of 41 different non-tuberculous mycobacteria (NTM) was additionally tested (Supplementary Table S4). Moreover, Nocardia paucivorans, Escherichia coli and Streptococcus suis were included in this exclusivity panel in order to test for any non-specific signals.
Table 4.
MTBC positive samples used for the development of the HRM method.
| Species | Host | No. of isolates |
|---|---|---|
| cultured material (n = 38) | ||
| M. tuberculosis | elephant | 3 |
| M. caprae | cow | 7 |
| M. bovis | cow | 15 |
| M. microti | cat | 7 |
| M. microti | alpaca | 3 |
| M. microti | llama | 2 |
| M. microti | wild boar (Spain) | 1 |
| clinical specimens (n = 25) | ||
| M. tuberculosis | elephant | 2 |
| M. caprae | cow | 5 |
| M. bovis | cow | 7 |
| M. microti | cat | 5 |
| M. microti | alpaca | 4 |
| M. microti | llama | 2 |
| Total | 63 | |
38 isolates obtained from cultured material, whereas 25 samples were clinical specimens directly extracted from tissue samples. 62 samples derived from Switzerland whereas one isolate originated from Spain.
Culture and DNA extraction
Sample preparation, culture and DNA extraction were proceeded as described previously6. Briefly, genomic DNA was extracted harvesting mycobacteria from 1.5 ml of MGIT subcultures by centrifugation for 10 min at 13,000 × g. The sediment was suspended in 180 µl ATL buffer (Qiagen), transferred onto a bead beating matrix in a 2 ml microtube (Omni International, Kennesaw, USA), heat inactivated and subjected to mechanical cell lysis using a TissueLyser II (Qiagen) and enzymatic digestion with Proteinase K (Qiagen). Automated DNA preparation was performed on the QIAcube instrument using the QIAamp cador Pathogen Mini Kit protocol (Qiagen). DNA concentration was measured using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, Reinach, Switzerland) and stored at −20 °C until use. DNA obtained from pure mycobacterial cultures were identified as MTBC using artus M. tuberculosis RG PCR Kit (Qiagen). Species identification of cultured samples was performed by GenoType MTBC test (Hain Lifescience). Clinical specimens were tested by Spoligotyping32 and multi-locus variable number tandem repeat analysis using an internationally established 24-loci panel33. Standard biosecurity procedures have been carried out for handling of samples. Cultures involving MTBC or NTM isolates were performed at the Biosafety Level 3 facility until heat deactivation. Sample preparation and DNA extraction were carried out under Biosafety Level 2 containment.
HRM development and optimisation
A primer pair was designed specifically targeting a conserved region for MTBC on the gyrB gene. The forward HRM_gyrB_for (5′-CGGCTCGAAGTCGAGATCAAG-3′) and reverse HRM_gyrB_rev (5′-TTCGAAAACAGCGGGGTCG-3′) primers flank a 144 base pair (bp) amplicon. In contrast, other closely and distantly related NTM have a greater variability in the primer region as well as in the whole 144 base pair amplicon (Fig. 3).
Figure 3.
Sequence alignment of the amplicon within gyrB generated by the real-time PCR of the high resolution melting. Primer regions are indicated in yellow. Red letters and dots represent conserved bases whereas blue letters show areas with substitutions. The two single nucleotide polymorphisms distinguishing the main members of the Mycobacterium tuberculosis complex detected by the high resolution melting assay are highlighted with green.
The HRM assay was performed on the Rotor-Gene Q system (Qiagen) with the Type-it HRM PCR Kit (Qiagen). The reaction was performed in a total volume of 15 µl. 1 µl of sample DNA was added to a reaction mixture containing 7.5 µl 2X Type-it HRM Mastermix containing EvaGreen DNA-binding dye (Qiagen), 0.5 µM final concentration of each primer (Microsynth AG, Balgach, Switzerland) and ultrapure water. The PCR thermocycling conditions were as follows: initial denaturation at 95 °C for 5 min, 40 cycles with denaturation at 95 °C for 10 s and annealing/extension at 55 °C for 30 s followed by a second cycling step at 95 °C for 10 s and 40 °C for 2 min followed by a HRM ramping from 80 °C to 93 °C. Fluorescence data were acquired at 0.1 °C increments every 2 s to generate specific melting curves. For each experiment, the three reference strains M. microti ATCC 19422, M. bovis BCG Pasteur ATCC 35734 and M. tuberculosis H37Rv were included as melting curve standards and positive controls. To exclude contaminations in the reaction mixture, ultrapure water was added as a negative control in each experiment.
Data analysis was performed using Rotor-Gene Q Software 2.3.1 (Qiagen). Normalized and difference plots were generated. To normalize the results, the pre-melt (initial fluorescence) and post-melt (final fluorescence) signals of all samples were set to uniform relative values from 100% to 0%. In order to generate difference plots, normalized fluorescence data of sample curves were subtracted from a reference curve of M. bovis BCG Pasteur ATCC 35734 to visually accentuate differences in a greater resolution. The threshold value for peak calling was set at 0.5 dF/dT. In order to alleviate false negative results due to inhibition, clinical specimens were tested in duplicate undiluted and as a 1:5 dilution. The cultured samples were tested at concentrations between 100 pg and 10 ng.
