Summary
The cerebral cortex has expanded in size and complexity in primates, yet the molecular innovations that enabled primate-specific brain attributes remain obscure. We generated cerebral cortex organoids from human, chimpanzee, orangutan, and rhesus pluripotent stem cells and sequenced their transcriptomes at weekly time points for comparative analysis. We used transcript structure and expression conservation to discover gene regulatory long non-coding RNAs (lncRNAs). Of 2,975 human, multi-exonic lncRNAs, 2,472 were structurally conserved in at least one other species and 920 were conserved in all. Three hundred eighty-six human lncRNAs were transiently expressed (TrEx) and many were also TrEx in great apes (46%) and rhesus (31%). Many TrEx lncRNAs are expressed in specific cell types by single-cell RNA sequencing. Four TrEx lncRNAs selected based on cell-type specificity, gene structure, and expression pattern conservation were ectopically expressed in HEK293 cells by CRISPRa. All induced trans gene expression changes were consistent with neural gene regulatory activity.
Keywords: stem cells, induced pluripotent stem cells, brain development, human evolution, neurogenesis, neural development, lncRNA, primate evolution, scRNA-seq, RNA-seq
Highlights
-
•
New orangutan and chimpanzee iPSC lines enable pan-primate gene expression analysis
-
•
Transiently expressed (TrEx) lncRNAs identified in primate cerebral cortex organoids
-
•
Conserved TrEx pattern correlates with cell-type specificity by single-cell RNA-seq
-
•
Four primate-conserved TrEx lncRNAs influence neural genes when ectopically expressed
In this article, Salama and colleagues identified transiently expressed (TrEx) lncRNAs from human, chimpanzee, orangutan, and rhesus pluripotent stem cell-derived cerebral cortex organoids and assessed their structural and expression conservation. Transient expression correlated with cell-type specificity as measured by single-cell RNA-seq. Ectopic expression of TrEx lncRNAs by CRISPRa modulated expression of neural genes in trans, suggesting regulatory function.
Introduction
Pluripotent stem cell (PSC)-derived cerebral cortex organoid (CO) cell cultures have allowed researchers to probe gene regulatory events that occur during the differentiation of early neocortical cell types using cell lines representing normal and disease states (Eiraku et al., 2008, Lancaster et al., 2013). These protocols closely recapitulate the cellular organization and gene expression events observed in fetal tissue (Camp et al., 2015, Fatehullah et al., 2016, Nowakowski et al., 2017). Comparisons of human with other primate COs have revealed subtle differences in the timing of cell divisions and differentiation events (Mora-Bermudez et al., 2016, Otani et al., 2016), although the mechanisms by which these changes are enacted are unknown.
Here we focus on one class of gene regulatory element, long non-coding RNAs (lncRNAs), which often show tissue-specific expression, account for a significant proportion of Pol II output, and are particularly enriched in neural tissues (Cabili et al., 2011, Derrien et al., 2012, Pauli et al., 2012). LncRNAs have diverse roles in gene regulation, including chromosome inactivation (Penny et al., 1996, Zhao et al., 2008), imprinting (Buiting et al., 2007, Leighton et al., 1995, Pandey et al., 2008), and developmental processes (Heo and Sung, 2011), and have been implicated in establishment of pluripotency (Guttman et al., 2011), stem cell maintenance (Rani et al., 2016), reprogramming (Loewer et al., 2010), and differentiation (Guttman et al., 2011). Nevertheless, most human lncRNAs have undetermined function (Hon et al., 2017, Lagarde et al., 2017) and lack sequence conservation among vertebrate species (Cabili et al., 2011, Kutter et al., 2012, Ulitsky et al., 2011). Their tissue-specific expression patterns and rapid sequence evolution make lncRNAs an attractive target as arbiters of lineage-specific gene regulation during development.
It has been suggested that exon structure conservation is more predictive of function than nucleic acid sequence alone (Ulitsky, 2016) and we postulate that expression pattern conservation during differentiation may imply a conserved role in gene regulation. Here, we used both aspects of conservation in equivalent developing tissues among closely related primates to identify gene regulatory lncRNAs active in human neural differentiation. We generated COs from human, chimpanzee, orangutan, and rhesus PSCs to recapitulate early events in cortical development and enable comparative molecular analysis of this process. RNA sequencing (RNA-seq) was performed at weekly time points to assess the conservation of lncRNA transcript structure and expression among primates. This enabled the discovery of transiently expressed (TrEx) lncRNAs in multiple species, which have potential roles in early cortical cell fate specifications, including the generation of neuroepithelium (NE), radial glia (RG), and early-forming Cajal-Retzius (CR) neurons. Single-cell RNA-seq (scRNA-seq) of time points relevant to major differentiation events was used to identify cell types associated with the expression of candidate TrEx lncRNAs. Finally, CRISPR activation (CRISPRa) in HEK293FT cells was used to express these transcripts out of context to probe whether TrEx lncRNAs can regulate genes related to corticogenesis.
Results
Generation and RNA-Seq of Primate COs
To study the transcriptional landscape of early cell-type transitions during primate cortical neuron differentiation, we subjected human, chimpanzee, orangutan, and rhesus macaque PSCs to a CO differentiation protocol based on Eiraku et al., 2008 (Figures 1A and 1B). Embryonic stem cell (ESC) lines were used for human (H9) and rhesus (LYON-ES1) time courses. Since ESCs are not available for great apes, we generated integration-free induced PSCs (iPSCs) for chimpanzee (Epi-8919-1A) and orangutan (Jos-3C1) from primary fibroblasts (Figure S1).
