Many biological species distinguish self from nonself by using different mechanisms. Higher animals recognize close kin via complex processes that often involve the five senses, cognition, and learning, whereas some microbes achieve self-recognition simply through the activity of a single genetic locus. Here we describe a single locus, traA, in myxobacteria that governs cell-cell recognition within natural populations. We found that traA is widespread across the order Myxococcales. TraA is highly polymorphic among diverse myxobacterial isolates, and such polymorphisms determine selectivity in self-recognition. Through bioinformatic and experimental analyses, we showed that traA governs many distinct recognition groups within Myxococcales. This report provides an example in which a single locus influences social recognition across a wide phylogenetic range of natural populations.
KEYWORDS: cell surface, cellular transfer, kin recognition, myxobacteria, polymorphism
ABSTRACT
Self-recognition underlies sociality in many group-living organisms. In bacteria, cells use various strategies to recognize kin to form social groups and, in some cases, to transition into multicellular life. One strategy relies on a single genetic locus that encodes a variable phenotypic tag (“greenbeard”) for recognizing other tag bearers. Previously, we discovered a polymorphic cell surface receptor called TraA that directs self-identification through homotypic interactions in the social bacterium Myxococcus xanthus. Recognition by TraA leads to cellular resource sharing in a process called outer membrane exchange (OME). A second gene in the traA operon, traB, is also required for OME but is not involved in recognition. Our prior studies of TraA identified only six recognition groups among closely related M. xanthus isolates. Here we hypothesize that the number of traA polymorphisms and, consequently, the diversity of recognition in wild isolates are much greater. To test this hypothesis, we expand the scope of TraA characterization to the order Myxococcales. From genomic sequences within the three suborders of Myxococcales, we identified 90 traA orthologs. Sequence analyses and functional characterization of traAB loci suggest that OME is well maintained among diverse myxobacterial taxonomic groups. Importantly, TraA orthologs are highly polymorphic within their variable domain, the region that confers selectivity in self-recognition. We experimentally defined 10 distinct recognition groups and, based on phylogenetic and experimental analyses, predicted >60 recognition groups among the 90 traA alleles. Taken together, our findings revealed a widespread greenbeard locus that mediates the diversity of self-recognition across the order Myxococcales.
INTRODUCTION
Bacteria are excellent team players and have evolved numerous ways to exploit group cooperation to survive in rapidly changing environments. Self-recognition is an important survival strategy that allows individuals to establish social contacts with close kin and conduct multicellular behaviors (1). Recognition in bacteria often relies on the matching of specific loci that indicate genetic relatedness between cells (2). Selectivity in self-recognition is enabled by genetic polymorphisms within these loci. In nature, bacteria are vastly diverse. However, little is known about the conservation and variability of bacterial recognition systems among wild populations. In addition, how polymorphic loci govern self-recognition across a wide phylogenetic range of natural isolates remains underexplored.
Bacteria use specific cues (e.g., diffusible chemicals, cell surface receptors) to communicate with their neighbors and recognize kin (1, 3). One example of self-recognition that we recently described is in a Gram-negative soil-dwelling bacterium, Myxococcus xanthus. Myxobacteria are well known for their sociality, which is exemplified by their ability to aggregate individuals from the environment and build multicellular fruiting bodies wherein cells differentiate into spores in response to starvation (4). To help modulate their social life, myxobacteria are equipped with recognition and discrimination systems. M. xanthus uses a polymorphic cell surface receptor, TraA, to recognize related individuals upon physical contact (5). This system is termed outer membrane exchange (OME), because bulk sharing of outer membrane (OM) components between cells occurs after recognition. The sharing of OM materials among related individuals facilitates cooperation, including the repair of damaged membranes or the phenotypic restoration of cellular defects caused by mutations (6). In contrast, if two individuals share compatible TraA receptors but are not true clonemates, then antagonistic interactions can ensue (7, 8).
Two OM proteins, TraA and TraB (TraA/B), play indispensable roles in OME (9). TraA/B function together as cell-cell adhesins. TraA is a cell surface receptor and, notably, is polymorphic within its variable domain (VD) across environmental isolates (5). These polymorphisms dictate selective cell-cell interactions, and only individuals bearing identical or nearly identical TraA receptors recognize others as self and undergo OME. Although TraB does not determine the specificity of self-recognition, it assists TraA to function in cell-cell binding and OME (10). We previously showed that coincubation of M. xanthus strains expressing different TraA receptors leads to selective cell-cell adhesion in mixed populations (10), indicating that TraA homotypic interactions govern self-recognition between cells. We also revealed the malleability of TraA recognition (10). Sequence changes within the VD can alter the TraA recognition specificity and subsequently reprogram how cells interact.
A simple and elegant recognition system that is derived from kin selection theory relies upon a single genetic locus and is called “greenbeard” recognition (11–13). The M. xanthus traA locus fulfills the criteria for identification as a greenbeard gene (14), as it (i) encodes a perceptible cue, (ii) allows recognition of others bearing the same cue, and (iii) facilitates preferential interactions among the cue-bearers. Polymorphic greenbeard loci are thought to be rare in nature, and one reason is that their genetic diversity is not easily maintained during evolution (15). In theory, the most common allele(s) is predicted to reach fixation because it provides a fitness advantage to the largest population, whereas rare alleles are eliminated because they provide fitness gains only infrequently. However, because OME among nonclonemates involves antagonistic outcomes, there is selective pressure for the maintenance and diversification of TraA recognition in OME, mediated by polymorphisms within the VD (7).
Our previous studies on 16 isolates of M. xanthus revealed six distinct recognition groups, named A through F (5, 10). Another study, using some of the same strains and additional strains all isolated from the same small patch of soil, inferred that those traA alleles belonged within the same recognition groups by bioinformatic analysis (16). Because those prior studies of TraA diversity were limited to a single species and were biased to include highly related strains, we hypothesized that the scope of TraA variability, and hence recognition diversity, was largely unexplored. Therefore, to expand our understanding of the TraA diversity landscape, we examined 90 wild-type alleles across the Myxococcales order. We evaluated the prevalence of OME in Myxococcales through the identification and, in some cases, the characterization of traAB loci from these isolates. Using phylogenetic and experimental approaches, we show that TraA is a highly polymorphic locus that governs self-recognition among the members of a diverse collection of natural myxobacterial isolates.
RESULTS
Identification of traAB alleles across the Myxococcales order.
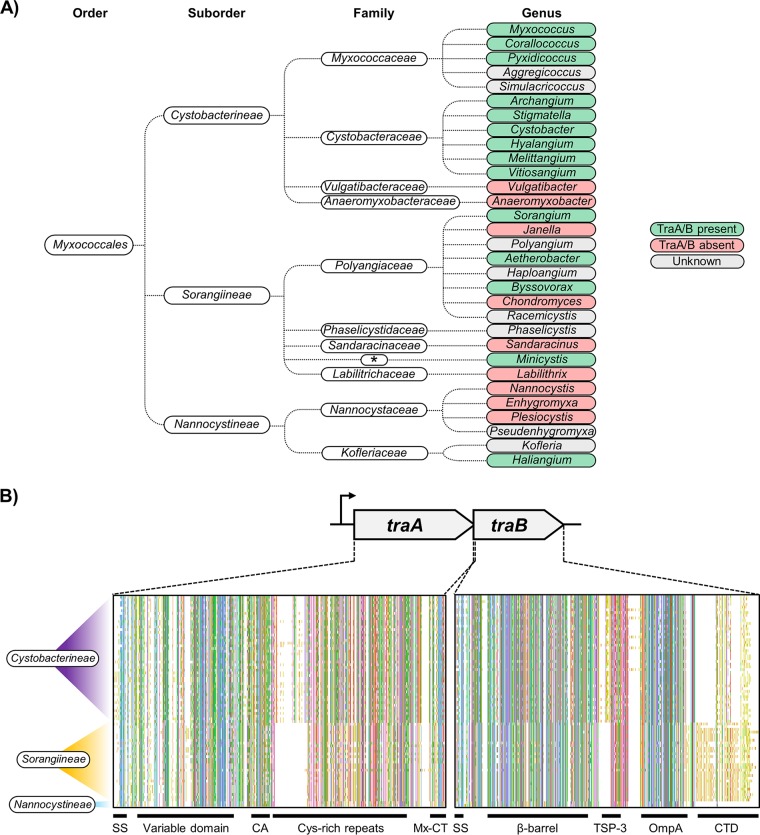
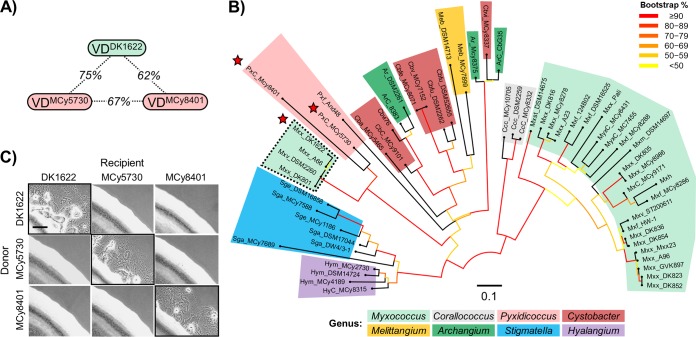
To date, orthologs of traAB have been found only within the order Myxococcales, a highly diverse group of social bacteria within the Deltaproteobacteria class. To assess the prevalence of OME in this order, we identified traAB orthologs from a wide variety of natural Myxococcales isolates from in-house and public databases. Currently, the Myxococcales order contains 31 known genera (62 species) (17–19), 23 of which have draft or complete genomic sequences that were usable in this study. Inspection of these genomes from over 100 isolates, combined with 12 traA alleles that we had previously sequenced (5), identified 90 traA genes (89 of which were unique). For all of the traA alleles found from genomic searches, we also identified the corresponding traB locus; 76 of the alleles were unique. These isolates encompassed all 3 suborders and included 4 families, 14 genera, 25 defined species, and 9 operational taxonomic units (currently with no species description) (Fig. 1A; see also Fig. S1 in the supplemental material).