To examine the intra- and inter-assay CV of the Tm, representing the repeatability of the developed HRM method, a randomly chosen subset of 22 cultured and 19 clinical specimens were tested in triplicates in three independent runs at three different days.
Analytical Specificity
In order to proof the analytical specificity of the primers an exclusivity panel including NTMs, Nocardia paucivorans, Escherichia coli and Streptococcus suis were tested.
Efficiency and limit of detection
The efficiency and the analytical sensitivity of the RT-PCR were evaluated by triplicate testing of a 10-fold serial dilution series of each of the three reference strains. With an estimated genome size of 4.4 Mb, a DNA quantity of 4.8 fg was calculated for one GE of MTBC. The limit of detection was determined as lowest dilution with amplification of all triplicates with a standard deviation of ≤0.5.
Supplementary information
Acknowledgements
We thank Lucía de Juan Ferré and Beatriz Romero Martinez from the European Union Reference Laboratory for Bovine Tuberculosis for kindly providing a M. microti wild boar isolate from Spain.
Author Contributions
P.L. & S.S. designed the study, performed the experiments and analysed the data. P.L. wrote the manuscript. S.S. & R.S. reviewed the manuscript. All authors read and approved the final manuscript.
Data Availability
Data generated during the study is presented in an analysed format in this manuscript. Raw datasets generated from the intra- and inter-assays are included in the Supplementary Information file. Additional raw data are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-38243-6.
References
- 1.Rodriguez-Campos S, Smith NH, Boniotti MB, Aranaz A. Overview and phylogeny of Mycobacterium tuberculosis complex organisms: Implications for diagnostics and legislation of bovine tuberculosis. Res. Vet. Sci. 2014;97:S5–S19. doi: 10.1016/j.rvsc.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 2.WHO | Tuberculosis. WHO Available at, http://www.who.int/mediacentre/factsheets/fs104/en/ (Accessed: 19th March 2018).
- 3.Michalak K, et al. Mycobacterium tuberculosis infection as a zoonotic disease: transmission between humans and elephants. Emerg. Infect. Dis. 1998;4:283–287. doi: 10.3201/eid0402.980217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribeiro MG, et al. Pre−Multidrug-Resistant Mycobacterium tuberculosis Infection Causing Fatal Enteric Disease in a Dog from a Family with History of Human Tuberculosis. Transbound. Emerg. Dis. 2016;64:e4–e7. doi: 10.1111/tbed.12513. [DOI] [PubMed] [Google Scholar]
- 5.Ocepek M, Pate M, Žolnir-Dovč M, Poljak M. Transmission of Mycobacterium tuberculosis from Human to Cattle. J. Clin. Microbiol. 2005;43:3555–3557. doi: 10.1128/JCM.43.7.3555-3557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghielmetti, G. et al. Epidemiological tracing of bovine tuberculosis in Switzerland, multilocus variable number of tandem repeat analysis of Mycobacterium bovis and Mycobacterium caprae. PLoS ONE12 (2017). [DOI] [PMC free article] [PubMed]
- 7.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger EC. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thierry D, et al. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasai H, Ezaki T, Harayama S. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 2000;38:301–308. doi: 10.1128/jcm.38.1.301-308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemann S, Harmsen D, Rüsch-Gerdes S, Richter E. Differentiation of Clinical Mycobacterium tuberculosis Complex Isolates by gyrB DNA Sequence Polymorphism Analysis. J. Clin. Microbiol. 2000;38:3231–3234. doi: 10.1128/jcm.38.9.3231-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek J, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jagielski, T. et al. Current Methods in the Molecular Typing of Mycobacterium tuberculosis and Other Mycobacteria. BioMed Res. Int. 2014 (2014). [DOI] [PMC free article] [PubMed]
- 13.Costa P, et al. Rapid identification of veterinary-relevant Mycobacterium tuberculosis complex species using 16S rDNA, IS6110 and Regions of Difference-targeted dual-labelled hydrolysis probes. J. Microbiol. Methods. 2014;107:13–22. doi: 10.1016/j.mimet.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Pinsky BA, Banaei N. Multiplex real-time PCR assay for rapid identification of Mycobacterium tuberculosis complex members to the species level. J. Clin. Microbiol. 2008;46:2241–2246. doi: 10.1128/JCM.00347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddington K, et al. SeekTB, a two-stage multiplex real-time-PCR-based method for differentiation of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 2012;50:2203–2206. doi: 10.1128/JCM.