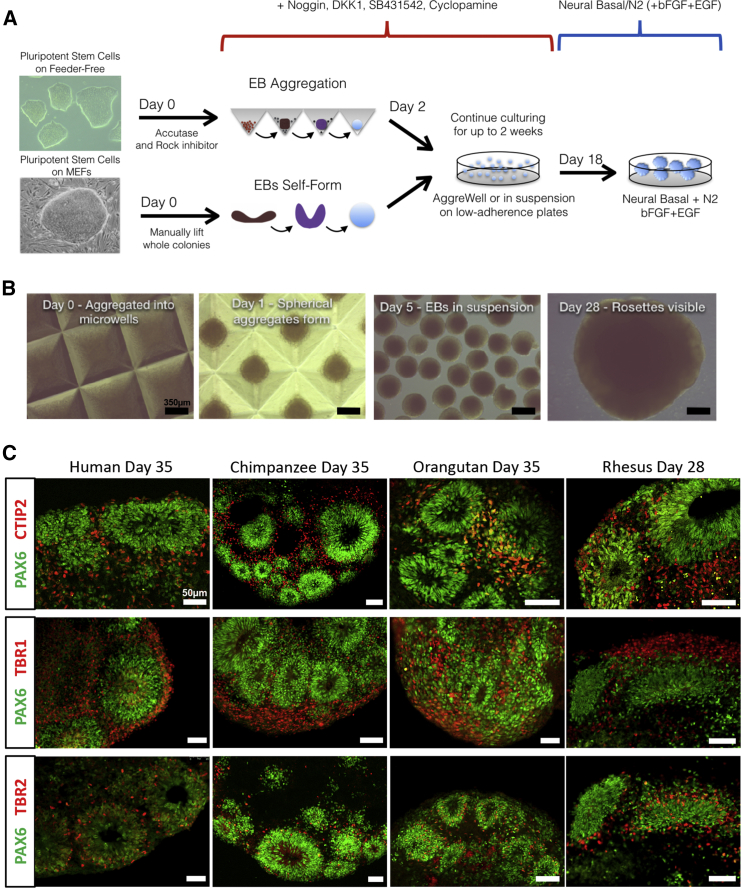
Figure 1.
Cerebral Organoid Differentiation Protocol
(A) Outline of the dorsal cerebral cortex neuron differentiation assay. EB, embryoid body; MEFs, mouse embryonic fibroblasts.
(B) An example of chimpanzee aggregation and differentiation at days 0, 1, 5, and 28. Scale bar, 350 μm.
(C) IF staining at 5 weeks (28 days in rhesus) for PAX6 (neural progenitors), CTIP2 and TBR1 (early deep-layer neurons), and TBR2 (intermediate progenitors or early migrating neurons). Scale bar, 50 μm.
See also Figures S1 and S2.
The performance of these PSC lines in our CO assay was evaluated by immunofluorescence (IF) staining at day 35 (or the equivalent day 28 for rhesus), showing efficient production of RG, intermediate progenitors, and deep-layer cortical neurons (Figure 1C) in highly structured neural rosettes as described previously (Eiraku et al., 2008, Lancaster et al., 2013, Camp et al., 2015). RNA samples were collected from at least two replicates of PSCs and weekly time points over 5 weeks of differentiation in each species and used to create strand-specific RNA-seq libraries. Due to their shorter gestational period and faster cell division rates, rhesus samples had adjusted time points with ∼5 day weeks (Figure S2; Experimental Procedures). In all, 49 libraries averaged 41 million uniquely mapping reads per library with a minimum of 46 million unique reads across replicates per species time point. After mapping to the appropriate genome (Experimental Procedures) DESeq (Love et al., 2014) was used to assess relative gene expression for known genes (Table S1). The generation of on-target dorsal cortical tissue was confirmed by profiling marker genes (Figure 2A). Pluripotency markers such as OCT3/4 were down-regulated by week 1, while early neural stem cell markers, including PAX6, were up-regulated and deep-layer neuron markers such as TBR1 were strongly expressed by week 5 in all species (Figure 2A). Overall, there was strong induction of early neural and dorsal forebrain markers with little expression of markers of other brain regions (Figure 2A).
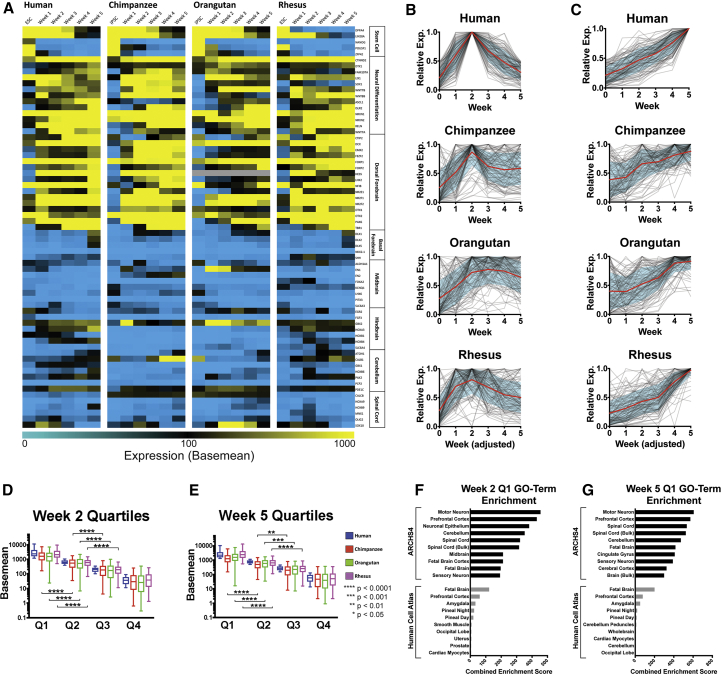
Figure 2.
Analysis of Differentiation Accuracy, Efficiency, and Kinetics
RNA-seq data are represented as the mean of 2 biological replicates/time points (A–E). (A) Heatmap of marker gene expression (DESeq2 expression values). (B) Top 100 “week 2 genes” (n = 3,431) or (C) “week 5 genes” (n = 3,838) identified in human are displayed for each species (gray lines) with centroid curves (red) plus or minus SD (blue shading). (D) Week 2 genes (857–858 genes per quartile) or (E) week 5 genes (959–960 genes per quartile) were ranked into quartiles by expression in human (blue), and the same genes are displayed for chimpanzee (red), orangutan (green), and rhesus (purple), excluding genes with base mean <10 in human and those not expressed in another species. Boxplot whiskers show 5th to 95th percentile. Significance was calculated by one-way ANOVA. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. GO term analysis of the top quartiles from (F) week 2 genes and (G) week 5 genes using Enrichr (Kuleshov et al., 2016) is shown. The top 10 enriched GO terms from ARCHS4 (Lachmann et al., 2018; based on publicly available RNA-seq data from human and mouse) and Human Cell Atlas (Su et al., 2004; based on microarrays of human and mouse tissues) are ranked by their combined enrichment score. See also Table S1.
Comparability of Time Points across Species
We next sought to establish criteria for performing cross-species analysis at each time point. We selected two sets of genes with clear expression pattern trends in the human time course: (1) “week 2 genes,” the genes peaking at week 2 and below 50% maximal expression at weeks 0 and 5 (Figure 2B), and (2) “week 5 genes,” the genes maximally expressed at week 5 but below 50% maximal expression at week 0 (Figure 2C). The categories “week 2 genes” and “week 5 genes” contain 3,431 and 3,838 genes, respectively. The top 100 are displayed in Figures 2B and 2C. All of them are displayed in Figures 2D and 2E. When plotting the top 100 genes fitting these profiles, all species consistently show the highest expression for human week 5 genes at their corresponding week 5, confirming an appropriate progression to this endpoint for all species (Figure 2C). Human week 2 genes show weaker, though overall, correspondence, peaking at week 2 or 3 in other species (Figure 2B). Importantly, human and chimpanzee plots show strong correspondence (Figures 2B and 2C), showing that conserved features of neurogenesis can be seen despite comparing ESCs (human) and iPSCs (chimpanzee). Orangutan samples appear to maintain high expression of the human-classified week 2 genes into later time points, perhaps indicating a slower or delayed transition into later differentiation events, although it is challenging to attribute this as a bona fide cross-species difference with only a single orangutan iPSC line.
To ensure that the relative amplitude of gene expression was similar across species at these time points, we performed quartile analysis of protein-coding genes fitting the above expression profiles, labeled “week 2 quartiles” and “week 5 quartiles,” respectively (Figures 2D–2G). We required a minimum of 10 base mean-normalized reads in human and non-zero expression in all other species to minimize annotation bias. Human protein-coding genes were then sorted into expression quartiles and the same genes are shown in each other species (Figures 2D and 2E). Although chimpanzee and orangutan appear to have lower overall expression in the top quartile versus human in both gene sets, sorting in this way significantly segregates genes in the top three quartiles in all species by one-way ANOVA, suggesting a similar relative ranking of the gene expression common to each animal. Gene Ontology (GO) term analysis of the first quartiles from the week 2 and week 5 gene sets using Enrichr (Kuleshov et al., 2016) showed significant enrichment of terms associated with neural development, including prefrontal cortex and fetal brain (Figures 2F and 2G). Week 2 was also particularly enriched with genes associated with neuronal epithelium, which is absent in the week 5 gene set, indicating that those cultures progressed to a more differentiated stage.
Expression and Gene Structure Conservation of Primate LncRNAs
To assemble unannotated transcripts in each species, Cufflinks v.2.0.2 (Trapnell et al., 2012) was used, and the Cuffmerge tool combined gene models across time points in each species using FANTOM5 lv3 (Hon et al., 2017) as a reference annotation. CAT (Fiddes et al., 2018) was used to project the FANTOM lv3 set through a progressive Cactus whole-genome alignment (Paten et al., 2011, Stanke et al., 2008) to each of the other primate genomes. Guided by the Cufflinks annotation set in each genome, these projections were assigned a putative gene locus. RSEM v.1.3.0 (Li and Dewey, 2011) was used to calculate expression values of these gene models in each primate species.
Conservation of exon boundaries within an lncRNA gene can be indicative of functional transcripts (Ulitsky, 2016). Gene structure conservation of expressed transcripts among our primate species was assessed using homGeneMapping in the AUGUSTUS toolkit (Konig et al., 2016). This tool makes use of Cactus alignments to project annotations in all pairwise species comparisons, providing an accounting of features found in other genomes. homGeneMapping was given both the Cufflinks transcript assemblies and the expression estimates derived from the combination of all RNA-seq time points in all species. The results of this pipeline were combined with the Cactus alignment-based transcript projections to ascertain a set of gene loci that appear to have human-specific expression, human-chimp-specific expression, great ape-specific expression, and expression in all primates (Figures 3A and 3B, Tables S2 and S3). Transcript models with at least 50% intron junction support in human were considered conserved in a non-human genome if that genome had RNA-seq read support for any of its intron junctions and the gene cluster had a transcripts per million (TPM) value greater than 0.1. Single-exon transcripts were filtered out. Using these parameters, 2,975 human poly-exonic lncRNA gene clusters were identified in human. Five hundred three lncRNAs were observed only in human, while 457 were seen in human and chimp, 586 were seen in all great apes, and 920 were confirmed as primate conserved (Figure 3B, Tables S2 and S3). Although these figures serve as an underestimate of how conserved these transcripts are due to the lack of cell line replicates, they show higher overlap in species separated by less evolutionary distance as would be expected. Among the primate-conserved category are the previously described mammalian conserved lncRNAs MALAT1, NEAT1, H19, PRWN1, and CRNDE (Tables S2 and S3). Three hundred forty-seven previously unannotated human gene clusters were also found by Cufflinks, 160 of which were found only in human, and 164 were conserved in chimp, 105 in great apes, and 79 across all of the represented primates (Figure S3, Table S4), showing a distribution similar to that of annotated lncRNAs. Five hundred eighty chimpanzee-specific, 1,709 orangutan-specific, and 593 rhesus-specific gene loci were also detected (Table S4), further supporting a relatively fast turnover of lncRNAs, though we suspect that the orangutan estimates are inflated due to its relatively poor genome assembly and, consequently, poor alignment to the other genomes. Comparing these figures to protein-coding genes, 14,453 coding genes were found to be expressed in human (Table S2) and 12,474 (86%) of these coding genes were expressed and shared intron boundaries among all species (Figure 3A). This confirms a much higher degree of structural conservation of mRNAs by these strict metrics. Given the experimental limitations of using one cell line per species and to avoid cell-line-specific effects, we focused our study on transcripts with conserved gene structure in at least two species.
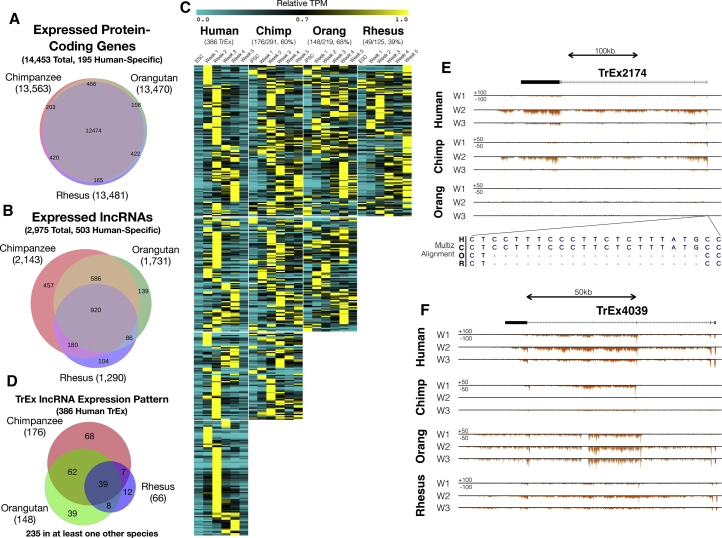
Figure 3.
LncRNA Structure and Expression Pattern Conservation
(A and B) Venn diagrams show intron boundary conservation of human (A) protein-coding genes and (B) lncRNAs in each species.
(C) A heatmap with TPM (mean, 2 biological replicates/time point) normalized to the maximum value in each species for TrEx lncRNAs.
(D) Venn diagram of TrEx lncRNA expression pattern conservation between species.
(E and F) UCSC Genome Browser screenshots showing (E) TREX2174 and (F) TREX4039. The Multiz alignment just upstream of the transcription start site for TREX2174 has a 19 bp insertion that is specific to human and chimpanzee among extant great apes (E).
LncRNAs have previously been reported to exhibit dynamic expression in developing tissues (Amaral and Mattick, 2008, Pauli et al., 2012). TrEx lncRNAs could contribute to the rapid evolution of regulatory networks in developing tissues that underlie important phenotypes, like the expansion of the cerebral cortex over the human lineage. Here we define TrEx lncRNAs as those with maximal expression between weeks 1 and 4, and less than 50% of their maximal expression at weeks 0 and 5. Using these metrics, we identified 386 human TrEx lncRNAs, most of which were expressed primarily at one weekly time point (Figures 3C and 3D, Table S4). We next assessed if these transcripts were also TrEx in other species, requiring that they also have maximal expression at weeks 1–4 in that species. One hundred seventy-six had a conserved TrEx pattern in chimpanzee (61% of 291 transcripts with conserved structure), 148 (68% of 219) in orangutan, 66 (53% of 125) in rhesus, and 39 (31% of 125 transcripts with conserved structure in all four species) had a TrEx pattern in all four species (Figures 3C and 3D, Table S4). Even with the observed timing differences of week 2 protein-coding genes observed in orangutan (Figure 2B), human TrEx lncRNAs still retain similar temporal expression patterns to a much higher degree in orangutan than in the more distant rhesus.
Several examples highlight the general features of these TrEx lncRNAs and illustrate their potential evolutionary impact. TREX2174 (RP11-314P15) is notable in its week 2-specific expression, which is also observed in chimpanzee (Figure 3E), but not in orangutan or rhesus at any time point. Interestingly, TREX2174 has a 19 bp insertion overlapping its transcription start site that is specific to human and chimpanzee. This suggests TREX2174 may be a recently evolved lncRNA or has a recently evolved expression pattern. Among the lncRNAs that were observed in all four of our species, TREX4039 (overlapping AC011306 and MIR217HG) (Figure 3F) peaks at week 1 or 2 in all species and is extinguished by week 5. Chimpanzee appears to express an isoform of this transcript that is not shared with human or rhesus, but can be seen expressed early in orangutan. While chimpanzee ceases expression from this locus at week 2, orangutan appears to switch to the longer isoform observed in the other two species at week 2. This demonstrates how, even among transcripts that share structural elements across species, expression regulation can be diverse.
TrEx LncRNAs Show Cell-Type-Specific Expression Patterns
An scRNA-seq study in fetal brain has shown in more mature neural tissue that lncRNAs are often restricted to specific cell-type clusters, having higher expression in individual cells than it would appear from bulk RNA-seq (Liu et al., 2016a). To explore the possibility that TrEx RNAs could be restricted to transitory cell states found during cortical development, we performed 10× Chromium 3′ end scRNA-seq on human ESCs (hESCs) and COs at weeks 0, 1, 2, and 5. In all, nearly 800 million reads were obtained from 14,086 cells, averaging 56,600 reads per cell. The total number of genes detected per library ranged from 28,000 in week 5 COs to 36,000 at week 1, averaging between 1,702 and 4,978 genes per cell.
t-distributed stochastic neighbor embedding (tSNE) plots generated with Cell Ranger (10× Genomics) identified increasing cell heterogeneity as differentiation progressed (Figure S4). Using a combination of k-means clustering, graphical clustering, and visual inspection, we manually curated clusters of cells with gene expression profiles matching NE, RG, and CR cells in our week 2 libraries (Figures 4A and S5; Table S4). NE cells were identified by expression of HES3 and NR2F1, forming a cluster of 1,261 cells (29%) (Figure S5C). CR cells expressed TBR1, EOMES, LHX9, and NHLH1, comprising a cluster of 356 cells (8%) (Figure S5E). The largest cluster strongly expressed cortical RG markers SOX2, EMX2, NNAT, PTN, and TLE4, making up 2,593 cells (59%) (Figure S5D). One hundred seventy-six cells (4%) showed no strong association with these clusters and had no significant distinguishing genes. We determined that they likely represented cell doublets and their prevalence is consistent with theoretical estimates based on the number of cells we captured per library. At week 5, cells expressing NE markers were virtually absent and instead additional clusters expressing neuronal markers emerged (Figure S4).
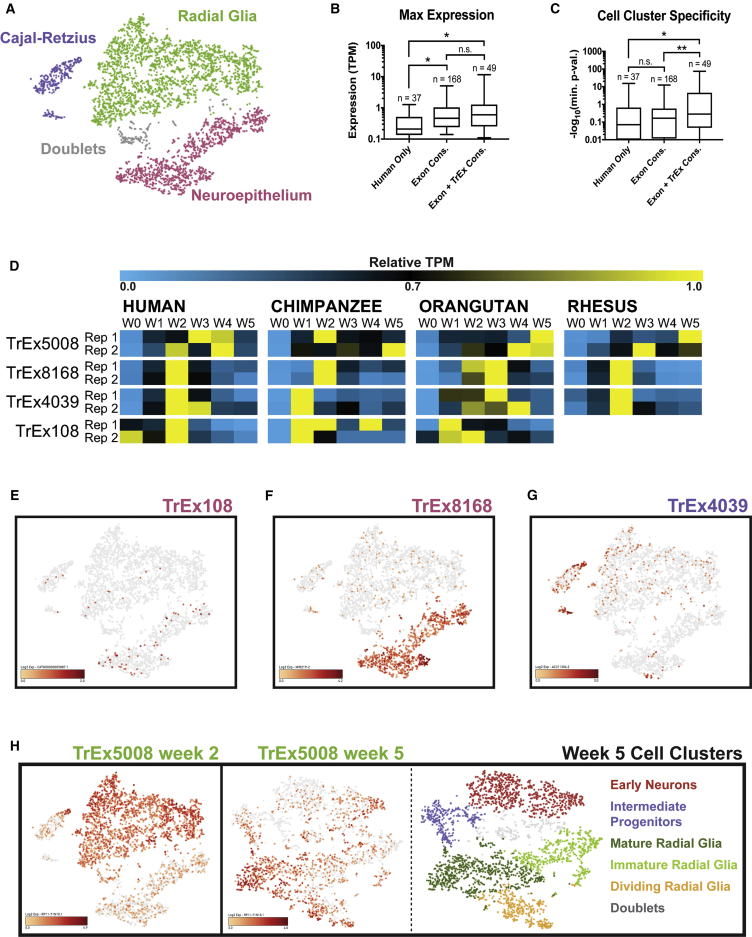
Figure 4.
TrEx LncRNAs Associate with Specific Cell Subtypes in Single-Cell RNA-Seq
(A–C) (A) A tSNE plot of week 2 scRNA-seq. 4,386 cells were manually curated into clusters with gene expression consistent with CR (8%, violet), RG (59%, green), NE (29%, pink), and cell doublets (4%, gray) using the Loupe Browser (10× Genomics). (B and C) Human week 2 TrEx lncRNAs were categorized by conserved exonic structure in all species (Exon Cons.), both conserved exonic structure and conserved TrEx expression pattern (Exon + TrEx Cons.), or present only in human (Human only) (B and C). (B) Maximum expression values are plotted for each category. (C) Cell-type specificity was determined by the Loupe Browser's (10× Genomics) “locally distinguishing genes” function. The −log(minimum p value) is shown for each comparison. Significance values were calculated by one-way ANOVA (∗p < 0.01, ∗∗p < 0.001) (B and C).
(D) Heatmap showing relative TPM across each species' time course.
(E–H) tSNE plots show lncRNA expression (red). (E) TREX108 and (F) TREX8168 were enriched in NE cells. (G) TREX4039 is expressed in a subpopulation of CR cells. (H) TREX5008 is expressed in RG at week 2 (left) and week 5 (center). 3,240 week 5 organoid cells were manually curated into clusters consistent with early neurons (26%, red), intermediate progenitors (11%, violet), mature RG (26%, dark green), immature RG (18%, light green), dividing RG (12%, orange), and cell doublets (8%, gray) (right).
See also Figures S4 and S5, Tables S1 and S4.
Next we addressed whether the TrEx pattern of lncRNAs observed in bulk tissue corresponded to an increased likelihood of cell-type specificity (Figures 4B and 4C). LncRNAs were separated into three conservation categories: those observed only in human samples (“human only”), those with observed exon boundary conservation in all species but no evidence of TrEx expression pattern conservation (“exon conserved”), and those with observed exon boundary and TrEx expression pattern conservation in all species (“exon + TrEx conserved”). Higher transcript conservation by exon boundaries correlates well with higher expression in bulk RNA-seq (Figure 4B). Adding the criteria of TrEx conservation does not significantly bolster this trend. However, using the “Globally Distinguishing Genes” tool in the Loupe Cell Browser (10× Genomics) on our manually curated cell types, we see that lncRNAs with a conserved TrEx pattern are much more likely to be cell-type specific where exon structure conservation alone has little predictive power (Figure 4C). Overall, these results suggest that many TrEx lncRNAs may be associated with short-lived cell-type intermediates and thus warrant further investigation as biomarkers of specific cell states.
Out-of-Context Activation of LncRNAs Modulates Neural Gene Expression
Since many lncRNAs have been implicated in gene regulatory function either in cis (Leighton et al., 1995, Orom et al., 2010, Pandey et al., 2008, Penny et al., 1996, Wang et al., 2011, Zhao et al., 2008) or in trans (Guttman et al., 2011, Khalil et al., 2009, Loewer et al., 2010, Nagano et al., 2008, Pandey et al., 2008), we assessed potential TrEx lncRNA gene regulatory function by CRISPRa using dCas9-VP64 to drive transcription at the endogenous locus in HEK293FT cells (Konermann et al., 2014), thus allowing detection of either mode of action. We chose four lncRNAs that are TrEx in human (Figure 4D), have conserved exonic structure through great apes, are detectable in our single-cell data, are expressed predominantly in one cell type (Figures 4E–4H and Table S4), and lack expression in HEK293FT cells. TREX108 (FANTOM CATG00000005887) and TREX8168 (overlapping MIR219-2) are highly expressed in NE and absent in week 5 single-cell data (Figures 4E and 4F). TREX4039 (AC011306/MIR217) is slightly expressed in NE and RG, but concentrated in a portion of CR cells (Figure 4G). TREX5008 (RP11-71N10) is restricted to the large RG cluster at week 2 (Figure 4H). Interestingly, while TREX5008 is TrEx in human bulk RNA-seq data, this pattern is not retained in other species (Figure 4D) and it is still highly expressed in a subset of RG in week 5 scRNA-seq data (Figure 4H). This suggests that some transcripts identified as TrEx lncRNAs by bulk RNA-seq methods are instead restricted to one cell subtype that persists, but whose relative abundance declines as more cell types are generated.
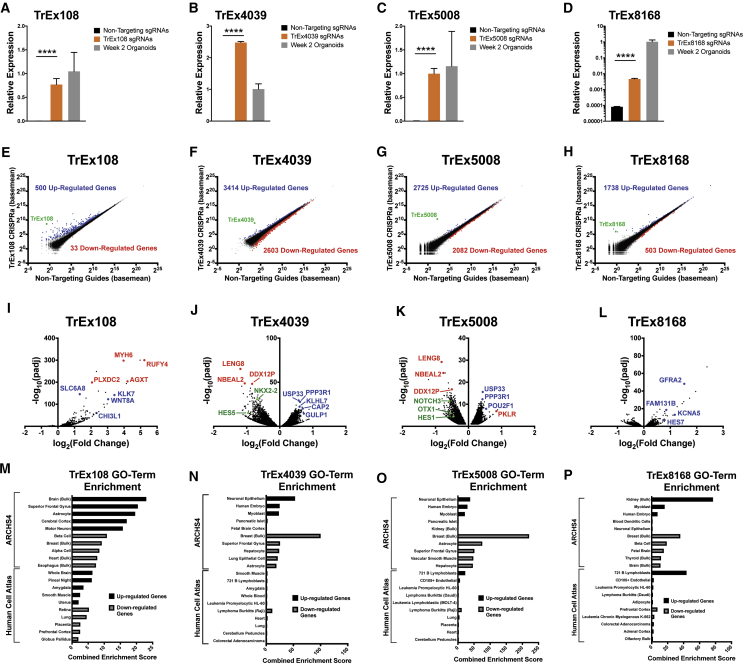
Five CRISPR single-guide RNAs (sgRNAs) were designed 50–450 bp upstream of each target TrEx lncRNA and co-transfected into HEK293FT with dCas9-VP64. We achieved activation of all four TrEx lncRNAs (140- to 8,600-fold increase over non-targeting scrambled sgRNA controls [NTCs]), with all but TREX8168 activated to a similar or higher expression level compared with bulk week 2 human CO RNA (Figures 5A–5D). To assess the regulatory potential of the activated TrEx lncRNAs, we used RNA-seq to measure their effect on protein-coding genes against NTCs (Figure 5). Amazingly, we found that all four TrEx lncRNAs had robust effects on gene expression, with none significantly affecting their immediately neighboring genes. This shows that CRISPRa specifically induced expression of our intended targets and their gene regulatory effects were largely in trans. TREX108 and TREX8168 predominantly showed activating activity, while TREX4039 and TREX5008 showed similar numbers of induced and repressed genes (Figures 5E–5L).
Figure 5.
CRISPRa of TrEx LncRNAs Regulates Genes Associated with Brain Development
(A–D) qRT-PCR of CRISPRa-induced expression of (A) TREX108, (B) TREX4039, (C) TREX5008, and (D) TREX8168 in HEK293 cells relative to non-gene-targeting controls and expression in human week 2 COs (data, mean ± SD of 4 biological replicates, ∗∗∗∗p < 0.0001).
(E–H) Scatterplots showing RNA-seq data for the indicated TrEx lncRNA versus non-targeting controls (3 biological replicates). Significantly up-regulated (blue) genes, down-regulated (red) genes, and target TrEx lncRNA (green) as determined by DESeq are highlighted (adj. p < 0.01).
(I–L) Volcano plots of significant genes (adj. p < 0.01) for each activated TrEx lncRNA. Log2 fold change calculated versus non-targeting controls. A selection of neural stem cell (green), neural (blue), and endoderm/mesoderm (red)-associated genes is highlighted.
(M–P) The top 5 GO terms from ARCHS4 (Lachmann et al., 2018) and Human Cell Atlas (Su et al., 2004) associated with the significantly (adj. p < 0.01) up-regulated (black) and down-regulated (gray) genes in each CRISPRa experiment ranked by combined enrichment score calculated by Enrichr (Kuleshov et al., 2016).
See also Table S1.
The significantly up- and down-regulated genes associated with activation of each TrEx lncRNA were compared with the ARCHS4 (Lachmann et al., 2018) and Human Cell Atlas (Su et al., 2004) gene sets using GO term analysis by Enrichr (Kuleshov et al., 2016) (Figures 5M–5P). Both libraries contain gene sets representing adult and embryonic human and mouse tissues. Genes associated with whole brain, superior frontal gyrus, and cerebral cortex were greatly enriched in those activated by TREX108, suggesting a role in general neural gene networks (Figure 5M). TREX4039 and TREX5008 (associated with CR and RG, respectively) both induced genes enriched in the ARCHS4 neural epithelium gene set and repressed expression of those associated with superior frontal gyrus and astrocytes (Figures 5N and 5O). The genes most changed by their activation, neural progenitor-associated genes such as HES1, HES5, NOTCH3, and OTX1, are significantly reduced (Figures 5J and 5K), perhaps indicating a role in differentiation or CR specification. Finally, although we see the neural-associated genes GFRA2 and HES7 strongly activated by TREX8168 (Figure 5L), GO term analysis shows enrichment of mesoderm germ layer markers with fetal brain and prefrontal cortex-associated genes appearing in the GO terms from down-regulated genes (Figure 5P). It is difficult to speculate what role TREX8168 may have in NE cells as we are at a disadvantage in detecting repressive gene regulatory function in HEK293 cells. Overall, it is promising that we see such drastic gene expression changes involving neural genes when expressing these TrEx lncRNAs in a non-neuronal cell type.
Discussion
The lncRNA field has been mired in controversy over the functional relevance of the tens of thousands of human transcripts (Gingeras, 2012, Hon et al., 2017, Kowalczyk and Higgs, 2012), with claims that most represent non-functional transcription from enhancer elements or spurious transcriptional noise (Ponjavic et al., 2007, Struhl, 2007) due to their low sequence conservation across vertebrates (Babak et al., 2005, Kutter et al., 2012, Pang et al., 2006, Ponjavic et al., 2007) or low levels of expression in bulk tissues (Cabili et al., 2011). It has also been suggested that tissue-specific lncRNAs are less conserved than those expressed in multiple tissues (Ulitsky, 2016), but we found that many human lncRNA transcripts expressed during early cortical neuron differentiation have structural conservation through primates. Of the 2,975 lncRNAs expressed in human cortical neuron differentiation, 72% had conserved structure through chimp, 58% through orangutan, and 43% through rhesus. Fifty-one percent was conserved in the great ape species and 31% had evidence of conserved structure in all tested species, much greater than the estimates of sequence conservation through mouse (Babak et al., 2005, Pang et al., 2006, Ponjavic et al., 2007). Striking among these transcripts were those expressed transiently in COs. Three hundred eighty-six TrEx lncRNAs were observed in human and had a remarkably conserved expression pattern in great ape species, with at least 223 (58%) retaining TrEx patterns in chimpanzee or orangutan. While TrEx patterns are less conserved than exonic structure, we are likely undersampling relevant time points in each species for optimal detection, considering many TrEx lncRNAs were primarily expressed at a single time point. Our analysis revealed that having a conserved pattern of expression across primates strongly correlates with tissue specificity in scRNA-seq, opening the possibility of a role for TrEx lncRNAs in establishing transient developmental cell states.