FIG 1.
Identification of traAB orthologs across the order Myxococcales. (A) The monophyletic order Myxococcales currently contains three well-defined suborders, 10 families, and 31 genera. Genera that harbor or lack traAB orthologs are indicated, whereas genera with unavailable genomic sequences are shaded in gray. *, family affiliation not designated. (B) Schematics of multiple-sequence alignments of 78 TraA/B orthologs across the order Myxococcales. Amino acid residues are displayed using the Clustal X default scheme. The traAB operon is shown at the top. Domain architectures of TraA/B are labeled as follows: SS, type I signal sequence; CA, Cys-A region; Mx-CT, MYXO-CTERM (TIGR03901); TSP-3, thrombospondin type 3 (Pfam02412); CTD, carboxyl-terminal domain. Suborders of the aligned TraA/B sequences are indicated on the left.
Phylogenies of TraA and TraB orthologs identified across the order Myxococcales. Maximum likelihood trees are shown for 90 TraA orthologs (top) and 79 TraB orthologs (bottom). Different suborders, families, and genera are labeled with distinct border lines and colors. Allele prefixes indicate taxonomic origin (refer to Table 1). The A/P205 molecular switch residues are also labeled. Red stars highlight the alleles tested in Fig. 2. Branch supports are color coded. Scale bars represent the number of substitutions per residue. Download FIG S1, TIF file, 1.8 MB (1.9MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Some myxobacteria lacked traAB orthologs. For example, Anaeromyxobacter and Vulgatibacter isolates lacked traAB, but as species from these genera have significantly reduced genome sizes—about half the size of the genomes of typical myxobacteria—and have lost many of their social traits (20–23), their absence from these genera was not surprising. In other cases, myxobacteria that have large genomes and complex social behaviors (e.g., Chondromyces crocatus) also lacked traAB orthologs (24). As we considered it plausible that analogs of TraA/B could function in OME, we experimentally tested whether the C. crocatus Cm c5 strain could undergo OME with clonemates. However, using a lipophilic dye transfer assay (9), we did not detect transfer (Fig. S2), which, in conjunction with the absence of TraA/B, suggests that this strain does not undergo OME.
Fluorescent marker transfer assay performed with M. xanthus, S. cellulosum, and C. crocatus cells. (A) Cells labeled with Cy3 (donors) were mixed with the same strain labeled with CFDA-SE (recipients). M. xanthus strains DK8615 and DW1415 served as experimental positive and negative controls, respectively. Cells for which transfer of Cy3-labeled material occurred are indicated with arrowheads. (B) A lipid dye transfer assay (9) was used to test for OME in C. crocatus cells. Donor cells were labeled with red fluorescent DiD lipid dye, and recipient cells were labeled with CFDA-SE. Scale bar, 3 µm. Download FIG S2, TIF file, 4.2 MB (4.3MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The discovery of traAB across a wide spectrum of Myxococcales natural isolates suggests that OME was relatively well maintained during evolution (Fig. 1A). To explore the evolutionary relationships of these orthologs, we constructed phylogenetic trees of TraA and TraB (Fig. S1). Prefixes were assigned to traAB allele in the trees to indicate their taxonomic origins (see Table 1). TraA/B orthologs from within and related species were found to cluster together, and the TraA/B phylogenies were generally congruent with myxobacteria taxonomy (refer to Fig. 1A), suggesting that the traAB genes were primarily inherited by vertical transmission.
TABLE 1.
Abbreviations used as prefixes to traAB alleles that indicate taxonomic origins
| Prefix | Species |
|---|---|
| Aef | Aetherobacter fasciculatus |
| Aer | Aetherobacter rufus |
| AeCa | Aetherobacter clade |
| Ar | Archangium gephyra |
| ArCa | Archangium clade |
| Byc | Byssovorax cruenta |
| Ccc | Corallococcus coralloides |
| CcCa | Corallococcus clade |
| Cba | Cystobacter armeniaca |
| Cbfe | Cystobacter ferrugineus |
| Cbfu | Cystobacter fuscus |
| Cbv | Cystobacter velatus |
| Cbvi | Cystobacter violaceus |
| CbCa | Cystobacter clade |
| Hao | Haliangium ochraceum |
| Hym | Hyalangium minutum |
| HyCa | Hyalangium clade |
| Meb | Melittangium boletus |
| Mir | Minicystis rosea |
| Mxf | Myxococcus fulvus |
| Mxh | Myxococcus hansupus |
| Mxm | Myxococcus macrosporus |
| Mxst | Myxococcus stipitatus |
| Mxv | Myxococcus virescens |
| Mxx | Myxococcus xanthus |
| MxCa | Myxococcus clade |
| MyxCa | Myxococcaceae clade |
| Pxf | Pyxidicoccus fallax |
| PxCa | Pyxidicoccus clade |
| SoSa | Sorangiineae suborder |
| Soce | Sorangium cellulosum |
| Sga | Stigmatella aurantiaca |
| Sge | Stigmatella erecta |
Unclassified species that is assigned to a defined clade (C) or suborder (S) based on 16S rRNA sequence similarities.
The prototypic TraA proteins from M. xanthus isolates harbor an N-terminal VD (a distant homolog of the PA14 domain [Pfam07691]) followed by a more conserved C-terminal region that contains cysteine-rich repeats and a putative protein sorting motif called MYXO-CTERM (9). The prototypic TraB proteins harbor an N-terminal OM β-barrel domain and a C-terminal region that contains thrombospondin type 3 repeats and an OmpA domain (10). A multiple-sequence alignment of distant TraA/B orthologs shows that they have the same domain architectures as the M. xanthus TraA/B proteins (Fig. 1B). However, the patterns of sequence conservation of TraA/B orthologs did show some variation across the Myxococcales order, which prompted us to evaluate whether they share functions similar to those of the M. xanthus TraA/B proteins.
Functional analysis of traAB alleles from distant myxobacterial isolates.
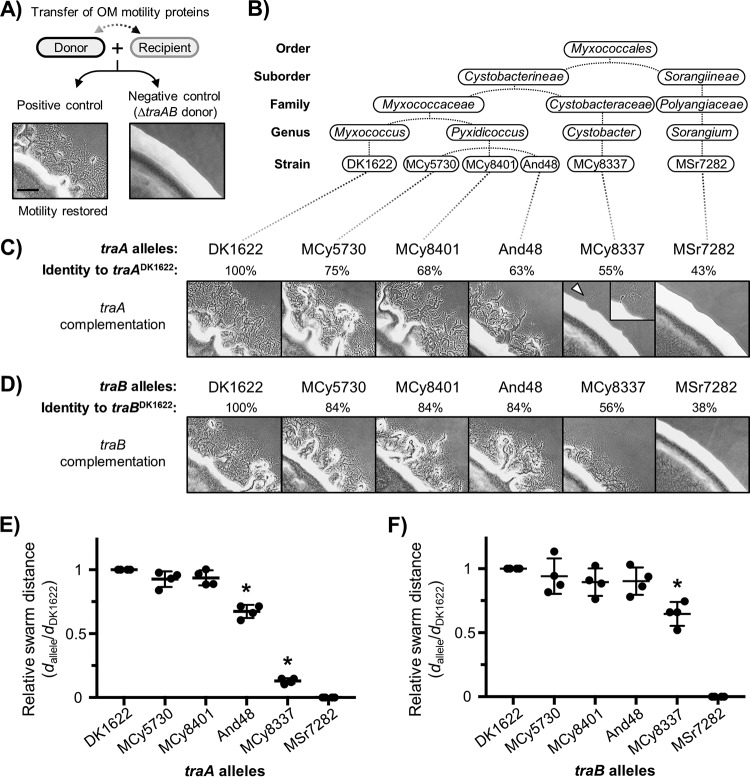
To examine whether TraA/B orthologs are functional in OME, we selected five distinct allele sets for heterologous expression in isogenic M. xanthus strains that lack their native traA or traB genes. An extracellular complementation assay named “stimulation” was used to assess whether these divergent alleles can restore OME to these M. xanthus mutants (9). In brief, this assay monitors the outcome of OME between two mutants with different motility defects (25, 26). Here, the transfer of OM lipoproteins from a nonmotile donor strain to a nonmotile recipient strain led to the restoration of motility to the recipients by providing the missing motility proteins, which was observed as emergent flares from the edge of the mixed colony (Fig. 2A). First, we expressed divergent traA alleles in the ΔtraA donor and a cognate ΔtraA recipient strain (note that these tra mutants were derived from the M. xanthus DK1622 laboratory strain) and tested for functional complementation (see Fig. 2B and Table 2 for details). As shown in Fig. 2C, TraAMCy5730, TraAMCy8401, TraAAnd48, and TraAMCy8337 were all functional in this assay whereas TraAMSr7282 was nonfunctional.
