Abstract
The development of sensitive measurements to analyze individual cells is of relevance to elucidate specialized roles or metabolic functions of each cell under physiological and pathological conditions. Lipids play multiple and critical roles in cellular functions and the application of analytical methods in the lipidomics area is of increasing interest. In this work, in vitro maturation of porcine oocytes was studied. Two independent sources of chemical information (represented by mass spectra in the positive and negative ion modes) from single oocytes (immature oocytes, 24-hour and 44-hour in vitro matured oocytes) were acquired by using desorption electrospray ionization-mass spectrometry (DESI-MS). Low and mid-level data fusion strategies are presented with the aim of better exploring the large amount of chemical information contained in the two mass spectrometric lipid profiles. Data were explored by principal component analysis (PCA) within the two multi-block approaches to include information on free fatty acids, phospholipids, cholesterol-related molecules, di- and triacylglycerols. After data fusion, clearer differences among immature and in vitro matured porcine oocytes were observed, which provide novel information regarding lipid metabolism throughout oocyte maturation. In particular, changes in TAG composition, as well as increase in fatty acid metabolism and membrane complexity were evidenced during the in vitro maturation process. This information can assist the improvement of in vitro embryo production for porcine species.
Keywords: Desorption Electrospray Ionization Mass Spectrometry, Porcine oocyte, In vitro maturation, Data fusion, Principal Component Analysis
Graphical abstract

1. Introduction
Lipids play multiple and critical roles in many biological processes, such as oxidative stress, signal transduction, gamete development, and epithelial morphogenesis. Together with environmental perturbations, pathological conditions and other external factors, variable expression of lipids and other cellular components (e.g., chromosomal material and metabolites) causes heterogeneity of biological systems at a cellular and even sub-cellular level [1–4]. Thus, the investigation of lipid content in single cells and their internal compartments may elucidate specific functionality of cell populations and provide new insights into complicated biological phenomena [5–7]. Several research efforts have been focused lately on mammalian reproductive biotechnologies [3, 8–11]. Indeed, these single cells are characterized by high complexity regarding the array of intracellular molecules [10]. The knowledge of the lipid content in individual oocytes and embryos is of interest in fundamental studies of embryology, in the attempts to understand cellular pluripotency, and in efforts to optimize in vitro culture conditions necessary for the improvements of assisted reproductive techniques [11]. Due to the relevant metabolic similarities between oocytes and embryos with cancer cells [12], the acquired knowledge on cell metabolism may be of interest for stem cell and cancer research as well.
The development of sensitive measurements for single cell analysis is a current need in analytical chemistry. Over time, traditional analytical protocols based on fluorescence microscopy, gas chromatography (GC) or liquid chromatography (LC) combined with mass spectrometry (MS) have been progressively substituted by direct MS systems, due to the necessity to scale-down analytical methods to a single cellular level [4, 6, 7, 10, 13]. Indeed, chemical analysis of lipids via traditional methods involves the pooling of large amount of cells that is unachievable for most studies and prevents analyzing individual variability [1, 8–9]. Moreover, such methods require complex clean-up and extraction procedures, as well as labeling or derivatization steps. On the contrary, direct MS techniques, such as matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) [14], secondary ion mass spectrometry (SIMS) [15], and ambient MS techniques, including desorption electrospray ionization (DESI) [16], liquid extraction surface analysis (LESA) [1], and laser ablation electrospray ionization (LAESI) [17], offer rapid and sensitive fingerprinting approaches to obtain lipid profiles from intact cells [9]. Preliminary chemical manipulation – of different extent, depending on the technique – is usually required. On the other hand, these direct MS protocols are less useful in performing quantitation and identification of lipids with similar structures (e.g., same nominal masses) [18], even though the utilization of high mass accuracy m/z measurements or tandem mass spectrometry helps for identification purposes [8,18]. Many applications for MS lipid detection and single cell analysis have been reviewed recently and details can be found elsewhere [6,7]. Briefly, MALDI and SIMS are usually employed for imaging experiments, since they provide adequate spatial resolution [6, 7, 18,19,20]. In comparison to these vacuum systems, ambient techniques offer the advantage of operating in open environment [2] making them suitable for live cell analysis, but they are limited in spatial resolution and, hence, usually adopted for MS profiling. A first attempt to perform live single-cell MS analysis was reported by Mizuno et al, which adopted a nano-ESI tip as a way to trap single mammalian cells and a means of ionization to detect numerous compounds by high-resolution MS [21].
A common aspect to all these direct MS analyses is the acquisition of untargeted lipid profiles to include a broad range of species, which can be used as fingerprints of cells or cellular compartments. As with the adoption of different ionization sources, the use of different mass analyzers and data acquisition systems can provide deeper structural insights into lipids of biological interest, resulting in an information-rich dataset [22]. For example, in the case of ion mobility spectrometry (IMS), an additional dimension of information is available by using chemical characterization [22,23].
Multivariate data analysis is normally used for pattern recognition purposes, in order to efficiently compress high-dimensional data and extract informative chemical features [1, 2, 6, 7, 10]. A wide portfolio of methods is available for two-way and even multi-way analysis, thus following the needs of the acquired MS data structures.
Whenever MS analyses do not alter the morphology of the cell, multiple sources of information can be acquired by repeatedly analyzing the same cell with either different MS techniques or the same one operating under different conditions (e.g., acquisition in positive and negative ion mode mass spectra). Based on the assumption that a broader range of lipids is detected in this way (e.g., different ionization sources may enhance or suppress desorption / ablation / ionization of specific compounds), the exploration of the multi-block sources of information allows a deeper characterization of the cell. By analyzing the same cell, stronger correlations among lipid species detected by multi-block experiments can be derived, and this may help defining biological pathways and processes [1]. To our knowledge, just two recent studies on single cells describe multi-block experiments to widen the acquisition of several classes of lipids from the same bovine embryo. They involve the adoption of MALDI with the utilization of a binary matrix [9], and DESI [11] with a so-called “morphologically friendly” solvent for the spray system [24,25]. While the first study only lists the lipids detected in bovine embryos, in the second one, a data fusion (i.e., multi-block) analysis was adopted to chemically characterize them [11], comprehensively considering the lipids detected in positive and negative ion mode mass spectra. Indeed, data fusion is capable to merge different data sources and analyze them to obtain a comprehensive exploration of the whole information content and model the data in a more robust way [26–30]. Data fusion was previously and extensively applied to merge datasets acquired by MS coupled with LC, GC or ICP – for elemental as opposed to molecular analysis [27] – and NMR, UV-Vis or IR spectroscopy [26,27,31–36], whereas the DESI-MS study on bovine oocytes and embryos is the only report on direct MS analysis.
The strategy of merging two MS profiles (±), instead of tackling them separately, revealed chemical differences between bovine oocytes developed in vivo and in vitro, highlighting inefficiencies of the current in vitro embryo production systems, with possible deleterious effects on the health status of the offspring [11]. Therefore, in the present study, an analogous dual mode DESI(±)-MS analysis, followed by principal component analysis (PCA) within a data fusion strategy, was adopted to explore the lipid content of porcine oocytes during the in vitro maturation (over 44 hours). The oocyte quality, which may affect developmental capacity, can only be poorly appreciated by morphological examination under a stereomicroscope and the investigation of chemical, metabolic, and genomic fingerprints is necessary instead. By contrast, the visual inspection of the cell is the gold-standard technique used to check the stage of development with nearly 100% confidence. That is, the unbiased exploration (i.e., the exploration of the data not guided by a priori categorization of samples, as it would be the case for supervised techniques like PLS-DA) of the information content enclosed into MS spectra by PCA is the initial interest.
Porcine oocytes have never been investigated before for lipid profiling by ambient MS. However, they present the highest content of lipids among domestic species [37–39], and pig is considered a reliable model in comparison to mice and cattle to study human diseases and stem cells [40]. To date, porcine in vitro embryo production and cryopreservation present poor success compared to mouse and bovine [41], and the efficiency of porcine in vitro maturation remains low. Its improvement may positively influence biomedical technologies, such as the production of transgenic pigs [42].
This work represents a further step of our ongoing research on dynamic lipid characterization of female gametes in the field of reproductive biology.
2. Materials and Methods
2.1. Porcine oocytes retrieval
Porcine oocytes obtained from ovaries collected in a local slaughterhouse were matured in vitro, as described in detail elsewhere [43]. Briefly, cumulus-oocyte-complexes (COCs) were placed into TCM-199 medium (50 COCs per 500 μL drop) supplemented with 0.1 mg mL−1 cysteine, 0.5 IU mL−1 LH, 0.5 IU mL−1 FSH and 10 ng mL−1 epidermal growth factor at 39 °C in 5% CO2. The complete maturation of the oocytes was performed for the 44-hour in vitro matured oocytes by visual inspection of the first polar body under a stereomicroscope, after removal of the cumulus cells by vortexing in 0.1% hyaluronidase.
At different times during in vitro maturation (0, 24 and 44 hours), the COCs were removed from the maturation medium, the surrounding cumulus cells removed by vortexing in 0.1% hyaluronidase and the denuded oocytes were washed three times in PBS supplemented with 0.1% polyvinyl alcohol (PVA) and in methanol:ultra pure water (1:1 v/v). The oocytes were individually placed onto the surface of a glass slide (Gold Seal, Portsmouth, NH, USA). The glass slides were frozen at –80°C until DESI-MS analysis, for which they were allowed to come to room temperature for 15 min and dried.
2.2. DESI-MS
The DESI protocol allows a straightforward analysis of intact individual oocytes and does not require any chemical manipulation, only the manual deposition of the cell onto glass slides is needed. DESI-MS spectra were acquired in positive and then negative ion modes from the same samples. For data acquisition, the background (glass slide) spectrum and then data for up to 10 samples were recorded (~30 s for each oocyte) in one chronogram [11]. The MS data for a total number of 96 immature oocytes (n = 31), 24-hour in vitro matured oocytes (n = 25) and 44-hour in vitro matured oocytes (n = 40) were collected in one working day, randomizing the order of analysis but never changing the order of acquisition between positive and negative ions.
An LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) was used for the experiments. DESI-MS profiles were acquired using a lab-built stage with the DESI spray positioned at ~2 mm from the glass slide containing the oocytes at an incident angle of 50° [11]. In positive ion mode, the DESI spray was nebulized by applying 5 kV to the stainless steel needle syringe and using a nitrogen gas pressure of 180 psi. Other instrument parameters were: injection time = 100 ms, 2 microscans, m/z range = 600–1250, capillary voltage = 0 V and tube lens voltage = 55 V. Acetonitrile (ACN) doped with 3.0 μg mL−1 silver nitrate at 5.0 μL min−1 flow rate was used as the DESI spray system. The use of AgNO3 in the DESI spray allows obtaining lipid silver adducts, which are easily recognized by the characteristic 1:1 abundance ratio of the 107Ag:109Ag isotopes [44]. In the negative mode, the voltage applied to the stainless steel needle syringe was –5 kV. Other operating conditions were: nitrogen gas pressure = 180 psi, injection time = 500 ms, 2 microscans, m/z range = 250–1000, capillary voltage = −27 V and tube lens voltage = −10 V. The solvent combination ACN and DMF (1:1 v/v) was used at 1.0 μL min−1 flow rate, as already reported elsewhere [10, 24].
Both acetonitrile doped with AgNO3 and the combination ACN-DFM (1:1 v/v) are “morphologically friendly” solvent systems. The term refers to operating with spray solvents that do not cause damage to the tissue. The use of morphologically compatible solvents is mandatory to acquire DESI mass spectra of individual oocytes because they allow samples to be sprayed for enough so that both positive and negative mass spectra can be acquired, without causing damage. The mechanism behind DESI-MS is a spot-by-spot micro-extraction of molecules from the sample. The extraction process occurring with morphologically friendly solvents is comparable to the fixative procedures commonly used in histology for morphological examination: lipids are removed whereas the intracellular and extracellular proteins stained in the histochemical treatment are preserved. Further details can be found elsewhere [24,25].