We focused on TrEx lncRNAs associated with a specific cell type at the week 2 time point, where there was a clear distinction between RG, NE, and CR cells in our scRNA-seq data (Figures 4 and S5). We used a strict definition of transcript conservation requiring both exon boundary and expression pattern conservation between human and at least one other species, reasoning that those have the highest likelihood of regulatory function. Cellular context is vitally important for lncRNA function (Liu et al., 2016b), but still we see significant effects on distal genes upon activation of these TrEx lncRNAs, indicating a robust regulatory function even out of their normal biological context. TREX108, TREX4039, and TREX5008 showed induction of gene sets associated with neurons, while TREX8168 activation yielded significant repression of fetal brain-associated genes as well as modulation of genes associated with non-neural cell types.
The RNA-seq data generated in this study provide a valuable resource for comparative studies aimed at understanding human, chimpanzee, orangutan, and rhesus cortical development. These tissues provide insight into early differentiation stages largely inaccessible in vivo and could shed light on what makes great apes and humans unique from each other and other species. Further, while human, chimpanzee, and rhesus have been studied with COs previously (Camp et al., 2015, Eiraku et al., 2008, Liu et al., 2016a, Mora-Bermudez et al., 2016, Otani et al., 2016), to our knowledge, we provide the first look at orangutan. Pairing weekly bulk RNA-seq across species with analysis of the cell type composition of these heterogeneous cultures by scRNA-seq in human provides additional insight into the expression events underlying the formation of early neural cell types, allowing the identification of lncRNAs associated with cell types present transiently during human development. Further detailed analysis of this dataset and the lncRNAs identified in this study promises to provide important insights into the transcriptional programs underlying primate-specific features of brain development.
Experimental Procedures
Cerebral Organoid Generation
The Eiraku et al. (2008) protocol was optimized for use with hESCs, rhesus ESCs, chimpanzee iPSCs, and orangutan iPSCs. PSCs were either manually lifted and allowed to self-form into embryoid bodies on low-attachment plates (Corning) in KSR medium or aggregated using AggreWell-800 plates in AggreWell medium (STEMCELL Technologies). DKK1 (Peprotech), NOGGIN (R&D Systems), SB431542 (Sigma), and cyclopamine, V. californicum (VWR), were added for the first 18 days of differentiation. Neurobasal with N2 (Thermo Fisher) and cyclopamine was used starting on day 18. Chimpanzee and orangutan cultures were also supplemented with bFGF and EGF. After day 26, all cultures were grown in Neurobasal/N2 medium without added factors. Total RNA was extracted at weekly time points for each species. For protocol details, including the rhesus time point adjustment and iPSC line generation, see Supplemental Information.
Primate Genome Alignment and Annotation
A progressive Cactus (Paten et al., 2011) whole-genome alignment was generated between the human hg19, chimpanzee panTro4, orangutan ponAbe2, and rhesus macaque rheMac8 assemblies and used as input to the Comparative Annotation Toolkit (Fiddes et al., 2018). FANTOM5 (Hon et al., 2017) annotations and RNA-seq obtained from SRA (https://www.ncbi.nlm.nih.gov/sra) were used to help guide the annotation process.
RNA-Seq Library Preparation
Total RNA was collected from organoid cultures by TRIzol (Thermo Fisher) extraction and depleted of rRNA by Ribo-Zero (Epicentre). Bulk strand-specific total-transcriptome RNA-seq libraries were prepared using dUTP during second-strand synthesis either with the TruSeq Stranded Total RNA Library Prep Gold kit (Illumina) or with home brew components (Parkhomchuk et al., 2009).
RNA-Seq Analysis
Paired-end Illumina reads were mapped with STAR v.2.5.1b (Dobin et al., 2013) to hg19 (human, Genome Reference Consortium GRCh37, 2009), panTro4 (chimpanzee, CGSC Build 2.1.4, 2011), ponAbe2 (orangutan, WUSTL Pongo albelii-2.0.2, 2007), and rheMac8 (rhesus macaque, Baylor College of Medicine HGSC Mmul_8.0.1, 2015). DESeq2 v.1.14.1 (Love et al., 2014) was used for differential expression analysis across the time course in each species (see Supplemental Information).
LncRNA Annotation Analysis
Cufflinks v.2.0.2 suite (Trapnell et al., 2010, Trapnell et al., 2012) was used to assemble lncRNA transcript predictions and combine them with FANTOM5 annotations in each species. These were then projected through the Cactus alignment (Stanke et al., 2008) to each other genome. RSEM v.1.3.0 (Li and Dewey, 2011) was used to provide TPM expression values for these newly generated transcripts (Table S1). Expressed lncRNAs were assessed using the “homGeneMapping” tool from the AUGUSTUS toolkit (Konig et al., 2016) to provide an accounting of features found in each genome (Tables S2 and S3, Supplemental Information).
3′ Single-Cell RNA-Seq
H9 hESCs were grown on vitronectin with E8-Flex medium (Thermo Fisher). COs were prepared with the aggregation method (Supplemental Information). Single-cell suspensions from hESCs as well as week 1, week 2, and week 5 COs were prepared for 10× Genomics Chromium scRNA-seq with TrypLE (Thermo Fisher) according to the 10× protocol RevA or RevB. Data were analyzed by Cell Ranger v.1.2 (10× Genomics). Cell clusters were identified and manually curated by expression of canonical cell markers using a combination of graphical and k-means clustering as a guide (Table S4). See Supplemental Information for further details.
CRISPRa Assay
The CRISPRa assay was based on Konermann et al. (2014) using a combination of five custom sgRNAs per target. Transfected cells were selected at 24 hr by puromycin and harvested at 48 hr with TRIzol reagent (Thermo Fisher). qPCR was performed with Quantitect SYBR Green RT-PCR (Qiagen). RNA-seq libraries were prepared with the NEXTflex Rapid Directional qRNA-Seq Library Prep Kit (PerkinElmer). Differential expression analysis was performed as described above (Table S1, Supplemental Information).
Author Contributions
Conceptualization, A.R.F., S.R.S., and D.H.; Methodology, A.R.F. and F.M.J.J.; Investigation, A.R.F., F.M.J.J., A.P.R.P., A.M.R.-O., E.L., L.W., V.M., J.L.R., and M.O.; Formal Analysis, A.R.F., I.T.F., M.H., and S.K.; Writing – Original Draft, A.R.F.; Writing – Review & Editing, S.R.S. and D.H.; Funding Acquisition, A.R.F., F.M.J.J., S.R.S., and D.H.; Supervision, S.R.S. and D.H.
Acknowledgments
This work was supported by CIRM Predoctoral (A.R.F.), Postdoctoral (F.M.J.J.), and Human Frontier Science Program Postdoctoral (F.M.J.J.) Fellowships, CIRM Center of Excellence for Stem Cell Genomics (Stanford), CIRM Center for Big Data in Translational Genomics (SALK), and NIH/NIGMS R01 GM109031 grants. D.H. is an Investigator of the Howard Hughes Medical Institute. We thank Florence Wianny and Colette Dehay for LYON-ES1; Oliver Ryder and the San Diego Frozen Zoo Project of the San Diego Zoo Institute for Conservation Research for Sumatran orangutan fibroblasts; Robert Diaz and Karen Shaff (Applied Stem Cell) for help generating chimpanzee iPSCs; Bryan King and Kristof Tigyi for animal handling; Susan Carpenter and Sergio Covarrubias for plasmids and expertise in designing CRISPRa experiments; Daniel Kim and Pablo Cordero for discussions on experimental design and scRNA-seq; Tom Nowakowski and Alex Pollen for advice on curating scRNA-seq clusters; Bari Nazario (UCSC Institute for the Biology of Stem Cells), Nader Pourmand (UCSC Genome Sequencing Center), Ben Abrams (UCSC Life Science Microscopy Center), and Shana McDevitt (UC Berkeley QB3 GSL) for excellent technical support; Jason Fernandes and all Haussler Lab members for helpful discussions and support.
Published: January 10, 2019
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.12.006.
Accession Numbers
GEO: GSE106245, bulk RNA-seq across cortical neuron differentiation in all species and scRNA-seq from human COs. GEO: GSE120702, bulk RNA-seq from CRISPRa experiments. These data can be visualized on the UCSC Genome Browser as a Track Hub in the Public Hubs section with Hub Name Primate x4 NeuroDiff and Human CRISPRa.
Supporting Citations
The following references appear in the Supplemental Information: Amit et al., 2000, Fluckiger et al., 2006, Langmead and Salzberg, 2012, Locke et al., 2011, Okita et al., 2011, Prokhorova et al., 2009, Quinlan and Hall, 2010, Smit et al., 2013-2015, Workman et al., 2013.
Supplemental Information
References
- Amaral P.P., Mattick J.S. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Amit M., Carpenter M.K., Inokuma M.S., Chiu C.P., Harris C.P., Waknitz M.A., Itskovitz-Eldor J., Thomson J.A. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev. Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- Babak T., Blencowe B.J., Hughes T.R. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genomics. 2005;6:104. doi: 10.1186/1471-2164-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K., Nazlican H., Galetzka D., Wawrzik M., Groβ S., Horsthemke B. C15orf2 and novel noncoding transcript from the Prader-Willi/Angelman syndrome region show monoallelic expression in fetal brain. Genomics. 2007;89:588–595. doi: 10.1016/j.ygeno.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ES cells and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- Fiddes I.T., Armstrong J., Diekhans M., Nachtweide S., Kronenberg Z.N., Underwood J.G., Gordon D., Earl D., Keane T., Eichler E.E. Comparative Annotation Toolkit (CAT)–simultaneous clade and personal genome annotation. Genome Res. 2018;28:1029–1038. doi: 10.1101/gr.233460.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckiger A.C., Marcy G., Marchand M., Négre D., Cosset F.L., Mitalipov S., Wolf D., Savatier P., Dehay C. Cell cycle features of primate embryonic stem cells. Stem Cells. 2006;24:547–556. doi: 10.1634/stemcells.2005-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T.R. Patience is a virtue. Nature. 2012;482:6–7. [Google Scholar]
- Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Hon C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig S., Romoth L.W., Gerischer L., Stanke M. Simultaneous gene finding in multiple genomes. Bioinformatics. 2016;32:3388–3395. doi: 10.1093/bioinformatics/btw494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M.S., Higgs D.R. RNA discrimination. Nature. 2012;482:6–7. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C., Watt S., Stefflova K., Wilson M.D., Goncalves A., Ponting C.P., Odom D.T., Marques A.C. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma’ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018;9:1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde J., Uszczynska-Ratajczak B., Carbonell S., Davis C., Gingeras T.R., Frankish A., Harrow J., Guigo R., Johnson R. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing (CLS) bioRxiv. 2017:1–26. doi: 10.1038/ng.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton P.A., Ingram R.S., Eggenschwiler J., Efstratiadis A., Tilghman S.M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., Nowakowski T.J., Pollen A.A., Lui J.H., Horlbeck M.A., Attenello F.J., He D., Weissman J.S., Kriegstein A.R., Diaz A.A., Lim D.A. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D., Attenello F.J., Villalta J.E., Cho M.Y., Chen Y. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2016 doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D.P., Hillier L.W., Warren W.C., Worley K.C., Nazareth L.V., Muzny D.M., Yang S.P., Wang Z., Chinwalla A.T., Minx P. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Bermudez F., Badsha F., Kanton S., Camp J.G., Vernot B., Köhler K., Voigt B., Okita K., Maricic T., He Z. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife. 2016;5:1–24. doi: 10.7554/eLife.18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Mitchell J.A., Sanz L.A., Pauler F.M., Ferguson-Smith A.C., Feil R., Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Orom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T., Marchetto M.C., Gage F.H., Simons B.D., Livesey F.J. 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell. 2016;18:467–480. doi: 10.1016/j.stem.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pang K.C., Frith M.C., Mattick J.S. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B., Earl D., Nguyen N., Diekhans M., Zerbino D., Haussler D. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 2011;21:1512–1528. doi: 10.1101/gr.123356.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A., Schier A.F. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C.P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova T.A., Harkness L.M., Frandsen U., Ditzel N., Schrøder H.D., Burns J.S., Kassem M. Teratoma formation by human embryonic stem cells is site dependent and enhanced by the presence of Matrigel. Stem Cells Dev. 2009;18:47–54. doi: 10.1089/scd.2007.0266. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani N., Nowakowski T.J., Zhou H., Godshalk S.E., Lisi V., Kriegstein A.R., Kosik K.S. A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron. 2016;90:1174–1188. doi: 10.1016/j.neuron.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F.A., Hubley R., Green P. RepeatMasker Open-4.0. 2013-2015. http://www.repeatmasker.org
- Stanke M., Diekhans M., Baertsch R., Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman A.D., Charvet C.J., Clancy B., Darlington R.B., Finlay B.L. Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci. 2013;33:7368–7383. doi: 10.1523/JNEUROSCI.5746-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.