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halse TA, Escuyer VE, Musser KA. Evaluation of a single-tube multiplex real-time PCR for differentiation of members of the Mycobacterium tuberculosis complex in clinical specimens. J. Clin. Microbiol. 2011;49:2562–2567. doi: 10.1128/JCM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pounder JI, et al. Mycobacterium tuberculosis complex differentiation by genomic deletion patterns with multiplex polymerase chain reaction and melting analysis. Diagn. Microbiol. Infect. Dis. 2010;67:101–105. doi: 10.1016/j.diagmicrobio.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Vossen RHAM, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum. Mutat. 2009;30:860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- 19.Jeffery N, Gasser RB, Steer PA, Noormohammadi AH. Classification of Mycoplasma synoviae strains using single-strand conformation polymorphism and high-resolution melting-curve analysis of the vlhA gene single-copy region. Microbiol. Read. Engl. 2007;153:2679–2688. doi: 10.1099/mic.0.2006/005140-0. [DOI] [PubMed] [Google Scholar]
- 20.Robertson T, et al. Characterization of Chlamydiaceae species using PCR and high resolution melt curve analysis of the 16S rRNA gene. J. Appl. Microbiol. 2009;107:2017–2028. doi: 10.1111/j.1365-2672.2009.04388.x. [DOI] [PubMed] [Google Scholar]
- 21.Stephens AJ, Inman-Bamber J, Giffard PM, Huygens F. High-resolution melting analysis of the spa repeat region of Staphylococcus aureus. Clin. Chem. 2008;54:432–436. doi: 10.1373/clinchem.2007.093658. [DOI] [PubMed] [Google Scholar]
- 22.Winchell JM, Wolff BJ, Tiller R, Bowen MD, Hoffmaster AR. Rapid identification and discrimination of Brucella isolates by use of real-time PCR and high-resolution melt analysis. J. Clin. Microbiol. 2010;48:697–702. doi: 10.1128/JCM.02021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteves LM, et al. Diagnosis of Human Leptospirosis in a Clinical Setting: Real-Time PCR High Resolution Melting Analysis for Detection of Leptospira at the Onset of Disease. Sci. Rep. 2018;8:9213. doi: 10.1038/s41598-018-27555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, et al. Rapid detection of isoniazid, rifampin, and ofloxacin resistance in Mycobacterium tuberculosis clinical isolates using high-resolution melting analysis. J. Clin. Microbiol. 2011;49:3450–3457. doi: 10.1128/JCM.01068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadav R, et al. Rapid detection of rifampicin, isoniazid and streptomycin resistance in Mycobacterium tuberculosis clinical isolates by high-resolution melting curve analysis. J. Appl. Microbiol. 2012;113:856–862. doi: 10.1111/j.1365-2672.2012.05379.x. [DOI] [PubMed] [Google Scholar]
- 26.Anthwal D, et al. Direct Detection of Rifampin and Isoniazid Resistance in Sputum Samples from Tuberculosis Patients by High-Resolution Melt Curve Analysis. J. Clin. Microbiol. 2017;55:1755–1766. doi: 10.1128/JCM.02104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Issa R, et al. High resolution melting analysis for the differentiation of Mycobacterium species. J. Med. Microbiol. 2014;63:1284–1287. doi: 10.1099/jmm.0.072611-0. [DOI] [PubMed] [Google Scholar]
- 28.Perng C-L, et al. Identification of non-tuberculous mycobacteria by real-time PCR coupled with a high-resolution melting system. J. Med. Microbiol. 2012;61:944–951. doi: 10.1099/jmm.0.042424-0. [DOI] [PubMed] [Google Scholar]
- 29.Khosravi AD, Hashemzadeh M, Hashemi Shahraki A, Teimoori A. Differential Identification of Mycobacterial Species Using High-Resolution Melting Analysis. Front. Microbiol. 2017;8:2045. doi: 10.3389/fmicb.2017.02045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright CA, Hoek KGP, Marais BJ, van Helden P, Warren RM. Combining fine-needle aspiration biopsy (FNAB) and high-resolution melt analysis to reduce diagnostic delay in Mycobacterial lymphadenitis. Diagn. Cytopathol. 2010;38:482–488. doi: 10.1002/dc.21223. [DOI] [PubMed] [Google Scholar]
- 31.Costa P, Botelho A, Couto I, Viveiros M, Inácio J. Standing of nucleic acid testing strategies in veterinary diagnosis laboratories to uncover Mycobacterium tuberculosis complex members. Front. Mol. Biosci. 2014;1:16. doi: 10.3389/fmolb.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruettger A, et al. Rapid spoligotyping of Mycobacterium tuberculosis complex bacteria by use of a microarray system with automatic data processing and assignment. J. Clin. Microbiol. 2012;50:2492–2495. doi: 10.1128/JCM.00442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supply P, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated during the study is presented in an analysed format in this manuscript. Raw datasets generated from the intra- and inter-assays are included in the Supplementary Information file. Additional raw data are available from the corresponding author on reasonable request.