FIG 2.
Functional characterization of diverse traAB orthologs in Myxococcales. (A) Schematic of the stimulation assay that restores motility to certain mutants (recipients) by the transfer of wild-type motility proteins from nonmotile donors. A positive control (donor and recipient, with both harboring traABDK1622) and a negative control (donor lacking traAB) are shown. (B) Taxonomic origins of the traAB alleles analyzed. (C) Stimulation assays testing for functional complementation by heterologously expressing different traA alleles in isogenic ΔtraA M. xanthus strains. Each micrograph shows a mixture of donors and recipients bearing identical traA alleles. Allele names and their percentages of identical amino acids relative to traADK1622 (full length) are shown. The arrowhead highlights a small emergent flare at the edge of the colony. The inset shows an enlarged view of stimulated cells. (D) Stimulation assays testing for complementation of heterologous traB alleles in isogenic ΔtraB strains. Allele names and allele identities to traBDK1622 are indicated. Stimulation efficacy was calculated by measuring the distance (d) of the movement of emergent flares from colony edges. Data representing the relative swarming distances determined in traA and traB complementation experiments are shown in panels E and F, respectively. Four experimental replicates were done. Error bars represent standard deviations from the means. Significant differences between the DK1622 group and other groups (i.e., functionally distant alleles) are indicated by asterisks (P < 0.05 [t test]). Strain details are given in Table 2 (see also Table S1). Scale bar, 200 µm.
TABLE 2.
Taxonomic origins of the traAB alleles tested by stimulation assay
| Allele | Taxonomic description |
Strainsa | Experimental use | |
|---|---|---|---|---|
| Genus | Species | |||
| DK1622 | Myxococcus | xanthus | traADK1622 D (DK8601), traADK1622 R (DW1466) | Fig. 2A, C, and D, 5C, and 6A |
| DK816 | Myxococcus | xanthus | traADK816 D (DW1468), traADK816 R (DW2221) | Fig. 6A |
| Pali | Myxococcus | xanthus | traAPali D (DW1471), traAPali R (DW2224) | Fig. 6A |
| A96 | Myxococcus | xanthus | traAA96 D (DW1469), traAA96 R (DW2222) | Fig. 6A |
| DK805 | Myxococcus | xanthus | traADK805 D (DW2212), traADK805 R (DW2234) | Fig. 6A |
| Mf(HW-1) | Myxococcus | fulvus | traAMf D (DW1470), traAMf R (DW2223) | Fig. 6A |
| MCy5730 | Pyxidicoccus | Unclassified | traAMCy5730 D (DW2243), traAMCy5730 R (DW2248), traBMCy5730 D (DW2257) | Fig. 2C and D, 5C, and 6A |
| MCy8401 | Pyxidicoccus | Unclassified | traAMCy8401 D (DW2244), traAMCy8401 R (DW2249), traBMCy8401 D (DW2258) | Fig. 2C and D, 5C, and 6A |
| And48 | Pyxidicoccus | fallax | traAAnd48 D (DW2245), traAAnd48 R (DW2254), traBAnd48 D (DW2259) | Fig. 2C and D and 6A |
| MCy8337 | Cystobacter | violaceus | traAMCy8337 D (DW2246), traAMCy8337 R (DW2255), traBMCy8337 D (DW2260) | Fig. 2C and D |
| MSr7282 | Sorangium | cellulosum | traAMSr7282 D (DW2247), traAMSr7282 R (DW2256), traBMSr7282 D (DW2261) | Fig. 2C and D |
Strains tested in stimulation assays were constructed by expressing different traA or traB alleles in isogenic ΔtraA or ΔtraB strains derived from M. xanthus DK1622. D, donor; R, recipient. See Table S1 for additional details.
Plasmids and strains used in this study. Download Table S1, DOCX file, 0.03 MB (30.6KB, docx) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next evaluated the corresponding traB alleles by expressing them in a M. xanthus ΔtraB donor strain that harbors its native traADK1622 allele. After mixing of the donors (refer to Table 2 for strain details) with the traABDK1622 recipient, TraBMCy5730, TraBMCy8401, TraBAnd48, and TraBMCy8337 were all found to complement the OME defect of the ΔtraB strain (Fig. 2D). As previously reported (10), this result confirms that TraB does not contribute to the specificity of recognition, because the donor and recipient strains contained different traB alleles but the same traA allele. In contrast to the other four traB alleles, TraBMSr7282 was nonfunctional, as was similarly found for TraAMSr7282. Because traAB from Sorangium cellulosum MSr7282 was more divergent from the other four myxobacteria isolates (Fig. S1), we postulated that the S. cellulosum TraA/B proteins may not interact with the cognate M. xanthus DK1622 counterparts. To address this possibility, we cloned the entire traAB operon from S. cellulosum MSr7282 and placed it in ΔtraAB M. xanthus donor and recipient strains. However, OME was again not restored (data not shown).
The abilities of traAB orthologs to functionally complement OME differed, as judged from the degree of stimulation (Fig. 2E and F). Such variation correlated with the phylogenetic distance between these orthologs and traAB from DK1622 (Fig. S1). That is, the traABMCy8401 and traABMCy5730 allele sets were the most similar to traABDK1622, and they restored OME in M. xanthus to a level comparable to that seen with the native alleles. A more distant allele, traAMCy8337, showed poor functional complementation in M. xanthus (Fig. 2C, inset). To clearly assess specificity in recognition (see descriptions of experiments below), a chimeric allele was created where VDMCy8337 was fused with the C terminus of TraADK1622 (Fig. S3), with the goal of improving activity. As predicted, the resulting allele showed robust functional complementation in testing against itself in M. xanthus, indicating that the C terminus of TraA, although not involved in recognition specificity, is important for OME and for interaction with host components. Consistent with the traAMCy8337 finding, the complementation of OME by traBMCy8337 was also relatively poor compared with that seen with the other functional alleles (Fig. 2F).
Construction of a chimeric allele that harbors VDMCy8337. (A) Schematics showing the parental TraADK1622 and TraAMCy8337 alleles and the chimeric allele for which the VD from TraAMCy8337 was swapped into TraADK1622. Dashed underlines indicate the TraA fragments used to create the chimeric allele. Protein lengths are shown on the right. (B) Stimulation assays testing both parental alleles and the chimeric allele in which each allele was placed in isogenic donor/recipient strain sets. White arrowheads indicate small emergent flares. The inset shows an enlarged view of emergent flares. The chimeric traAMCy8337/DK1622 allele displayed enhanced stimulation compared with traAMCy8337 (a second copy of traBDK1622 was also introduced into the donor/recipient strains to increase stimulation levels without altering recognition specificity). Strain details are given in Table S1. Scale bar, 200 µm. Download FIG S3, TIF file, 0.9 MB (943.8KB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. cellulosum traABMSr7282 represents one of the alleles that are most distant from traABDK1622, and it failed to complement OME in M. xanthus. Importantly, M. xanthus and S. cellulosum are in different suborders (Fig. 1A), and they have major physiological differences (27). For instance, S. cellulosum has an atypical OM that lacks lipopolysaccharides (LPS) (28), whereas M. xanthus and other species within the Cystobacterineae suborder have OMs that contain LPS. Because TraA resides on the cell surface and TraB resides in the OM, these differences in OM lipid composition complicate their functional analysis. Therefore, to test for OME, we developed an alternative strategy to detect transfer in S. cellulosum by directly labeling the surface proteins with the fluorescent Cy3 dye. As an experimental control, M. xanthus cells showed robust transfer of Cy3-labeled surface proteins to recipient cells in a tra-dependent manner (Fig. S2). We tested two S. cellulosum isolates under similar conditions, but no transfer was conclusively detected (Fig. S2). These findings suggest that S. cellulosum conducts OME only under particular/regulated conditions or that the traAB genes have an alternative function in this species.
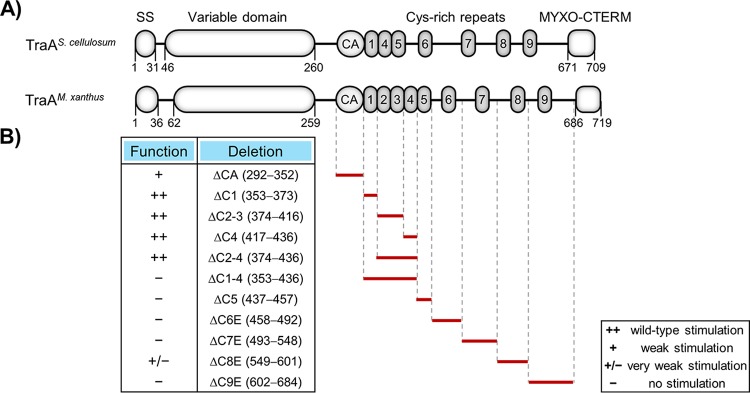
Sequence comparisons showed that S. cellulosum TraA orthologs lacked the C2 and C3 Cys-rich repeats (∼20 amino acids each) that are present in the TraA proteins from Myxococcus isolates (Fig. 1B; see also Fig. 3A and Fig. S4). Their absence raised the issue of whether these repeats were required for TraA function. We hypothesized that the Cys-rich repeats form a “stalk” to help display the recognition domain (VD) on the cell surface (4). To test for functional requirement of these and other Cys-rich repeats, we constructed a series of 11 in-frame deletions in TraA (Fig. 3B). As suggested from the S. cellulosum sequences, the first four tandem Cys-rich repeats were all individually dispensable, and, in fact, when two and even three of these repeats (but not four repeats) were deleted, full function was retained in the stimulation assay. In contrast, the distal repeats were all essential or nearly so for TraA function (Fig. 3B). We also identified an invariant sequence (SCNCCP) within tandem repeat C5 that is present in all 90 TraA orthologs (Fig. S5), suggesting that it plays a critical role. Supporting this notion, deletion of C5 abolished TraA function (Fig. 3B).
FIG 3.
Deletion analysis of the Cys-rich region within TraADK1622. (A) Domain architectures of TraA from a S. cellulosum strain (MSr7282) and a M. xanthus strain (DK1622). Cys-rich repeats are numerically labeled; note that S. cellulosum is missing C2 and C3. (B) Schematic depicting markerless in-frame deletion mutants (deleted residues listed) within the Cys-rich region of TraADK1622. The ability of these mutants to complement a ΔtraA mutant was assessed by a stimulation assay. Stimulation efficacy was judged as swarming distance of emergent flares from colony edges compared with a positive control (as described in the Fig. 2D and E legends). Wild-type stimulation, >75% efficacy; weak stimulation, ∼20% efficacy; very weak stimulation, <5% efficacy. See Table S1 for strain details.
Sequence comparison of TraASoce_MSr7282 and TraAMxx_DK1622. Domains are shaded as indicated. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Invariant adjacent cysteine residues among all TraA orthologs. The domain architecture is shown at the top. The alignment of a subregion (encompassing C4 and C5) of diverse TraA orthologs is shown at the bottom. Only one representative ortholog from each myxobacterial species is included for simplicity. Invariant sequence SCNCCP is shaded. Download FIG S5, TIF file, 0.7 MB (781.8KB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TraA recognition diversity among Myxococcales isolates.
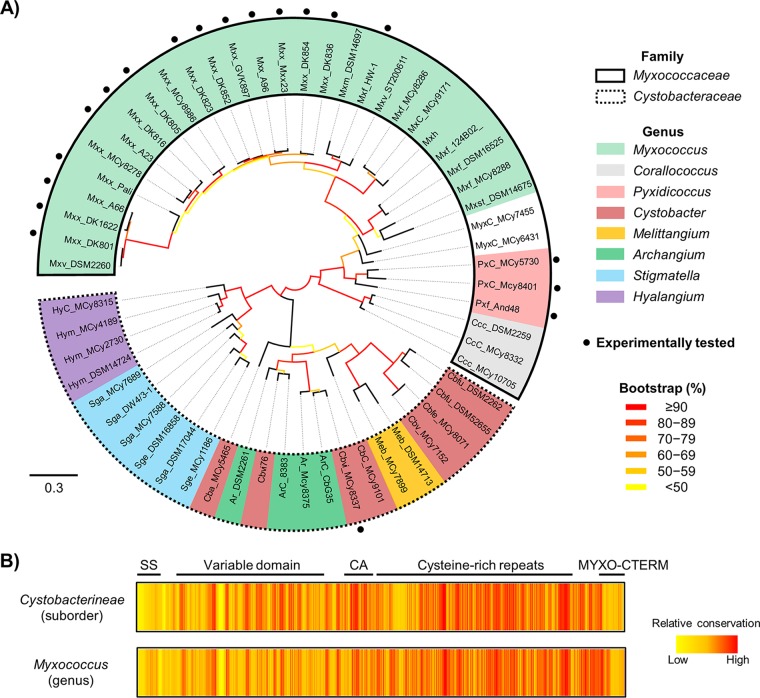
The VD in TraA dictates self-recognition among M. xanthus isolates (5). We tested whether this principle held true for TraA orthologs identified across the Myxococcales order. Because our prior work focused on TraA from the Cystobacterineae suborder, we started our analysis with this group. A phylogenetic tree of 59 full-length TraA orthologs from Cystobacterineae showed that TraA forms distinct clades that correlate with their taxonomic groupings (Fig. 4A). In addition, sequence analyses revealed that these TraA orthologs are highly divergent within their N-terminal regions, whereas their C-terminal regions are relatively conserved (Fig. 4B), supporting our findings that the VD confers recognition selectivity. Next, we experimentally analyzed two orthologs, TraAMCy8401 and TraAMCy5730, that contain VDs that are the most similar to the TraA from our M. xanthus DK1622 laboratory strain (5) (Fig. 5A and B). We note that the phylogenetic groupings of the VDs differ from the groupings of full-length TraA; e.g., we found that TraADK1622 and the corresponding recognition group A members no longer resided within the Myxococcus clade (Fig. 5B). This suggests that horizontal gene transfer had contributed to the diversification of the VD. The interactions among these three alleles were analyzed experimentally by a stimulation assay, and they displayed three distinct recognition specificities (Fig. 5C), indicating that TraA governs self-recognition beyond the Myxococcus clade (Fig. 4A).
FIG 4.
Bioinformatic analyses of TraA orthologs from the suborder Cystobacterineae. (A) Maximum likelihood tree showing the relationships among diverse Cystobacterineae TraA orthologs (full length). Families and genera are indicated by outlines and colors, respectively. Each allele prefix indicates its taxonomic origin (see Table 1 for details). TraA orthologs that were functionally characterized are marked with black dots. The scale bar represents the number of amino acid substitutions per residue. Bootstrap values (%) are color coded. (B) Heat maps showing sequence conservation of TraA orthologs from the suborder Cystobacterineae and the genus Myxococcus. TraA domain organization is indicated.
FIG 5.
TraA orthologs harboring related VDs display allele-specific recognition. (A) Pairwise amino acid sequence identity among three related VDs. (B) Maximum likelihood tree of the VDs of 59 Cystobacterineae TraA orthologs. Red stars highlight the three related VDs shown in panel A. Each allele prefix shows the taxonomic origin (see Table 1 for details). Different genera are indicated by colors. The dashed border indicates members of recognition group A. Bootstrap values are indicated with color. The scale bar represents the number of substitutions per amino acid site. (C) Stimulation assays showing specific recognition among these traA alleles. Strain details are given in Table 2 (see also Table S1). Scale bar, 200 µm.
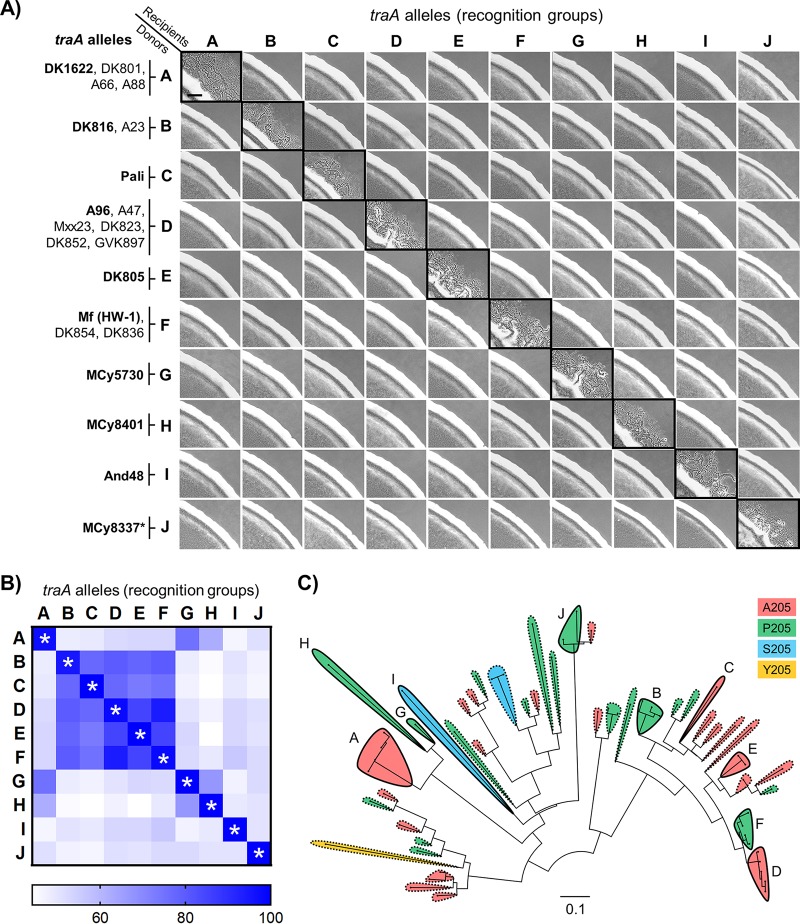
To explore the phylogenetic breadth of TraA recognition across the order Myxococcales, we expanded this analysis to the distant orthologs TraAAnd48 and TraAMCy8337. Their VDs display 46% and 51% identity to the VD of TraADK1622, respectively. Using the stimulation assay, we systematically tested the recognition specificity among the aforementioned four new environmental alleles and representative traA alleles from our prior groupings (5, 10). Pairwise interactions between isogenic donor and recipient strains bearing a collection of traA alleles revealed that they fall into 10 distinct recognition groups (Fig. 6A; strain details are given in Table 2). This result shows that traA alleles from a wide range of myxobacterial isolates display diverse specificities in cell-cell recognition.
FIG 6.
Self-recognition among a wide range of myxobacteria is governed by the traA locus. (A) Stimulation assays showing specific recognition among 10 traA alleles (names are shown in bold on the left) in an isogenic set of strains. Black borders highlight 10 distinct recognition groups (groups A to J). Additional group members that have been functionally characterized (5, 10) are also listed on the left. The asterisk indicates a chimeric allele harboring VDMCy8337 (see Fig. S3 for details). Scale bar, 200 µm. (B) Pairwise plot of (%) identity among VDs of the TraA orthologs tested in panel A (asterisks indicate self-recognition). (C) Same tree as that shown in Fig. 5B, where allele names are given. Shaded areas highlight distinct recognition groups, solid lines indicate characterized recognition groups (letters indicate group names), and dashed lines show predicted recognition groups. Groups are color coded according to the specificity-determining residue at position 205. The scale bar indicates the number of substitutions per amino acid residue.
The A/P205 molecular switch.
The malleable nature of TraA may have facilitated its diversification into distinct recognition groups (10). For instance, we previously identified a molecular switch at residue 205 [according to TraAMf(HW-1) numbering] within the VD (10). Single substitutions (e.g., A205→P or P205→A) can reprogram the selectivity of cell-cell recognition and OME. Notably, residue A/P205 is highly conserved across all TraA alleles (Fig. S1), and closely related alleles typically contain the same residue, suggesting that this molecular switch may broadly influence recognition among diverse isolates. Within this larger collection of alleles, there were, however, rare exceptions in which a serine (S) or tyrosine (Y) was located at the position corresponding to A/P205. To assess the potential roles of these alternative residues in recognition, we used TraA receptors that harbor an A or P at position 205 as templates to replace the original residue with S or Y. On the basis of results from a stimulation assay, S205 showed specificity similar to that shown by A205 (Fig. S6A), suggesting that S205 and A205 display similar TraA conformations involved in recognition. In contrast, Y substitutions at A205 or P205 abolished TraA function (Fig. S6B), indicating that Y205 might be tolerated in only a limited number of alleles. Despite the key role of A/P205, other residues are also involved in TraA specificity. Further studies are needed to elucidate the molecular and structural basis in which various VD residues govern TraA recognition.
The roles of S205 and Y205 in TraA recognition. TraA orthologs from Mf and A96 were used as templates for the construction of single amino acid substitutions. (A and B) The specificity determinant residues of TraAMf (P205) and TraAA96 (A205) were changed to S205 (A) and Y205 (B). The effects of such substitutions on TraA function/recognition were analyzed by stimulation assays. The black borders in panel A highlight that S205 exhibited the same specificity as A205. A cartoon illustration of how single residue substitutions changed the recognition specificity of TraA is shown at the bottom. The black borders in panel B show that Y205 substitutions abolished TraA function. Strain details are given in Table S1. Scale bar, 200 µm. Download FIG S6, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Finally, we sought to estimate how many distinct recognition groups were present in our collection of Cystobacterineae alleles. We first calculated the pairwise identities between VDs of all the traA alleles that were functionally characterized (Fig. 6B). Among the traA alleles that belonged to the same recognition groups, the VD identities ranged from 95% to 100%. In addition, compatible alleles contained no indels within their VDs and harbored the same residue at position 205. In contrast, VD identities in different (experimentally defined) recognition groups typically range from 45% to 85%, can contain different residues at position 205, and can contain indels. It is also worth noting that the recognition incompatibility between groups D and F is due to a single residue difference at position 205 (10) (Fig. S6), even though the sequence identity of their VDs can be as high as 94%. On the basis of these findings, we considered two uncharacterized traA alleles to be compatible if their VDs (i) displayed ≥90% identity (a conservative value between the experimentally determined values of 95% and 85%), (ii) contained no indels, and (iii) harbored identical residues at position 205. From these assumptions, we inferred that the 59 TraA orthologs from Cystobacterineae fall into 42 distinct recognition groups (Fig. 6C). In addition, we conducted similar analyses on S. cellulosum TraA alleles. As shown in Fig. S7, TraA S. cellulosum sequences were also polymorphic within their VDs, implying that they are involved in molecular recognition. Using the criteria outlined above, we predict that the 22 TraA S. cellulosum orthologs form 12 distinct recognition groups. These combined analyses highlight the polymorphisms and diversity of TraA receptors in homotypic self-recognition among wild populations of myxobacteria.
Prediction of recognition diversity among S. cellulosum TraA orthologs. (A) Heat map showing sequence conservation of 22 TraA alleles from S. cellulosum. (B) Maximum likelihood tree of the VDs from 22 S. cellulosum TraA orthologs. Dashed border lines highlight predicted recognition groups. All of the S. cellulosum TraA alleles harbored a P at position 205. Branch support data are color coded. The scale bar indicates the number of substitutions per amino acid residue. Download FIG S7, TIF file, 0.7 MB (753KB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Here we provide bioinformatic and experimental evidence that a single traA locus is sufficient to determine the recognition specificity for the sharing of cellular goods across a broad phylogenetic range of myxobacteria. Examples of single alleles governing social interactions have been described previously in a few other microbial species. For example, FLO1 promotes flocculation in yeasts, and, under certain conditions, only cells bearing this adhesin can congregate into flocs (29). However, the sequence variability of FLO1 among yeast isolates is limited and most likely determines only the competency of flocculation rather than partner specificity (29, 30). In Dictyostelium discoideum (31) and Proteus mirabilis (32), specific recognition is achieved through heterotypic interactions between two variable proteins. In contrast, our report provides a rare example in which a single polymorphic receptor is sufficient to modulate specificity in social behaviors through homotypic interactions.
We previously showed that variable TraA receptors enable cells to adhere differentially to only those cells that bear the identical receptor (10), demonstrating that homotypic binding governs selectivity. We also hypothesize that single-residue (A/P205) changes in TraA alter the conformation of a loop and hence recognition specificity (10). How TraA sequence variation generates many different binding interfaces that allow specificity in recognition remains an intriguing question. As mentioned, although TraB does not contribute toward specificity, it is required to form a functional adhesin with TraA and for OME.
What are the sources of genetic diversity within the traA locus? We suspect that the malleable nature of TraA has enabled it to tolerate de novo mutations that explore different conformations involved in homotypic recognition specificity, which in turn has led to distinct recognition groups. In addition to spontaneous mutations, horizontal gene transfer of traA has been suggested to occur between isolates (5). Horizontal acquisition of new alleles and homologous recombination between alleles likely contribute to genetic variability in traA. One example in which allele shuffling may have occurred is shown in Fig. 5B. Here, members from recognition group A formed a clade based on their VD sequences with the Pyxidicoccus isolates. However, when the full TraA sequence was used for analysis, group A members instead formed a clade with other Myxococcus isolates (Fig. 4A). In contrast to the high level of polymorphisms found within the VD, the C-terminal region of TraA displays less sequence variability. This region likely has conserved and essential roles in OME that restrict sequence variation.
How polymorphisms at a greenbeard locus are selected and maintained during evolution is an interesting puzzle (2, 14, 15). Wild populations of myxobacteria display extensive strain diversity, including at the subspecies level (33, 34). TraA polymorphisms are likely to have a role in creating social barriers that limit the cooperative behavior exemplified by sharing of cellular goods with clonemates and avoid possible adverse interactions with nonself cells (7, 8). As a greenbeard gene, traA itself determines OME compatibilities between cells, regardless of genetic relatedness at other loci. However, myxobacteria also contain a second discrimination layer to ensure that cells engaged in OME are truly clonemates or at least have a recently shared common ancestor (7). That is, a suite of polymorphic lipoprotein toxins are exchanged between cells bearing compatible TraA receptors, along with the bulk exchange of lipids and other proteins. Distinct isolates harbor different repertoires of tra-dependent polymorphic toxins and antitoxins, the latter of which are not transferred. Consequently, partners involved in OME must produce cognate antitoxins to transferred toxins to avoid a lethal outcome, which indeed occurs during OME between clonemates. We hypothesize that this antagonism provides selective pressure that drives and maintains traA allele diversity in wild populations. For example, the antagonistic interactions between cells bearing compatible TraA receptors would favor the selection of variants that display different specificities and would thus avoid antagonism with the dominant population. In turn, a novel traA recognition allele allows those individuals to cooperate among themselves and insulates them from detrimental interactions with other populations.
The wide distribution of traAB genes within the Myxococcales order supports the idea of the prevalence of OME among diverse isolates. OME is likely beneficial to myxobacteria as a consequence of promoting cooperation among clonemates and antagonism toward nonkin, which in turn promotes population diversity and resilience (6, 7, 35). Given the important roles of OME in modulating social behaviors, we were struck by the fact that some myxobacteria, e.g., C. crocatus, lack traAB. One obvious reason why some species lack traAB is that the exchange of toxins creates intense interstrain competition. This antagonism is alleviated by removing TraA/B function, and, indeed, this rapidly occurs when two strains compete by OME under laboratory conditions (8, 36). In addition, OME is energetically costly because it involves OM fusion and bulk cellular exchange. Loss of OME may thus provide fitness benefits under certain conditions. Finally, as noted above, Anaeromyxobacter and Vulgatibacter isolates also lack traAB, which may be related to their smaller genomes and is consistent with the absence of a number of social traits in these species (20–23).
The TraA/B proteins from S. cellulosum were not functional under conditions of heterologous expression in M. xanthus. Given the unique OM lipid composition of S. cellulosum (28), we suspect that their TraA/B proteins evolved to function in a different physiochemical environment that is not compatible with OME function in the M. xanthus OM. However, the TraA VDs of S. cellulosum isolates are also highly polymorphic (see Fig. S7 in the supplemental material), suggesting that they function in molecular recognition. Given that the S. cellulosum TraA/B domain architecture is like that of other TraA/B proteins (Fig. 3A), the S. cellulosum TraA/B proteins likely reside on the cell surface and function as adhesins. Nevertheless, further studies are needed to elucidate the function of TraA/B in S. cellulosum and to test whether OME is a regulated process that occurs only under certain conditions or during specific developmental stages.
Taken together, our results have shown the prevalence and diversity of the traAB genes, and hence the widespread occurrence of OME, in the order Myxococcales. From the analyses of the 90 TraA sequences, we predict that they represent >60 distinct TraA recognition groups (42 from Cystobacterineae, 12 from S. cellulosum, and the remaining number from other taxonomic groups shown in Fig. S1). To our knowledge, there are no other loci involved in cooperative social interactions that offer the diversity in recognition that has been reported here for TraA. Given that the extent of Myxococcales taxonomic diversity is currently unknown but clearly is vast (17, 33), our 90 representative alleles thus represent a great underestimation of the global diversity of traA. On the basis of this, we think there are many more distinct TraA recognition groups in nature that influence social interactions within the Myxococcales order.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table S1 in the supplemental material. M. xanthus was cultured in CTT medium (1% [wt/vol] Casitone, 10 mM Tris-HCl [pH 7.6], 1 mM KH2PO4, 8 mM MgSO4) in the dark at 33°C with shaking. S. cellulosum was maintained on VY/2 (37) agar medium in the dark at 33°C. To propagate S. cellulosum in liquid culture, soluble medium M [0.5% soy peptone, 1% maltose-monohydrate, 0.1% CaCl2·2H2O, 0.1% MgSO4·7H2O, 22 µM EDTA iron(III) sodium salt, 1.2% HEPES, pH 7.2] (38) was used. Escherichia coli was cultured in LB at 37°C. To generate solid medium for plate culture, 1.5% (wt/vol) agar (Criterion agar; Hardy Diagnostics) was added to the medium. For selection, 50 µg/ml kanamycin (Km) and 50 µg/ml zeocin (Zeo) were added to the media as needed.
Searches for traAB orthologs.
BLASTP searches were performed to identify orthologs of traAB across the Myxococcales order. Sequences of the TraA and TraB proteins from M. xanthus DK1622 were used as queries. The traAB orthologs with E values of <1 × 10−100 and query coverage values of >80% were retained for further analysis. In addition to searching for orthologs in the nonredundant NCBI protein database and the integrated microbial genomes (IMG) database (39), we also searched myxobacterial genomes from our in-house database (Müller laboratory) by following the same criteria. Next, we determined whether these sequences were traAB orthologs by (i) comparing their predicted domain architectures to that of TraA/BDK1622 and (ii) confirming that the corresponding parental strains harbored both traA and traB orthologs. In all cases examined, the corresponding genes overlapped in an operon. We also included an additional 12 TraA sequences from our prior work (5), in which traB was not sequenced. In one case (Cbfu_MCy9118), only a TraB ortholog was identified, probably because the genome sequence was not complete. Finally, we manually inspected each ortholog to correct for annotation errors, particularly analyzing whether the most likely start codon had been chosen. The final sequence files included 90 TraA orthologs and 79 TraB orthologs.
Bioinformatic analysis.
Sequences of TraA/B orthologs were aligned in MUSCLE (40). To create Fig. 1B, the alignments were visualized in Jalview (41) using the Clustal X default color scheme. To determine the boundaries of the VDs, orthologs were compared to TraADK1622 in alignments. The VD in TraADK1622 encompasses amino acids 62 to 259, and the sequences of other TraA orthologs were trimmed accordingly.
For phylogenetic analyses, maximum likelihood (ML) phylogenetic trees were constructed in PhyML 3.0 (42). We first predicted the best-fitting evolutionary models based on the corrected Akaike information criterion in ProtTest 3 (43). To generate phylogeny for the 59 TraA orthologs from the Cystobacterineae suborder (Fig. 4A), the best model (WAG+I+G+F) chosen by ProtTest was used, and 1,000 bootstrap replicates were performed. Regions encompassing the VDs of Cystobacterineae TraA orthologs were also used to generate a ML tree (Fig. 5B; see also Fig. 6C), based on a JTT+I+G model and 1,000 bootstrap replicates. We constructed a phylogenetic tree for the VDs of S. cellulosum TraA orthologs by using a WAG+I+G model and 1,000 bootstrap replicates (see Fig. S7 in the supplemental material). In addition, all the TraA and TraB sequences from the Myxococcales order were used to generate ML trees (Fig. S1) using a WAG+I+G model (300 bootstrap replicates) and a LG+I+G+F model (500 bootstrap replicates), respectively. Phylogenetic trees were visualized in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
To analyze residue conservation of diverse TraA orthologs, we first aligned the desired sequences (alignment gaps were removed) and scored residue conservation using Jensen-Shannon divergence (44). The corresponding heat maps were generated in Microsoft Excel. Protein sequence similarity analyses were conducted using SIM (45). A heat map showing the pairwise sequence identity of diverse VDs (Fig. 6B) was generated in Prism GraphPad (GraphPad Software, Inc.).
Plasmid and strain construction.
Plasmids and primers used in this study are listed in Table S1 and Table S2. The traA- and traB-containing plasmids (pPC26 to pPC35) were constructed by inserting appropriate alleles downstream of the pilA promoter (PpilA) in the pDP22 vector (between XbaI and HindIII) as described previously (10). To generate the traA chimeric allele (pPC36), desired subregions of traADK1622 and traAMCy8337 were ligated into pDP22 (linearized by EcoRI and XbaI digestion) using Gibson Assembly Master Mix (New England Biolabs) as described previously (10). Site-directed mutagenesis was done by Gibson assembly as described previously (10) to generate pPC37-40. To create markerless in-frame deletions within the Cys-rich region of TraA, different traA fragments harboring the desired deletions were synthesized (GenScript) and subcloned into traA-containing plasmid pDP27 (linearized using SacI and NgoMIV or NgoMIV and HindIII) to create pXW8 to pXW11 and pXW15 to pXW17. The rest of the Cys-rich deletion mutants (pXW12 to pXW14 and pXW18) were generated through Gibson assembly. To create pXW7, the neoR/kanR genes were first removed from pCR-XL-TOPO (Invitrogen) by PCR and blunt-end self-ligation. Subsequently, traBDK1622 and a fragment for integration at the Mx9 attachment site were introduced into the multiple cloning sites of this plasmid.
Primers used in this study. Download Table S2, DOCX file, 0.03 MB (27.7KB, docx) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All plasmids were verified by PCR, restriction enzyme digestion and/or by DNA sequencing. Plasmids were then transformed into M. xanthus cells and selected with appropriate antibiotics. All cloning vectors were integrated at the Mx8 attachment site of the M. xanthus genome, except for pXW7 (Mx9 attachment site).
Stimulation assay.
This assay was done as described previously (10). Briefly, M. xanthus cells were cultured overnight to mid-log phase, washed, and resuspended in TPM buffer (10 mM Tris-HCl [pH 7.6], 1 mM KH2PO4, 8 mM MgSO4) to a calculated density of ∼2.5 × 109 cells/ml. Mixtures (1:1 ratio) of desired strain combinations were then spotted onto 1/2 CTT (Casitone reduced to 0.5%) agar plates supplemented with 2 mM CaCl2. The edges of colonies were imaged after overnight incubation at 33°C.
Stimulation efficacy was estimated by measuring the distance of each swarm of emergent flares from the inoculum edges. Cell mixtures subjected to this analysis were spotted onto the same agar plate and incubated for the same time before imaging. The averaged swarm distance (dallele) was determined for each colony in ImageJ software (https://imagej.nih.gov/ij/). Swarm distances (dallele) of different mixtures were normalized against the swarm distance of the control group (dDK1622) on the same agar plate. Four experimental replicates were performed. Data analyses, including unpaired and two-tailed t tests, were done in Prism GraphPad.
Transfer assay.
We developed a method to label myxobacterial OM proteins with Cy3 (Lumiprobe) to test for transfer between cells. Live cells were grown to mid-log phase, washed, and incubated with Cy3 dyes in appropriate liquid medium as follows: for M. xanthus, 50 µg/ml Cy3 and 1-h incubation in CTT; for S. cellulosum, 1 µg/ml Cy3 and 15-min incubation in medium M. After incubation, the stained cells were washed three to five times in fresh medium and resuspended to ∼5 × 108 cells/ml. Recipient cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFDA-SE; Invitrogen) as described previously (5). Cy3-labeled donors and CFDA-SE-labeled recipients were then mixed (1:1 ratio) and incubated on agarose pads consisting of 1.2% agarose in the appropriate medium. Cell mixtures were incubated in the dark at 33°C for 0.5 h for M. xanthus cells and for ≥4 h for S. cellulosum cells before imaging was performed. To test for OME in C. crocatus Cm c5, a lipid dye transfer assay was done as described previously (9). Donor cells were labeled with a red fluorescent DiD lipid dye (lipophilic tracer sampler kit; Invitrogen), whereas the recipients were labeled with CFDA-SE (Invitrogen).
Microscopy.
Images of stimulation experiments were acquired using a Nikon E800 phase contrast/fluorescence microscope equipped with a 10× lens objective. Fluorescence microscopy of transfer assays was done with a 60× oil lens objective and fluorescein isothiocyanate (FITC) or Texas Red filter sets.
Data accessibility.
The new traAB DNA sequences used in this study have been deposited into the GenBank database under accession numbers MK091159 to MK091255.
ACKNOWLEDGMENTS
We thank Wei Hu for a strain.
This work was supported by the National Institutes of Health grant GM101449 and Wyoming INBRE (2P20GM103432) to D.W.
Footnotes
Citation Cao P, Wei X, Awal RP, Müller R, Wall D. 2019. A highly polymorphic receptor governs many distinct self-recognition types within the Myxococcales order. mBio 10:e02751-18. https://doi.org/10.1128/mBio.02751-18.
REFERENCES
- 1.Troselj V, Cao P, Wall D. 2018. Cell-cell recognition and social networking in bacteria. Environ Microbiol 20:923–933. doi: 10.1111/1462-2920.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wall D. 2016. Kin recognition in bacteria. Annu Rev Microbiol 70:143–160. doi: 10.1146/annurev-micro-102215-095325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao P, Dey A, Vassallo CN, Wall D. 2015. How myxobacteria cooperate. J Mol Biol 427:3709–3721. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pathak DT, Wei X, Dey A, Wall D. 2013. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet 9:e1003891. doi: 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, Wall D. 2015. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci U S A 112:E2939–E2946. doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vassallo CN, Cao P, Conklin A, Finkelstein H, Hayes CS, Wall D. 2017. Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria. eLife 6:e29397. doi: 10.7554/eLife.29397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dey A, Vassallo CN, Conklin AC, Pathak DT, Troselj V, Wall D. 2016. Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. J Bacteriol 198:994–1004. doi: 10.1128/JB.00964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. 2012. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet 8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao P, Wall D. 2017. Self-identity reprogrammed by a single residue switch in a cell surface receptor of a social bacterium. Proc Natl Acad Sci U S A 114:3732–3737. doi: 10.1073/pnas.1700315114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawkins R. 1976. The selfish gene. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 12.Dawkins R. 1983. The extended phenotype: the gene as the unit of selection (1982 ed) Freeman, New York, NY. [Google Scholar]
- 13.Haig D. 1996. Gestational drive and the green-bearded placenta. Proc Natl Acad Sci U S A 93:6547–6551. doi: 10.1073/pnas.93.13.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner A, West SA. 2010. Greenbeards. Evolution 64:25–38. doi: 10.1111/j.1558-5646.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- 15.Crozier R. 1986. Genetic clonal recognition abilities in marine invertebrates must be maintained by selection for something else. Evolution 40:1100–1101. doi: 10.1111/j.1558-5646.1986.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 16.Wielgoss S, Fiegna F, Rendueles O, Yu YTN, Velicer GJ. 2018. Kin discrimination and outer membrane exchange in Myxococcus xanthus: a comparative analysis among natural isolates. Mol Ecol 27:3146–3158. doi: 10.1111/mec.14773. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann T, Krug D, Bozkurt N, Duddela S, Jansen R, Garcia R, Gerth K, Steinmetz H, Müller R. 2018. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat Commun 9:803. doi: 10.1038/s41467-018-03184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landwehr W, Wolf C, Wink J. 2016. Actinobacteria and myxobacteria—two of the most important bacterial resources for novel antibiotics, p 273–302. In How to overcome the antibiotic crisis. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 19.Awal RP, Garcia R, Gemperlein K, Wink J, Kunwar B, Parajuli N, Müller R. 2017. Vitiosangium cumulatum gen. nov., sp. nov. and Vitiosangium subalbum sp. nov., soil myxobacteria, and emended descriptions of the genera Archangium and Angiococcus, and of the family Cystobacteraceae. Int J Syst Evol Microbiol 67:1422–1430. doi: 10.1099/ijsem.0.001829. [DOI] [PubMed] [Google Scholar]
- 20.Sanford RA, Cole JR, Tiedje JM. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol 68:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas SH, Wagner RD, Arakaki AK, Skolnick J, Kirby JR, Shimkets LJ, Sanford RA, Löffler FE. 2008. The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria. PLoS One 3:e2103. doi: 10.1371/journal.pone.0002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto E, Muramatsu H, Nagai K. 2014. Vulgatibacter incomptus gen. nov., sp. nov. and Labilithrix luteola gen. nov., sp. nov., two myxobacteria isolated from soil in Yakushima Island, and the description of Vulgatibacteraceae fam. nov., Labilitrichaceae fam. nov. and Anaeromyxobacteraceae fam. nov. Int J Syst Evol Microbiol 64:3360–3368. doi: 10.1099/ijs.0.063198-0. [DOI] [PubMed] [Google Scholar]
- 23.Huntley S, Hamann N, Wegener-Feldbrügge S, Treuner-Lange A, Kube M, Reinhardt R, Klages S, Müller R, Ronning CM, Nierman WC, Søgaard-Andersen L. 2011. Comparative genomic analysis of fruiting body formation in Myxococcales. Mol Biol Evol 28:1083–1097. doi: 10.1093/molbev/msq292. [DOI] [PubMed] [Google Scholar]
- 24.Zaburannyi N, Bunk B, Maier J, Overmann J, Müller R. 15 January 2016. Genome analysis of the fruiting body forming myxobacterium Chondromyces crocatus reveals high potential for natural product biosynthesis. Appl Environ Microbiol doi: 10.1128/AEM.03011-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pathak DT, Wall D. 17 February 2012. Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J Bacteriol doi: 10.1128/JB.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakobczak B, Keilberg D, Wuichet K, Søgaard-Andersen L. 2015. Contact-and protein transfer-dependent stimulation of assembly of the gliding motility machinery in Myxococcus xanthus. PLoS Genet 11:e1005341. doi: 10.1371/journal.pgen.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, Bode HB, Bolten CJ, Choudhuri JV, Doss S, Elnakady YA, Frank B, Gaigalat L, Goesmann A, Groeger C, Gross F, Jelsbak L, Jelsbak L, Kalinowski J, Kegler C, Knauber T, Konietzny S, Kopp M, Krause L, Krug D, Linke B, Mahmud T, Martinez-Arias R, McHardy AC, Merai M, Meyer F, Mormann S, Muñoz-Dorado J, Perez J, Pradella S, Rachid S, Raddatz G, Rosenau F, Rückert C, Sasse F, Scharfe M, Schuster SC, Suen G, Treuner-Lange A, Velicer GJ, Vorhölter F-J, et al. 2007. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25:1281. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- 28.Keck M, Gisch N, Moll H, Vorhölter F-J, Gerth K, Kahmann U, Lissel M, Lindner B, Niehaus K, Holst O. 2011. Unusual outer membrane lipid composition of the gram-negative, lipopolysaccharide-lacking myxobacterium Sorangium cellulosum So ce56. J Biol Chem 286:12850–12859. doi: 10.1074/jbc.M110.194209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé J-P, Fink GR, Foster KR, Verstrepen KJ. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goossens KV, Ielasi FS, Nookaew I, Stals I, Alonso-Sarduy L, Daenen L, Van Mulders SE, Stassen C, Van Eijsden RG, Siewers V. 2015. Molecular mechanism of flocculation self-recognition in yeast and its role in mating and survival. mBio 6:e00427-15. doi: 10.1128/mBio.00427-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruenheit N, Parkinson K, Stewart B, Howie JA, Wolf JB, Thompson CR. 2017. A polychromatic ‘greenbeard’ locus determines patterns of cooperation in a social amoeba. Nat Commun 8:14171. doi: 10.1038/ncomms14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardarelli L, Saak C, Gibbs KA. 2015. Two proteins form a heteromeric bacterial self-recognition complex in which variable subdomains determine allele-restricted binding. mBio 6:e00251-15. doi: 10.1128/mBio.00251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohr K. 2018. Diversity of myxobacteria—we only see the tip of the iceberg. Microorganisms 6:84. doi: 10.3390/microorganisms6030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vos M, Velicer GJ. 2009. Social conflict in centimeter-and global-scale populations of the bacterium Myxococcus xanthus. Curr Biol 19:1763–1767. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Bayona L, Comstock LE. 2018. Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 36.Dey A, Wall D. 2014. A genetic screen in Myxococcus xanthus identifies mutants that uncouple outer membrane exchange from a downstream cellular response. J Bacteriol 196:4324–4332. doi: 10.1128/JB.02217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichenbach H, Dworkin M. 1992. The myxobacteria, p 3416–3487. In The prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 38.Müller R, Gerth K. 2006. Development of simple media which allow investigations into the global regulation of chivosazol biosynthesis with Sorangium cellulosum So ce56. J Biotechnol 121:192–200. doi: 10.1016/j.jbiotec.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Chen I-MA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, Huntemann M, Varghese N, White JR, Seshadri R, Smirnova T, Kirton E, Jungbluth SP, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2019. IMG/M v. 5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 43.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capra JA, Singh M. 2007. Predicting functionally important residues from sequence conservation. Bioinformatics 23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 45.Huang X, Miller W. 1991. A time-efficient, linear-space local similarity algorithm. Adv Appl Math 12:337–357. doi: 10.1016/0196-8858(91)90017-D. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenies of TraA and TraB orthologs identified across the order Myxococcales. Maximum likelihood trees are shown for 90 TraA orthologs (top) and 79 TraB orthologs (bottom). Different suborders, families, and genera are labeled with distinct border lines and colors. Allele prefixes indicate taxonomic origin (refer to Table 1). The A/P205 molecular switch residues are also labeled. Red stars highlight the alleles tested in Fig. 2. Branch supports are color coded. Scale bars represent the number of substitutions per residue. Download FIG S1, TIF file, 1.8 MB (1.9MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fluorescent marker transfer assay performed with M. xanthus, S. cellulosum, and C. crocatus cells. (A) Cells labeled with Cy3 (donors) were mixed with the same strain labeled with CFDA-SE (recipients). M. xanthus strains DK8615 and DW1415 served as experimental positive and negative controls, respectively. Cells for which transfer of Cy3-labeled material occurred are indicated with arrowheads. (B) A lipid dye transfer assay (9) was used to test for OME in C. crocatus cells. Donor cells were labeled with red fluorescent DiD lipid dye, and recipient cells were labeled with CFDA-SE. Scale bar, 3 µm. Download FIG S2, TIF file, 4.2 MB (4.3MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids and strains used in this study. Download Table S1, DOCX file, 0.03 MB (30.6KB, docx) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of a chimeric allele that harbors VDMCy8337. (A) Schematics showing the parental TraADK1622 and TraAMCy8337 alleles and the chimeric allele for which the VD from TraAMCy8337 was swapped into TraADK1622. Dashed underlines indicate the TraA fragments used to create the chimeric allele. Protein lengths are shown on the right. (B) Stimulation assays testing both parental alleles and the chimeric allele in which each allele was placed in isogenic donor/recipient strain sets. White arrowheads indicate small emergent flares. The inset shows an enlarged view of emergent flares. The chimeric traAMCy8337/DK1622 allele displayed enhanced stimulation compared with traAMCy8337 (a second copy of traBDK1622 was also introduced into the donor/recipient strains to increase stimulation levels without altering recognition specificity). Strain details are given in Table S1. Scale bar, 200 µm. Download FIG S3, TIF file, 0.9 MB (943.8KB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence comparison of TraASoce_MSr7282 and TraAMxx_DK1622. Domains are shaded as indicated. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Invariant adjacent cysteine residues among all TraA orthologs. The domain architecture is shown at the top. The alignment of a subregion (encompassing C4 and C5) of diverse TraA orthologs is shown at the bottom. Only one representative ortholog from each myxobacterial species is included for simplicity. Invariant sequence SCNCCP is shaded. Download FIG S5, TIF file, 0.7 MB (781.8KB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The roles of S205 and Y205 in TraA recognition. TraA orthologs from Mf and A96 were used as templates for the construction of single amino acid substitutions. (A and B) The specificity determinant residues of TraAMf (P205) and TraAA96 (A205) were changed to S205 (A) and Y205 (B). The effects of such substitutions on TraA function/recognition were analyzed by stimulation assays. The black borders in panel A highlight that S205 exhibited the same specificity as A205. A cartoon illustration of how single residue substitutions changed the recognition specificity of TraA is shown at the bottom. The black borders in panel B show that Y205 substitutions abolished TraA function. Strain details are given in Table S1. Scale bar, 200 µm. Download FIG S6, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prediction of recognition diversity among S. cellulosum TraA orthologs. (A) Heat map showing sequence conservation of 22 TraA alleles from S. cellulosum. (B) Maximum likelihood tree of the VDs from 22 S. cellulosum TraA orthologs. Dashed border lines highlight predicted recognition groups. All of the S. cellulosum TraA alleles harbored a P at position 205. Branch support data are color coded. The scale bar indicates the number of substitutions per amino acid residue. Download FIG S7, TIF file, 0.7 MB (753KB, tif) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, DOCX file, 0.03 MB (27.7KB, docx) .
Copyright © 2019 Cao et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The new traAB DNA sequences used in this study have been deposited into the GenBank database under accession numbers MK091159 to MK091255.