The attribution of lipid species was made using high mass resolution DESI-MS analysis performed with a Thermo Scientific Exactive (San Jose, CA, USA) mass spectrometer, and collision-induced dissociation (CID) tandem MS experiments. Molecular formula matching and error calculations were performed using the instrument software Xcalibur v.1.0.1.03 (Thermo Fisher Scientific, San Jose, CA, USA) and online search for lipids was carried out in the LIPID MAPS [45] and METLIN [46] databases.
2.2. Principal component analysis (PCA) within a multi-block approach
For each sample, a list of m/z values and ion abundances from the averaged mass spectrum (~30 s of data acquisition) was imported into Matlab (The MathWorks, Inc., Natick, USA) and multivariate data processing was performed using in-house Matlab routines. All MS spectra were normalized by the standard normal variate (SNV) transform, so as to correct for both baseline shifts and global intensity variations [47,48].
Positive ions covered the mass range m/z 600–1250 (7801 m/z variables), whereas for the negative ion mode, each mass spectrum was composed by 6301 m/z variables over the mass range m/z 425–950. The acquisition step of m/z values was of m/z 0.0833 in both cases. The mass range of the negative ions was reduced from m/z 250–1000 to m/z 425–950 to exclude uninformative or confounding regions of the spectra in which signal from the background (lower masses) or only random noise (higher masses) was collected. Therefore, two data matrices were built: 96 rows (samples: immature oocytes, n = 31; 24-hour in vitro matured oocytes, n = 25; 44-hour in vitro matured oocytes, n = 40) and 7801 columns (m/z values) for positive ions, 96 rows and 6301 columns for negative ions.
PCA is commonly used for exploratory investigations of the complex information contained in full mass spectral datasets, to allow consideration of all the spectral variables and their inter-correlations simultaneously [49,50,51]. By means of PCA, the data information content of the original multivariate collinear m/z variables is reorganized and compacted in few principal components (PCs): that is, PCA can be used as an unsupervised data compression technique. When dealing with high-dimensionality data, the compression of the relevant information is a priority step in order to efficiently manage the data and extract useful information.
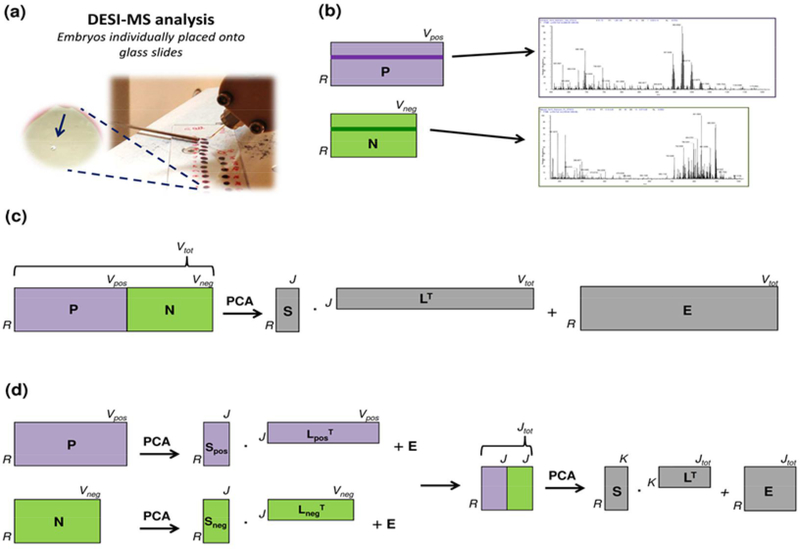
PCA was performed at two levels of data fusion to visualize relationships among samples and variables, taking into account the whole information content and inter-correlations among all variables. A graphical scheme of the different PCA strategies is shown in Fig. 1. Briefly, two different approaches of increasing complexity were adopted for data fusion, usually referred to as low and mid-level fusion, respectively [26–30]. In low-level fusion, raw signals obtained from different data sources are merely concatenated and then processed as a unique fingerprint of the samples (Fig. 1b). Usually, this type of approach requires suitable scaling or weighting of data, especially when the different data blocks are dimensionally unbalanced [28]. In the so-called mid-level fusion, informative features extracted from the raw signals of each analytical source – by means of variable selection procedures or latent-variable compression techniques (e.g., PCA itself) – are combined into a new (reduced) single set and then processed by further multivariate analysis (Fig. 1c).
Fig. 1.
(a) DESI-MS lipid profiling of single oocytes. (b) Schematics of two matrices enclosing positive (P) and negative (N) ions for individual porcine oocytes. Positive and negative mass spectra can be visualized for the same sample by selecting the same row Ri in both of the matrices (represented as dark violet and dark green rows in P and N, respectively). (c) PCA within a low-level data fusion strategy: concatenation of two matrices with R rows (96 samples) and V variables (positive ions: 7801; negative ions: 6301) - after block-scaling to equal variance - and then computation of scores (S), loadings (L), and residuals (E). (d) PCA within a mid-level data fusion strategy: PCA was performed individually on the original datasets with R rows and V variables (positive ions: 7801; negative ions: 6301), then the same number of PCs (5) was selected as for each original dataset, and the scores of the selected PCs were merged into a new data matrix, which underwent PCA again, after column autoscaling.
For this study, in the low-level fusion the two matrices of dimensions 96×7801 (positive ions) and 96×6301 (negative ions) were concatenated column-wise. Before concatenation, the two blocks were scaled to the equal variance (equal to 1) [52]. After concatenation, the matrix was column-centered to allow PC rotation around the global centroid of the data (Fig. 1b). In the mid-level fusion, PCA was applied separately to the two matrices. From both, the first PCs that cumulated about 90% of the total variance were selected, merged together in a new dataset, and used to apply a further PCA, after column autoscaling of the new data (Fig. 1c). Lastly, PCA was also performed on positive and negative ions separately to confirm the improved characterization achieved by data fusion in comparison to analyzing the datasets separately.
3. Results and discussion
In this work, a dual mode DESI(±)-MS protocol was used to analyze in a few minutes single porcine oocytes (about 150 μm in diameter), which contain about 156 nanograms of total lipids [53]. PCA was applied within two multi-block approaches (low and mid-level) on the DESI-MS spectra (±) obtained from single cells represented by individual porcine oocytes. The focus is on the porcine oocyte maturation dynamics, which involves a narrow time-window of about 2 days.
3.1. DESI-MS mass spectra obtained by direct analysis of oocytes
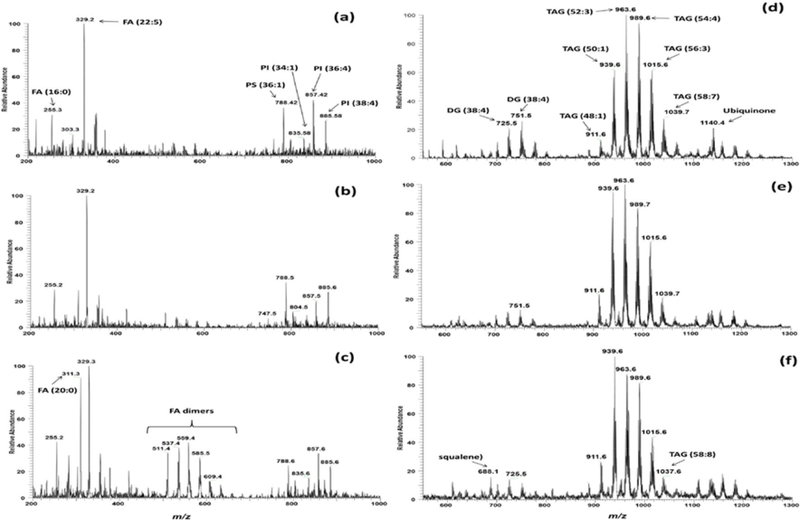
High-throughput protocols often compromise the quality of the chemical information that can be obtained from the samples. For example, high-throughput UHPLC-MS/MS methods often deal with scarce quantitative performances, partially sacrificed to expand the UHPLC-MS/MS multi-analyte capability to the utmost. On the other hand, when the high-throughput “morphologically compatible” DESI(±)-MS protocol is adopted to detect lipids from individual microscopic oocytes, a huge amount of complex chemical information can be acquired. Indeed, DESI is capable of softly ionizing lipids of several classes, including intact complex species (such as PLs, in the negative ion mode, and TAG, in positive ion mode). Figure 2 shows positive and negative ion mode mass spectra of three samples selected based on the outcomes of PCA performed within a mid-level data fusion scheme. Selection was made for the objects that were most distant (i.e., different) from each other and from the global centroid (i.e., the vector of the mean value for each variable) to show maximum differentiation. In the negative ion mode, free fatty acids were detected over the mass range of m/z 200–400, fatty acid dimers (resulting from the combination of free fatty acids in gas phase) were detected over the mass range of m/z 500700, and complex phospholipids (phosphatidylethanolamine, PE; phosphoatidylserine, PS; phosphatidylglycerol, PG; and phosphatidylinositol, PI) over the mass range of m/z 800–1000. Using similar experimental conditions, some lipid species have been previously detected in mouse [10] and bovine [11] oocytes and preimplantation embryos, proving that the chemical information detected by DESI-MS is consistent through oocytes and embryos of various animal species. However, differences can be revealed and are expected, since the lipid abundance in oocytes and embryos seems to be species-specific and time-dependent [54]. Lipid attributions based on high-resolution mass measurements and CID MS/MS experiments are listed in Table 1.
Fig. 2.
DESI mass spectra of porcine oocytes selected to show maximum differentiation. Negative ion mode mass spectra: (a) immature oocytes, (b) 24-hour in vitro matured oocytes, (c) 44-hour in vitro matured oocytes. Positive ion mode mass spectra: (d) immature oocytes, (e) 24-hour in vitro matured oocytes, (f) 44-hour in vitro matured oocytes. See Table 1 for tentative lipid class assignments of the major peaks.
Table 1.
Values of m/z with high resolution mass accuracy, predicted molecular formula, ion description, error in ppm (Delta ppm) and tentative attribution of lipids detected in negative and positive ion modes by DESI-MS.
| Ions | Predicted Molecular formula | Ion description | Delta ppma | Attributionb |
|---|---|---|---|---|
| 253.21610 | C16H29O2 | [M-H]− | −1.203 | 16:1c (palmitoleic acid) |
| 255.23173 | C16H31O2 | −1.224 | 16:0 (palmitic acid) | |
| 279.23163 | C18H31O2 | −1.324 | 18:2 (linoleic acid) | |
| 281.24728 | C18H33O2 | −1.324 | 18:1 (oleic acid) | |
| 283.26290 | C18H35O2 | −1.354 | 18:0 (stearic acid) | |
| 303.23150 | C20H31O2 | −1.454 | 20:4 (arachidonic acid) | |
| 311.29407 | C20H39O2 | −1.484 | 20:0 (eicosanoic acid) | |
| 329.24708 | C22H33O2 | −1.524 | 22:5 (docosapentaenoic acid) | |
| 339.32558 | C22H43O2 | −1.274 | 22:0 (docosanoic acid) | |
| 465.30219 | C27H45O4S | −2.214 | Cholesterol sulphate | |
| 511.47073 | C32H63O4 | −2.454 | FA dimer | |
| 537.48644 | C34H65O4 | −2.394 | FA dimer | |
| 539.50207 | C34H67O4 | −2.414 | FA dimer | |
| 559.47058 | C36H63O4 | −2.604 | FA dimer | |
| 585.50738 | C35H69O6 | −2.583 | FA dimer | |
| 609.48489 | C40H65O4 | −3.944 | FA dimer | |
| 788.54144 | C42H79O10NP | −3.267 | PS (36:1) | |
| 747.51486 | C40H76O10P | −2.201 | PG (34:1) | |
| 804.57281 | C43H83O10NP | −3.197 | PS (37:0) | |
| 835.53082 | C43H80O13P | −3.382 | PI (34:1) | |
| 857.51494 | C45H78O13P | −3.612 | PI (36:4) | |
| 885.54613 | C47H82O13P | −3.722 | PI (38:4) | |
| 887.56518 | C47H84O13P | −0.322 | PI (38:3) | |
| 686.18886 | C30H50O3NAg2 | [M+Ag2NO3]+ | 0.223 (for m/z | Squalene |
| 686.18803)d | ||||
| 725.42684 | C39H70O5Ag | [M+Ag]+ | −0.041 | DAG (36:2)c |
| 751.44284 | C41H72O5Ag | 0.426 | DAG (38:4) | |
| 909.60999 | C51H94O6Ag | 0.0445 | TAG (48:2) | |
| 911.62492 | C51H96O6Ag | −0.346 | TAG (48:1) | |
| 935.62574 | C53H96O6Ag | 0.540 | TAG (50:3) | |
| 937.64121 | C53H98O6Ag | 0.346 | TAG (50:2) | |
| 939.65570 | C53H100O6Ag | −0.889 | TAG (50:1) | |
| 961.64088 | C55H98O6Ag | −0.005 | TAG (52:4) | |
| 963.65635 | C55H100O6Ag | −0.192 | TAG (52:3) | |
| 965.65848 | C55H102O6Ag | 1.606 | TAG (52:2) | |
| 987.65692 | C57H100O6Ag | 0.390 | TAG (54:5) | |
| 989.67258 | C57H102O6Ag | 0.399 | TAG (54:4) | |
| 991.69092 | C57H104O6Ag | 3.111 | TAG (54:3) | |
| 1013.67296 | C59H102O6Ag | 0.764 | TAG (56:4) | |
| 1015.68910 | C59H104O6Ag | 1.245 | TAG (56:3) | |
| 1037.67308 | C61H102O6Ag | 0.862 | TAG (58:8) | |
| 1039.68994 | C61H104O6Ag | 2.024 | TAG (58:7) | |
| 1041.70459 | C61H106O6Ag | 1.060 | TAG (58:6) | |
| 1140.48205 | C59H90O7NAg2 | [M+Ag2NO3]+ | −0.612 (for m/z 1138.48184)d | Ubiquinone (Coenzyme Q10) |
Delta ppm column displays the difference between the specified mass and the calculated mass in ppm units.
Attribution of lipids is based on Lipid Maps (www.lipidmaps.org) and Metlin (http://metlin.scripps.edu) searches of the predicted molecular formulae and possible adducts.
(C:U) represents the number of carbon atoms (C) and the number of unsaturations (U) of the fatty acyl chains.
Abbreviations used: DAG – diacylglycerol; FA, fatty acid; PI- phosphatidylinositol; PS - phosphatidylserine; PG - Phosphatidylglycerol, TAG – triacylglycerol. The ‘o-’ suffix is used to indicate the presence of an alkyl ether substituent, whereas the ‘p-’ suffix is used for the 1Z-alkenyl ether (plasmalogen) substituent.
Due to the fact the squalene and ubiquinone were adducts presenting Ag2NO3, the exact mass has been calculated based on the monoisotopic mass as reported in PLoS One. 2013 Sep 20;8(9):e74981.
In the positive ion mode, the most abundant ions in the three experimental groups (immature oocytes, 24-hour and 44-hour in vitro matured oocytes) were triacylglicerol (TAG) lipids, which have 48 to 58 carbon atoms in the fatty acyl residues and one to five units of unsaturation (see Fig. 2 and Table 1). Indeed, TAGs have been reported to be the major intracellular lipids in porcine oocytes by conventional lipid extraction and chromatographic analysis of thousands of pooled porcine oocytes [55]. Other lipids detected in the DESI-MS profile were diacylglycerols (DAG), squalene and ubiquinone (see Fig. 2 and Table 1). Table 1 lists the major DAG and TAG species detected by DESI-MS in terms of the total number of carbon atoms and the unsaturation present in the fatty acyl residues (C:U). The unique identity of the fatty acyl residues in the DAG and TAG species can be specifically defined by MS/MS experiments.
3.2. PCA within a multi-block approach
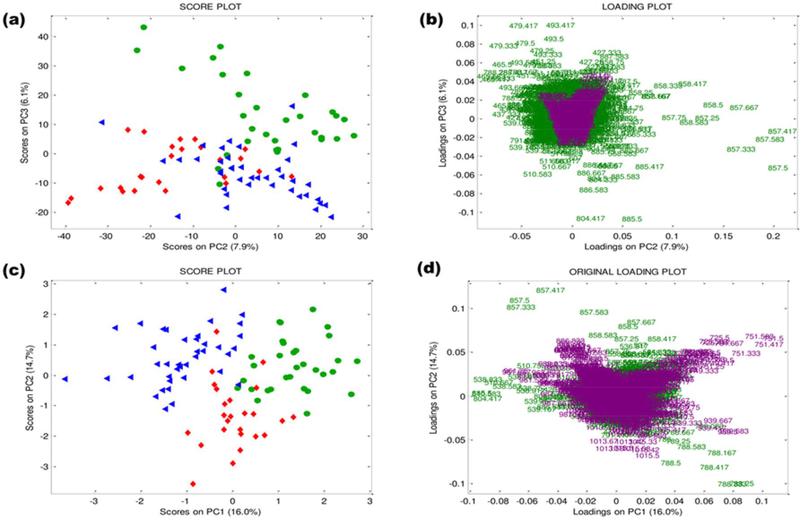
Multivariate analysis was performed to extract useful information from the porcine oocytes among the large amount of data embedded within the DESI-MS profiles. As graphically represented in Fig. 1, both low- and mid-level data fusion strategies were tried to compute PCA. For the two of them, Fig. 3 shows the score and loading plots of the lowest-order PCs along which the major separation among the objects (i.e., samples) is visible. In more detail, the score plots from the low and mid-level approaches, show separation among the color-labeled objects, which represent immature oocytes (green circles), 24-hour in vitro matured oocytes (red diamond) and 44-hour in vitro matured oocytes (blue triangles). Notably, the separation for the mid-level fusion is visible within the PC1 vs. PC2 score space (Fig. 3c) and appears clearer both in respect to the low-level fusion (Fig. 3a) and to the PCA performed on positive and negative ions separately (Fig. 4a and 4c). Moreover, in the latter cases the exploration of higher-order PCs is necessary to see groupings among the target categories (Fig 3a, Fig. 4a and 4c). This means that the inter-correlations among positive and negative blocks are informative and that the compression of the whole information content through a mid-level strategy is so efficient that the separation of interest shows up as the dominant fraction of variability into the MS data.
Fig. 3. PCA of the fused datasets from positive and negative ion mode mass spectra in a low-level approach:
(a) PC2 vs. PC3 score plot. (b) PC2 vs. PC3 original loading plots labeled in terms of m/z ratio. PCA of the fused datasets from positive and negative ion mode mass spectra in a mid-level approach: (c) PC1 vs. PC2 score plot. (d) PC1 vs. PC2 original loading plots labeled in terms of m/z ratio. Immature oocytes: green circles (n = 31), 24-hour in vitro matured oocytes: red diamonds (n = 25), 44-hour in vitro matured oocytes: blue triangles (n = 40). Negative ions: green; positive ions: violet.
Fig. 4. PCA of negative ion mode mass spectra:
(a) PC2 vs. PC5 score plot. (b) PC2 vs. PC5 original loading plots labeled in terms of m/z ratio. PCA of positive ion mode mass spectra: (c) PC2 vs. PC3 score plot. (d) PC2 vs. PC3 original loading plots labeled in terms of m/z ratio. Immature oocytes (green circles, n = 31), 24-hour in vitro matured oocytes (red diamonds, n = 25), 44-hour in vitro matured oocytes (blue triangles, n = 40).
The mid-level fusion also appears to be more informative in the attempts to extract the relevant chemical features from the loading plot with a balance among the information provided by both positive and negative ions (Fig 3d). Note that the original MS data are described by a higher number of variables for the positive ion mode and two orders of magnitude of difference for the absolute ion intensities (positive ions are more intense than negative ions). By contrast, in the low-level fusion, the loadings associated to the positive ion mode are compressed to the centroid and appear to be unbalanced in comparison to those of negative ions in computing the PCs (Fig 3b), although the two blocks of data have been block-scaled to equal variance [53]. Note that the loadings (L) originally coming from the dataset of positive ions were color-labeled in violet, while those of the dataset of negative ions were color-labeled in green, and computed through PCA from the relation:
where X is the concatenated mass dataset of positive and negative ions, S are the scores, and E the residuals (Fig. 1b).
In Fig. 3d, the PC1 vs. PC2 loading plot shows the contribution of the original variables in determining the final PCs, after mid-level data fusion. Loadings of the combined PCA and related to the original variables (O) have been computed by:
where F is obtained by merging column-wise the individual loading matrices obtained in the separated PCA (LposT and LnegT), and LT is the loading matrix obtained from the PCA performed onto the merged score matrices (Fig. 1c). In more detail, F is a Jtot × Vtot matrix, where Jtot is the total number of principal components from the individual PCA and Vtot is the number of original variables; LT has dimensions K × Jtot, where K is the number of PCs retained in the final PCA. Therefore, O has dimension K × Vtot.
The visualization of the original variable contribution from the two independent MS sources in determining the final PCs allows a straightforward interpretation of the differences in the chemical features among sample categories. From the loading plot, it is possible to extract information on the inter-correlation among lipids (based on the relative position of different ions in the loading plot) and to highlight ions that seem to contribute the most to PC computation and, thus, to the separation among sample categories that is visible along the same PCs in the score plot. In interpreting the loadings for this specific study, it is important to remark that, although SNV has the peculiarity to potentially shifting information across the signal range, so that the interpretation of the results referring to the original signals may be deceiving [47,48]. In the outcomes of the present study, the changes in the ion relative abundances highlighted by PCA are in accordance with the information shown by the averaged mass spectra of each sample category (see Fig. S1 of the Supplementary Material).
In more detail, in the loading plot of PC1 vs. PC2, the positive ions (violet m/z values in the loading plot) of m/z 725.5 (DAG 36:2), 751.5 (DAG 38:4) appear to be more abundant in the immature oocytes (green circles). Diacylglycerol is well characterized as a second messenger signaling lipid originating from the hydrolysis of the phospholipid phosphatidylinositol 4,5bisphosphate (PIP2) by the enzyme phospholipase C (PLC). In the same reaction, inositol trisphosphate (IP3) is produced. Although IP3 diffuses into the cytosol, diacylglycerol is hydrophobic and remains within the plasma membrane where it is believed to activate protein kinase C, thereby regulating calcium influx [56]. The increase in DAG in immature oocytes may be related to the active signaling processes needed to resume meiosis and promote maturation. By using MS, it is possible to obtain more details on the structure (number of carbon in the fatty acyl chains and unsaturation) of the DAG present in the oocytes and, if needed, MS/MS can be used to provide the identity of the fatty acyl residues. Diacylglycerols tend to be minor components of most tissues, in quantitative terms, but they are very important in animal tissues, because they function as second messengers in many cellular processes, modulating vital biochemical mechanisms. Indeed, mouse oocytes with higher rate of meiosis resumption presented increased content of DAG [57]. Also, alteration in lipid metabolism involving DAG species has been reported in adipose tissue of Bscl2-deficient mice [58].
The ions of m/z 1013.6 (TAG 56:4), 1015.6 (TAG 54:3), 939.6 (TAG 50:1) are more abundant in 24-hour in vitro matured oocytes (red diamonds), whereas the ions of m/z 886.6, 987.6 (TAG 54:5), 989.6 (TAG 54:4), 963.6 (TAG 52:3) are more abundant in the 44-hour in vitro matured oocytes (blue triangles). This outcome suggests that TAG metabolism occurring during oocyte maturation involves changes in the abundance of some TAG species and these changes can be monitored by DESI-MS. As a matter of fact, a decrease in TAG content reflects the massive uptake of TAG as energy source during oocyte maturation [59].
In contrast to bovine immature and in vitro matured oocytes [11], squalene (m/z 688.6) does not contribute to differentiate the samples according to the maturation state. On the other hand, ubiquinone (m/z 1140.4) is of low abundance in porcine oocytes in comparison to bovine oocytes [11]. This result may suggest that differences in cholesterol metabolism and mitochondrial activity during maturation exist between bovine and porcine oocytes.
In the negative ion mode (green m/z values in the loading plot), the relative abundance of the ions of m/z 788.5 (PS 36:1) is higher after 24 hours of maturation (red diamonds). Dimers of free fatty acids (over the mass range m/z 500–600) appear to be more abundant in the 44-hour in vitro matured oocytes (blue triangles), as well as in the ions of m/z 804.4 (PS 37:0), 857.5 (PI 36:4), 885.5 PI (38:4). The relative abundances of all these lipids appear to be lower in the immature oocytes (green circles). Therefore, as indicated by DESI-MS, the increase of free fatty acid concentration (probably by TAG metabolism) as well as membrane specialization for signaling and apoptosis processes should occur during porcine oocyte maturation. Higher abundance of heavier and more complex lipids has been previously characterized by DESI-MS lipid profiles of mouse and bovine oocytes after maturation [10, 11]. The wide range of PLs in in vitro matured oocytes compared with immature oocytes was related to a preparation for fertilization and reflects the acquisition of functional and structural specialization by cellular membrane [10].
It should be mentioned that limitations of direct lipid MS analysis in single cells exist and are mainly related to the measurement of the relative amount of lipids present in the samples instead of to absolute quantification, even though good agreement between relative signals in DESI and LC-MS/MS analyses has been reported [60]. This is due to the impossibility of adding internal standards and to the absence of sample extraction processing. Therefore, even though direct analysis in a complex mixture allows for sensitive and broad lipid characterization, it may introduce bias in the data. Nevertheless, the simplified information extracted by PCA, displayed in score and loading plots, highlights relevant chemical features and may operate as a screening step. Indeed, further MS analyses for quantification can be focused on the lipids that appear to be characterizing.
4. Conclusions
The ambient MS technique DESI was utilized to acquire two independent sources of chemical information – positive and negative ion mode mass spectra – from porcine oocytes through the in vitro maturation process. Data of both ion modes were acquired from the same microscopic samples by switching polarity and optimizing the DESI spray solvent system. Using a data fusion protocol, PCA was applied to extract and visualize the information content. The mid-level approach showed improvements in data analysis in comparison to working up the data separately. Through this methodology, it was possible to observe a dynamic change in TAG composition as well as an increase in fatty acid metabolism and membrane complexity during porcine oocyte maturation in vitro. These findings are coherent with previous knowledge about the mammalian oocyte maturation process. Remarkably, this dual mode DESI(±)-MS analysis followed by a multi-block approach for data analysis proved sensitive to even minor changes in the lipid content, and thus feasible to reveal dynamic difference in single cells, besides differences between oocytes and embryos at different developmental stages and between in vivo and in vitro counterparts [10,11].
With the assumption that multiple sources of chemical information may be joined to improve data analysis and the resulting biological interpretation, data fusion is feasible for single cell analysis using independent combinations of MS techniques; for example, DESI followed by MALDI [61], to combine lipid and protein profiles. Prospectively, these molecular fingerprints can be used jointly to model one or more classes of embryos and chemically compare them with counterparts developed under different conditions.
The expansion of this multi-block methodology to process MS images and multi-way datasets acquired on the same sample is also possible. For imaging MS, ambient techniques may be of preference, since neither spatial dislocation nor chemical modifications of endogenous compounds occur on the sample surface [62].
Supplementary Material
Highlights.
Repeated analysis by DESI(±)-MS of intact single oocytes for lipid characterization
Deployment of a data fusion strategy to merge positive and negative ion mode data
Enhanced interpretation of metabolic changes by more efficient analysis of spectral data
Discovery of increased fatty acid metabolism and membrane complexity during maturation
Assistance in the improvement of in vitro embryo production for porcine species
Acknowledgements
The Authors thank Prof. Heiner Niemann from the Institute of Farm Animal Genetics, FLI Mariensee (Germany) for his keen cooperation. AFGS was supported from the German Academic Exchange Service (DAAD) and the Fundación Gran Mariscal de Ayacucho (FUNDAYACUCHO); VP was supported from the Foundation L’Oreal / UNESCO for Women in Science (Italian Award L’Oreal / UNESCO for Women in Science 2013); CF was supported from the Purdue University Center for Cancer Research Small Grants and from the Brazilian National Council for Scientific and Technological Development (CNPq 237237/2012–1); RGC by NIH; Grant 1R21EB015722 all gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ellis SR, Ferris CJ, Gilmore KJ, Mitchell TW, Blanksby SJ, in het Panhuis M M, Direct lipid profiling of single cells from inkjet printed microarrays, Anal. Chem 84 (2012) 9679–9683. [DOI] [PubMed] [Google Scholar]
- [2].Shrestha B, Vertes A, High-Throughput Cell and Tissue Analysis with Enhanced Molecular Coverage by Laser Ablation Electrospray Ionization Mass Spectrometry Using Ion Mobility Separation, Anal. Chem 86 (2014) 4308–4315. [DOI] [PubMed] [Google Scholar]
- [3].Ferreira MS, de Oliveira DN, Gonçalves RF, Catharino RR, Lipid characterization of embryo zones by silica plate laser desorption ionization mass spectrometry imaging (SPLDI-MSI), Anal. Chim. Acta 807 (2014) 96–102. [DOI] [PubMed] [Google Scholar]
- [4].Gong X, Zhao Y, Cai S, Fu S, Yang C, Zhang S, Zhang X, Single Cell Analysis with Probe ESI-Mass Spectrometry: Detection of Metabolites at Cellular and Subcellular Levels, Anal. Chem 86 (2014) 3809–3816. [DOI] [PubMed] [Google Scholar]
- [5].Schober Y, Guenther S, Spengler B, Römpp A, Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Chem 84 (2012) 6293–6297. [DOI] [PubMed] [Google Scholar]
- [6].Trouillon R, Passarelli MK, Wang J, Kurczy ME, Ewing AG, Chemical Analysis of Single Cells, Anal. Chem 85 (2013) 522–542. [DOI] [PubMed] [Google Scholar]
- [7].Li M, Yang L, Bai Y, Liu H, Analytical Methods in Lipidomics and Their Applications, Anal. Chem 86 (2014) 161–175. [DOI] [PubMed] [Google Scholar]
- [8].Ferreira CR, Saraiva SA, Catharino RR, Garcia JS, Gozzo FC, Sanvido GB, Santos LF, Lo Turco EG, Pontes JH, Basso AC, Bertolla RP, Sartori R, Guardieiro MM, Perecin F, Meirelles FV, Sangalli JR, Eberlin MN, Single embryo and oocyte lipid fingerprinting by mass spectrometry, J. Lipid Res 51 (2010) 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tata A, Sudano MJ, Santos VG, Landim-Alvarenga FD, Ferreira CR, Eberlin MN, Optimal single-embryo mass spectrometry fingerprinting, J. Mass Spectrom 48 (2013) 844–849. [DOI] [PubMed] [Google Scholar]
- [10].Ferreira CR, Pirro V, Eberlin LS, Hallett JE, Cooks RG, Developmental phases of individual mouse preimplantation embryos characterized by lipid signatures using desorption electrospray ionization mass spectrometry, Anal. Bioanal. Chem 404 (2012) 2915–2926. [DOI] [PubMed] [Google Scholar]
- [11].González-Serrano AF, Pirro V, Ferreira CR, Oliveri P, Eberlin LS, Heinzmann J, Lucas-Hahn A, Niemann H, Cooks RG, Desorption electrospray ionization mass spectrometry reveals lipid metabolism of individual oocytes and embryos, PLOS ONE 8 (2013) e74981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smith DG, Sturmey RG, Parallels between embryo and cancer cell metabolism. Biochem. Soc. Trans 41 (2013) 664–669. [DOI] [PubMed] [Google Scholar]
- [13].Perdian DC, Cha S, Oh J, Sakaguchi DS, Yeung ES, Lee YJ, In situ probing of cholesterol in astrocytes at the single-cell level using laser desorption ionization mass spectrometric imaging with colloidal silver, Rapid Commun. Mass Spectrom 24 (2010) 1147–1154. [DOI] [PubMed] [Google Scholar]
- [14].Karas M, Hillenkamp F, Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem 60 (1988) 2299–2301. [DOI] [PubMed] [Google Scholar]
- [15].Benninghoven A, Rudenauer FG, Werner HW. (1987) Secondary ion mass spectrometry–basic concepts, instrumental aspects, applications and trends. New York: Wiley. 1277 p. [Google Scholar]
- [16].Cooks RG, Ouyang Z, Takats Z, Wiseman JM, Detection Technologies. Ambient mass spectrometry, Science 311 (2006) 1566–1570. [DOI] [PubMed] [Google Scholar]
- [17].Nemes P, Vertes A, Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal. Chem 79 (2007) 8098–106. [DOI] [PubMed] [Google Scholar]
- [18].Passarelli MK, Ewing AG, Winograd N, Single-cell lipidomics: characterizing and imaging lipids on the surface of individual Aplysia californica neurons with cluster secondary ion mass spectrometry. Anal. Chem 85 (2013) 2231–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM, Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS, J. Mass Spectrom 47 (2012) 1473–81. [DOI] [PubMed] [Google Scholar]
- [20].Passarelli MK, Ewing AG, Single-cell imaging mass spectrometry, Curr. Opin. Chem. Biol 17 (2013) 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mizuno H, Tsuyama N, Harada T, Masujima T, Live single-cell video-mass spectrometry for cellular and subcellular molecular detection and cell classification, J. Mass Spectrom 43 (2008) 1692–1700. [DOI] [PubMed] [Google Scholar]
- [22].Shah V, Castro-Perez JM, McLaren DG, Herath KB, Previs SF, Roddy TP, Enhanced data-independent analysis of lipids using ion mobility-TOFMSE to unravel quantitative and qualitative information in human plasma, Rapid Commun. Mass Spectrom 27 (2013) 2195–2200. [DOI] [PubMed] [Google Scholar]
- [23].Peng WP, Chou SW, Patil AA, Measuring masses of large biomolecules and bioparticles using mass spectrometric techniques, Analyst 139 (2014) 3507–3523. [DOI] [PubMed] [Google Scholar]
- [24].Ferreira CR, Eberlin LS, Hallett JE, Cooks RG, Single oocyte and single embryo lipid analysis by desorption electrospray ionization mass spectrometry, J. Mass Spectrom 47 (2012) 29–33. [DOI] [PubMed] [Google Scholar]
- [25].Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cheng L, Cooks RG, Nondestructive, histologically compatible tissue imaging by desorption electrospray ionization mass spectrometry, Chembiochem. 12 (2011) 2129–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Smolinska A, Blanchet L, Coulier L, Ampt KA, Luider T, Hintzen RQ, Wijmenga SS, Buydens LM, Interpretation and visualization of non-linear data fusion in kernel space: study on metabolomic characterization of progression of multiple sclerosis, PLoS One 7 (2012) e38163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Silvestri M, Bertacchini L, Durante C, Marchetti A, Salvatore E, Cocchi M, Application of data fusion techniques to direct geographical traceability indicators, Anal. Chim. Acta 769 (2013) 1–9. [DOI] [PubMed] [Google Scholar]
- [28].Bevilacqua M, Bucci R, Magrì AD, Marini F, Data fusion for food authentication. Combining near and mid infrared to trace the origin of extra virgin olive oils, NIR news 24 (2013) 12–15. [Google Scholar]
- [29].Smolinska A, Blanchet L, Buydens LM, Wijmenga SS, NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review, Anal. Chim. Acta 750 (2012) 82–97. [DOI] [PubMed] [Google Scholar]
- [30].Smilde AK, van der Werf MJ, Bijlsma S, van der Werff-van der Vat BJ, Jellema RH, Fusion of mass spectrometry-based metabolomics data, