Abstract
While the binding of biotin by streptavidin does not appear to be cooperative in the traditional sense of altered binding strength, it has been suggested that it may be cooperative in terms of differential structural changes in the protein. In this work we present intrinsic tryptophan fluorescence data as evidence of a cooperative structural change. The technique involves examination of the differences in fluorescence emission corresponding to distinct tryptophan populations accompanying protein-ligand binding. Specifically we note that the 335 nm emission population (i.e. more hydrophobic) saturates prior to the saturation of the 350 nm emission population commonly used in the standard binding activity assay. We also note that the wavelength of maximum emission, total integrated fluorescence emission and full width at half maximum during the titration of ligand into streptavidin also reach saturation before the expected 4:1 stoichiometric end point. This suggests that the binding of the first 3 biotins effect greater structural changes in the protein than the final ligand.
Keywords: Streptavidin, Tryptophan fluorescence, Protein-ligand binding, Photobleaching, Allosterism
Highlights
-
•
Structurally sensitive intrinsic Trp fluorescence changes upon biotin addition.
-
•
Key features of this fluorescence stops changing prior to full biotin saturation.
-
•
This suggests that ligand-induced structural changes are allosteric.
-
•
We introduce use of ‘cooperative allosterism’ to distinguish from traditional usage.
1. Introduction
Traditional definitions of cooperativity involve multiple binding sites where the binding of a ligand to the first site increases the affinity of subsequent ligands as measured by equilibrium binding constants. While one can create models to explain cooperativity without allosteric changes within the quaternary structure of a multimeric protein, allosterism often provides the structural basis of cooperativity. The reverse is not necessarily the case, however, as ligand binding may induce structural rearrangements across subunits without changing the intrinsic binding affinity.
When measuring binding affinity, one often follows an intrinsic signal from either the protein or the ligand to monitor binding. If the signal change is directly proportional to fraction of ligand bound it is straightforward to build binding models to fit equilibrium binding constants and cooperativity values [1]. The validity of the proportionality of signal change to fraction of ligand bound may be verified by a general method of analysis where equilibrium binding events are considered [2]. When the association constant is high enough, the binding is essentially stoichiometric and cannot be handled in that manner. Those binding plots of saturation vs. [ligand] are generally biphasic linear plots and cannot be used to determine binding affinity.
In the case of the specific binding of biotin (vitamin H) to streptavidin and the analogous animal protein, avidin, the affinity is essentially stoichiometric. Due to this high affinity interaction, these proteins have been the focus of much study and have been used in the development of a wide range of biotechnology and biomedical techniques. The tetrameric protein is known to be quite stable with respect to heat and chemical denaturants [3], [4], [5], [6], with a Tm greater than 75 °C in the absence of denaturant [4], [7]. Further, it has been shown that this structural stability is significantly enhanced upon the binding of biotin [4]. Structural studies of apo- and holo-streptavidin indicate that some of this added stability can be attributed to the movement of W120 across the dimer-dimer waistline, from one subunit into the binding pocket of a neighboring subunit upon binding of biotin [6], [8]. A few studies have provided evidence that there is a general tightening of the structure upon binding [6], [9], [10].
While the binding constants for biotin and some analogs with streptavidin have been measured, and there are x-ray structures for some of these complexes, there has been some ongoing discussion of the binding cooperativity, or lack thereof. Sano and Cantor originally proposed that the binding of biotin to streptavidin was cooperative, based on observations from electrophoretic mobility in the presence and absence of urea [11]. Later Jones and Kurzban came to the opposite conclusion based on chromatographic separation of streptavidin-biotin complexes formed by the equilibrium exchange of biotin between apo- and holo-streptavidin [12]. The data of Jones and Kurzban suggest that there is little, if any, difference in the affinity for the four binding sites in streptavidin [12]. However, since the binding is effectively stoichiometric, measurement of differences in affinity would be very hard to discern. Hyre et al. showed that hydrogen bonding between S45 and D128 with biotin is cooperative through calorimetry and x-ray diffraction [13]. It has also been suggested that biotin binding to streptavidin is positively cooperative, though, perhaps not in the classical sense where binding affinity is altered [6], [9]. This recent evidence indicates that the binding affinity itself may not be cooperative, but that the binding events are accompanied by cooperative structural changes of the tetramer [6], [9]. We shall henceforth refer to this as “cooperative allosterism.”
The streptavidin gene codes for a 159 residue subunit, though natural proteolysis can cleave residues 1–14 and 139–159 from the full length streptavidin, leading to ‘core’ streptavidin [14], [15]. Both full length and core streptavidin contain 6 tryptophan residues per subunit, which serve as structural probes of the protein conformation via intrinsic fluorescence. Upon excitation at 280 nm, the tryptophans in streptavidin exhibit an emission maximum at 333–335 nm [16], [17], indicating that they are relatively inaccessible to solvent (i.e. a more hydrophobic environment). At this excitation wavelength there is also excitation of intrinsic fluorescence of tyrosine (λem,max=303 nm), which will be addressed later in this work. Upon saturation with biotin a fluorescence blue shift of 4–5 nm accompanied by a 25–39% decrease in intensity at 350 nm has been reported [6], [17]. Upon unfolding, however, this emission red shifts to 350 nm as the tryptophan residues are fully exposed to the aqueous solvent [7], [16]. Others have shown that monitoring fluorescence intensity at 350 nm as a function of biotin added provides an assay for the binding of biotin to avidin [18], [19]. Monitoring this binding event should be independent of the wavelength used if the signal change originates from the local vicinity of the binding pocket where each binding event may be viewed as identical and independent.
2. Materials and methods
2.1. Materials
Streptavidin (SA 10) was obtained from Prozyme (San Leandro, CA) and used without further purification. This source was chosen due to the high degree of homogeneity and lot-to-lot consistency [7]. Protein stock solutions were prepared at roughly 3–6 mg/mL in 10 mM triethanolamine (TEA) at a pH 7.3 (titrated with HCl). Streptavidin concentration was determined using A280 with = 3.2 cm−1 [20]. Type I, 18 MΩ water was used to prepare all solutions. D-biotin was obtained from Fluka (Buchs, Switzerland) and Sigma (St. Louis, MO) and also dissolved in the 10 mM TEA buffer at pH 7.3.
2.2. Fluorescence spectroscopy
A Photon Technology International (PTI, Lawrenceville, NJ) QuantaMaster Dual-Emission Spectrofluorimeter with a thermostatted single sample cell was used for all fluorescence analyses. Excitation illumination at 280–290 nm was provided by a 75 W output power Xenon arc lamp. Bandpasses were 2 nm and 8 nm for excitation and emission, respectively. Photomultiplier tube (PMT) voltages were 980–1000 V. Upon each addition of protein or ligand to the solution a 2 min equilibration period was allowed. Samples were subjected to constant stirring.
In all cases the excitation shutter was closed between readings to avoid photobleaching. In timebased experiments where multiple emission wavelengths were examined, all emission wavelengths were examined on each instrument channel. Fluorescence emission intensity was corrected for background emission and dilution of streptavidin.
A nominal Streptavidin concentration of 0.8–1 μM provided emission maxima of approximately 1.5–3 × 106 counts/s, well under the 3.7 × 106 signal saturation point of the instrument. A standard 1 cm path length quartz cuvette was used for all experiments. Starting with a 2 mL volume of protein and titrating in 6 µL aliquots of 83–104 µM biotin solution. Concentration of biotin solutions was determined using the standard 350 nm emission titration curve [18], [19].
3. Results
3.1. Fluorescence emission spectra
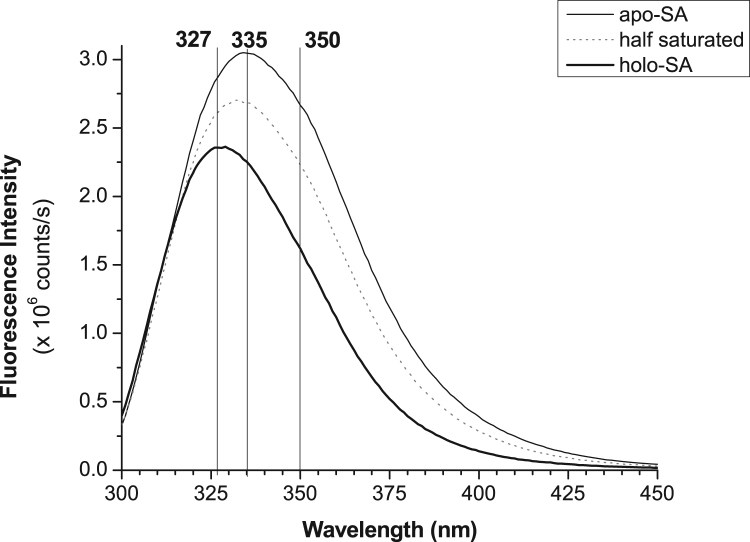
Emission scans were recorded for apo-streptavidin and again after the addition of 0.09–0.1 mM biotin aliquots. Emission spectra for apo-streptavidin, holo-streptavidin and approximately half saturated streptavidin are shown in Fig. 1.
Fig. 1.
Emission spectra of streptavidin at varying levels of biotin saturation. Only three spectra are shown for clarity.
Consistent with previous work, there is a blue shift as well as a quenching of fluorescence intensity upon binding of biotin [6], [17]. In this work, we find that the emission maximum blue shifts by 7–8 nm, from 335 nm to 327 nm and that the decrease in intensity is 30%, based on total integrated intensity. Consistent with Kurzban et al. we also observe a decrease in the full width at half maximum (FWHM), from approximately 57 to 50 nm, consistent with the apparent sharpening of the emission peak [17].
3.2. Emission scan titrations
In another set of experiments the emission was scanned from 328 to 337 nm on one channel and 346–355 nm on the second channel (0.5 nm/pt with 1 s integration time). The data from the first channel was fit to a single Gaussian function to determine the apparent wavelength of maximum emission. Plotting λem,max vs. [biotin]: [SA] ratio shows a similar titration curve to those shown in Fig. 2. As shown in Table 1 the apparent crossing point for these occurs at a [biotin]: [SA] ratio of 3.64 ± 0.16, which is 9 ± 4% lower than the 4:1 stoichiometric ratio observed for the 350 nm emission saturation.
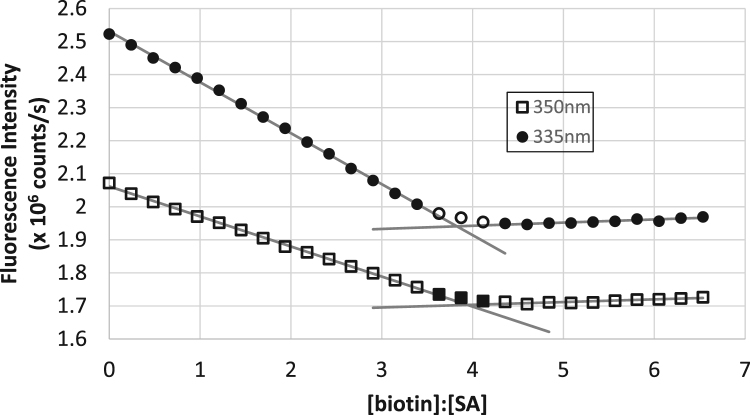
Fig. 2.
Fluorescence titration of biotin into streptavidin, monitoring emission at 335 nm and 350 nm (λex=280 nm). The three points in each curve with opposite filling are those not included in the linear fits.
Table 1.
Observed break points in various streptavidin fluorescence parameters upon titration with biotin aliquots. Parameters taken directly from fits to emission fluorescence spectra and plotted as a function of ligand to protein concentration ratio. Uncertainties are estimated using the standard deviation (N = 6).
| λmax(fit) | Area | FWHM | Height | |
|---|---|---|---|---|
| [biotin]: [SA] | 3.64 | 3.81 | 3.83 | 4.0 |
| Std. dev. | 0.16 | 0.14 | 0.10 | 0.2 |
| Avg. % chg. | 9 | 5 | 4 | |
| Std. dev. | 4 | 4 | 2 |
In an additional set of experiments the excitation wavelength was set to 280 nm while the emission wavelength was scanned from 290 to 470 nm (0.5 s/pt). Data were then integrated using Origin 6.0 software, obtaining the total integrated intensity, full width at half maximum (FWHM) and height of the emission peak. These variables were then plotted as a function of [biotin]: [SA] ratio, which gave saturation curves similar to those seen for fluorescence emission at 350 nm, saturating at a [biotin]: [SA] ratio of 4.0 ± 0.2. While the peak height tracks with the 350 nm single point emission curves, the peak area and FWHM both saturate before the 4:1 stoichiometric point. These signals each saturate at a [biotin]: [SA] ratio of 3.82 ± 0.12, or 5 ± 3% lower. These data are summarized in Table 1.
3.3. Dual wavelength emission titrations
To more efficiently follow the fluorescence titration of streptavidin with biotin, we collected emission at 335 nm and 350 nm with 4 s signal averaging. Because the two emission channels might have differing sensitivity and dark currents, 335 nm and 350 nm data were collected on each channel at each point in the titration. As seen in Fig. 2 there is clearly a break in the emission of both wavelengths (λex=280 nm) near the 4:1 stoichiometric ratio of biotin to streptavidin. The decay of both the 335 and 350 nm data is nearly linear, though the 335 nm data exhibits a small degree of nonlinearity, especially at lower biotin to streptavidin ratios. Also apparent in Fig. 2 is the difference in the slopes for the 335 nm and 350 nm emission data. The 350 nm emission slope is 1.5 times that of the 335 nm. The standard error and the standard deviation among slopes of multiple trials suggests an uncertainty of 4–8%.
When performing linear regression of the data, the points closest to the apparent signal saturation (highlighted on Fig. 2) were omitted to reduce potential bias near the equivalence point. However, these points are included on the plots and are seen to fall along the pre or post-saturation regression lines.
Using the breakpoint of the 350 nm emission as the 4:1 stoichiometric ratio [21], the 335 nm emission is consistently found to saturate earlier. The results of replicate measurements are shown in Table 2, showing the 335 nm emission signal saturating 6 ± 4% lower than the 4:1 ratio. This is consistent with the idea that distinct ensembles of tryptophan residues behave differently based on the level of ligand saturation.
Table 2.
Observed break points in streptavidin fluorescence emission signals upon addition of biotin. Uncertainties are estimated using the standard deviation.
| [biotin]: [SA] at saturation | ||||
|---|---|---|---|---|
| λem (nm) |
||||
| λex (nm) | 335 nm | 350 nm | % diff. | N |
| 280 | 3.76 ± 0.18 | 4.00 | 6 ± 4 | 6 |
| 290 | 3.82 ± 0.04 | 4.00 | 4.6 ± 1.1 | 9 |
| λem (nm) |
||||
| λex (nm) | 327 nm | 350 nm | % diff. | N |
| 280 | 3.81 ± 0.05 | 4.00 | 5.1 ± 1.2 | 7 |
| 290 | 3.69 ± 0.04 | 4.00 | 8.6 ± 1.3 | 16 |
Similar data were also collected with excitation at 290 nm. At 280 nm excitation there is significant absorption of radiation by tyrosine residues, as well as tryptophan [22]. Shifting the excitation wavelength to 290 nm eliminates the contribution of tyrosine fluorescence (λem=303 nm) [22]. As seen in Table 2 the saturation of the 335 nm signal is essentially unchanged by removal of tyrosine excitation (λex = 280 nm vs. 290 nm).
When excited at 290 nm vs. 280 nm the emission at λem,max=327 nm saturates at a statistically (P=0.0010) lower ligand:protein ratio than the 335 nm emission (λex=280 nm). The emission at 327 nm (λex=280 nm) saturates at the same ligand to protein ratio as the emission at 335 nm (λex=280 and 290 nm). The largest discrepancy in the observed breakpoint is found for λex=290 nm and λem=327 and 350 nm.
3.4. Photobleaching and quenching
To insure that the fluorescence changes observed were not an artifact of photobleaching, we carefully examined the fluorescence intensity as a function of illumination time for both apo- and holo-streptavidin. After 10 min of 280 nm illumination there was a 3.1 ± 1.0% decrease in the fluorescence intensity at 335 and 350 nm. After 10 min in the dark the initial fluorescence intensity was recovered. In another experiment a streptavidin solution was exposed to 280 nm excitation for 5 s then left in the dark for 4 min, similar to our fluorescence titration conditions (i.e. 2 min equilibration times). After a cumulative light exposure of 10 min there was no net loss of fluorescence intensity.
In contrast to the 3% photobleaching observed for apo-streptavidin, saturation of the biotin binding sites of streptavidin was found to reduce the extent of photobleaching to 1.3 ± 0.5% decrease in signal after 10 min of constant illumination.
In addition to the effect of biotin binding on the photobleaching of streptavidin, there is also a quenching of the fluorescence signal. Addition of biotin to streptavidin (6.5 to 1 M ratio) led to a 25.3 ± 0.9% decrease in fluorescence intensity at 335 nm and a 37.2 ± 0.6% decrease in 350 nm intensity. Integration of the emission spectra for apo and holo-streptavidin showed a 30% decrease in total fluorescence intensity upon biotin saturation, similar to that reported by Kurzban et al. [17].
4. Discussion
4.1. Fluorescence spectra
Qualitatively consistent with Kurzban et al. and Gonzalez et al. we observe a blue shift of the fluorescence emission upon biotin binding [6], [17]. Quantitatively they observed 4–5 nm shift, however we find a 7–8 nm blue shift of the emission maximum. We also see ~30% decrease in total intensity, a 25% decrease in 335 nm emission and 37% decrease in 350 nm emission. Kurzban et al. also reported a narrowing of the spectral width from 53 to 46 nm, while the current work found a 57–50 nm change in the FWHM [17].
The quenching of total fluorescence emission and the 7 nm decrease in FWHM upon biotin saturation are consistent between this work and that of Kurzban et al. [17]. However, we see enhancement of the blue shift in the emission maximum of 3 nm. Additionally, the FWHM we observed is 4 nm wider. This is likely due to differences between the full-length (WT) protein used by Kurzban et al. and the Prozyme streptavidin used here is more consistent with ‘core’ streptavidin (residues 13–139), as well as instrumental differences.
4.2. Fluorescence titration
Intrinsic tryptophan fluorescence emission at 327 nm, 335 nm or 350 nm show essentially linear decreases as one moves from apo to holo-streptavidin. The 335 nm and 350 nm emission corresponds to tryptophans in more hydrophobic and more solvent exposed (i.e. hydrophilic) environments, respectively [22]. Analysis of the 350 nm vs. 327 nm or 335 nm emission clearly shows a different rate of change and a difference in the saturation of the signal quenching compared to the saturation of the protein with ligand. The standard fluorescence assay for biotin binding using intrinsic fluorescence, suggested by Green, as well as Lin and Kirsch, uses the saturation of the 350 nm emission signal [18], [19].
In each subunit of streptavidin 4 of the 6 tryptophan residues (79, 92, 108, 120) are located in the binding site, interacting with biotin to differing extents [23], [24], [25]. Thus the observed changes in intrinsic tryptophan fluorescence are seen to accompany the binding of biotin. The early saturation of the more blue shifted tryptophan residues is consistent with the suggestions of previous work that biotin binding to streptavidin exemplifies cooperative allosterism (i.e. is positively cooperative, in a structural sense) [6], [9]. Relative to the standard measure of ligand saturation (i.e. emission at 350 nm), the more blue shifted residues reach their fluorescence saturation prior to the 4:1 stoichiometric binding. This is especially significant since the general trend is that the emission envelope of the entire tryptophan population blue shifts upon binding biotin.
The quench of the tryptophan emission suggests direct interaction of the residues with the biotin ligand (as shown in a number of x ray studies [23], [24], [25]) and/or altered energy transfer between tryptophan and neighboring residues. When the excitation is shifted to 290 nm, effectively eliminating absorbance by tyrosine, the emission at λem,max (327 nm) saturates at an even lower ligand to protein ratio. This is consistent with tyrosine fluorescence contributing in a significant manner to the more blue shifted fluorescence, which makes sense given the λem,max of 303 nm for tyrosine.
The blueshift of the tryptophan fluorescence is consistent with the ensemble of residues shifting to a more hydrophobic environment. This may be due to alteration of their proximity to the more hydrophobic residues and/or to the proximity of the more hydrophobic central region of the biotin molecule.
4.3. Photobleaching
Photobleaching is minimal under the experimental conditions used. Also, since most photobleaching is thought to occur via generation of diffusible reactive singlet oxygen by UV photons [22], the photosensitivity of the tryptophans would be expected to depend on their solvent accessibility. The observation that photobleaching is less extensive in holo- vs. apo-streptavidin is consistent with a tightening of the structure, as indicated by lower tryptophan solvent accessibility upon binding of biotin [6], [9], [10].
5. Conclusion
5.1. Proposed binding model
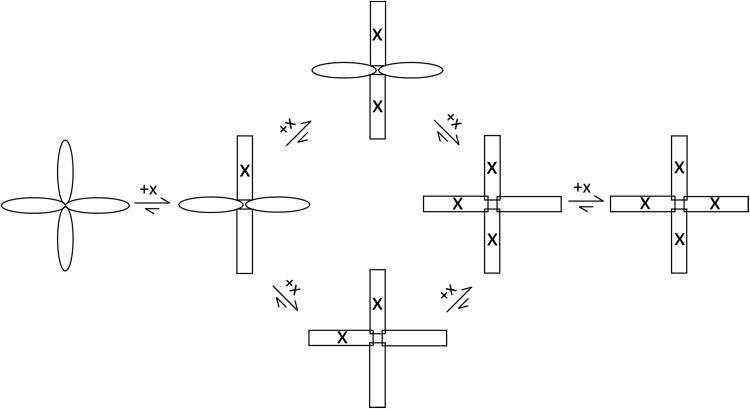
Fig. 3 presents a schematic model for biotin binding to streptavidin based on cooperative allosterism, exemplified by inducement of a structural change that varies with extent of binding. Previous studies provided evidence that the binding of biotin induces structural changes consistent with the Koshland, Nemethy and Filmer (KNF) sequential model of allosterism [6], [26]. Here we treat the situation where streptavidin acts as a dimer of dimers. This is in keeping with prior structural studies, where the binding of biotin leads to a structural change that causes W120 from an adjacent subunit to move into the biotin pocket, across the dimer-dimer interface [16], [19], [24]. Upon binding the first biotin, the structure of two subunits across the dimer-dimer interface are affected. The next ligand could bind to any of the three remaining sites, where two of these three are in their native (i.e. apo-) structural state (i.e. unaffected by biotin binding in that subunit, or one adjacent to it across the dimer-dimer interface). If there is no difference in binding affinity between the three available sites, consistent with previous studies [9], [12], then the lower route of Fig. 3 is statistically favored. Fairhead et al. showed in a recent work that the lower route is more stable, based on their design of streptavidin mutants with one or more ‘dead’ (i.e. non-biotin binding) subunits [27]. This means the most significant structural changes to the tetramer likely occur during the first two binding events. In cases where the second biotin binds directly across the dimer-dimer interface, the second binding results in a structural change affecting the overall structure and structural stability to a lesser extent, but the third binding would then involve a more significant change across the opposite side of the molecule. In any case the final binding event would lead to the least substantial overall structural change, which is consistent with the early saturation of the 335 nm and 327 nm emission, as well as other key fluorescence parameters (total integrated emission intensity, wavelength of maximum emission and FWHM) compared to the 350 nm emission.
Fig. 3.
Proposed model for structurally cooperative biotin (x) binding to a Streptavidin tetramer, where ellipses represent subunits unaffected by ligand induced structural change. Although both the upper and lower routes are possible, the lower route is the statistically favored path.
Acknowledgements
This work was supported by The John Carroll University College of Arts & Sciences. The fluorimeter was purchased using funds provided by The Camille and Henry Dreyfus Foundation, Inc., Special Grant Program (SG-01-056). We also wish to acknowledge Seema Patel for assistance in preparation for this work.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.12.011.
Appendix A. Transparency document
Supplementary material.
References
- 1.Lohman T.M., Mascotti D.P. Nonspecific ligand-DNA equilibrium binding parameters determined by fluorescence methods. Methods Enzymol. 1992;212:424–458. doi: 10.1016/0076-6879(92)12027-n. [DOI] [PubMed] [Google Scholar]
- 2.Lohman T.M., Bujalowski W. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: spectroscopic approaches to monitor binding. Methods Enzymol. 1991;208:258–290. doi: 10.1016/0076-6879(91)08017-c. [DOI] [PubMed] [Google Scholar]
- 3.Bayer E.A., Ehrlich-Rogozinski S., Wilchek M. Sodium dodecyl sulfate-polyacrylamide gel electrophoretic method for assessing the quaternary state and comparative thermostability of avidin and streptavidin - Bayer - - - Wiley online library. Electrophoresis. 1996;17(8):1319–1324. doi: 10.1002/elps.1150170808. [DOI] [PubMed] [Google Scholar]
- 4.González M., Argaraña C.E., Fidelio G.D. Extremely high thermal stability of streptavidin and avidin upon biotin binding. Biomol. Eng. 1999;16(1–4):67–72. doi: 10.1016/s1050-3862(99)00041-8. [DOI] [PubMed] [Google Scholar]
- 5.Bayer E.A., Ben-Hur H., Wilchek M. Isolation and properties of streptavidin. Methods Enzymol. 1990;184:80–89. doi: 10.1016/0076-6879(90)84262-f. [DOI] [PubMed] [Google Scholar]
- 6.González M., Bagatolli L.A., Echabe I., Arrondo J.L., Argaraña C.E., Cantor C.R., Fidelio G.D. Interaction of biotin with streptavidin. thermostability and conformational changes upon binding. J. Biol. Chem. 1997;272(17):11288–11294. doi: 10.1074/jbc.272.17.11288. [DOI] [PubMed] [Google Scholar]
- 7.Waner M.J., Navrotskaya I., Bain A., Oldham E.D., Mascotti D.P. Thermal and sodium dodecylsulfate induced transitions of streptavidin. Biophys. J. 2004;87(4):2701–2713. doi: 10.1529/biophysj.104.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sano T., Vajda S., Smith C.L., Cantor C.R. Engineering subunit association of multisubunit proteins: a dimeric streptavidin. Proc. Natl. Acad. Sci. USA. 1997;94(12):6153–6158. doi: 10.1073/pnas.94.12.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams D.H., Stephens E., Zhou M. Ligand binding energy and catalytic efficiency from improved packing within receptors and enzymes. J. Mol. Biol. 2003;329:389–399. doi: 10.1016/s0022-2836(03)00428-5. [DOI] [PubMed] [Google Scholar]
- 10.Sano T., Cantor C.R. Intersubunit contacts made by tryptophan 120 with biotin are essential for both strong biotin binding and biotin-induced tighter subunit association of streptavidin. Proc. Natl. Acad. Sci. USA. 1995;92(8):3180–3184. doi: 10.1073/pnas.92.8.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sano T., Cantor C.R. Cooperative biotin binding by streptavidin. electrophoretic behavior and subunit association of streptavidin in the presence of 6 M urea. J. Biol. Chem. 1990;265(6):3369–3373. [PubMed] [Google Scholar]
- 12.Jones M.L., Kurzban G.P. Noncooperativity of biotin binding to tetrameric streptavidin. Biochemistry. 1995;34(37):11750–11756. doi: 10.1021/bi00037a012. [DOI] [PubMed] [Google Scholar]
- 13.Hyre D.E., Le Trong I., Merritt E.A., Eccleston J.F., Green N.M., Stenkamp R.E., Stayton P.S. Cooperative hydrogen bond interactions in the streptavidin-biotin system. Protein Sci. 2006;15(3):459–467. doi: 10.1110/ps.051970306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Argaraña C.E., Kuntz I.D., Birken S., Axel R., Cantor C.R. Molecular cloning and nucleotide sequence of the streptavidin gene. Nucleic Acids Res. 1986;14(4):1871–1882. doi: 10.1093/nar/14.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayer E.A., Ben-Hur H., Hiller Y., Wilchek M. Postsecretory modifications of streptavidin. Biochem. J. 1989;259(2):369–376. doi: 10.1042/bj2590369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurzban G.P., Bayer E.A., Wilchek M., Horowitz P.M. The quaternary structure of streptavidin in urea. J. Biol. Chem. 1991;266(22):14470–14477. [PubMed] [Google Scholar]
- 17.Kurzban G.P., Gitlin G., Bayer E.A., Wilchek M., Horowitz P.M. Biotin binding changes the conformation and decreases tryptophan accessibility of streptavidin. J. Protein Chem. 1990;9(6):673–682. doi: 10.1007/BF01024762. [DOI] [PubMed] [Google Scholar]
- 18.Lin H.J., Kirsch J.F. A Rapid, Sensitive Fluorometric Assay for Avidin and Biotin. Vol. 62. Academic Press; Cambridge, MA: 1979. pp. 287–289. (In Methods in Enzymology; Vitamins and Coenzymes Part D) [DOI] [PubMed] [Google Scholar]
- 19.Green N.M. Avidin and Streptavidin. Vol. 184. Academic Press, Inc; Cambridge, MA: 1990. pp. 51–67. (In Avidin-BiotinTechnology) [DOI] [PubMed] [Google Scholar]
- 20.Suter M., Cazin J., Butler J.E., Mock D.M. Isolation and characterization of highly purified streptavidin obtained in a two-step purification procedure from streptomyces Avidinii grown in a synthetic medium. J. Immunol. Methods. 1988;113(1):83–91. doi: 10.1016/0022-1759(88)90384-5. [DOI] [PubMed] [Google Scholar]
- 21.Mock D.M., Horowitz P.M. Fluorometric Assay for Avidin-Biotin Interaction. Vol. 184. Academic Press, Inc; Cambridge, MA: 1990. pp. 234–240. (In Avidin-BiotinTechnology) [DOI] [PubMed] [Google Scholar]
- 22.R.Lakowicz. Joseph, Principles of Fluorescence Spectroscopy; Springer.
- 23.Katz B.A. Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at Low PH. J. Mol. Biol. 1997;274(5):776–800. doi: 10.1006/jmbi.1997.1444. [DOI] [PubMed] [Google Scholar]
- 24.Weber P.C., Ohlendorf D.H., Wendoloski J.J., Salemme F.R. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243(4887):85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 25.Weber P.C., Pantoliano M.W., Simons D.M., Salemme F.R. Structure-based design of synthetic azobenzene ligands for streptavidin. J. Am. Chem. Soc. 1994;116(7):2717–2724. [Google Scholar]
- 26.Koshland D.E., Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 27.Fairhead M., Krndija D., Lowe E.D., Howarth M. Plug-and-play pairing via defined divalent streptavidins. J. Mol. Biol. 2014;426(1):199–214. doi: 10.1016/j.jmb.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.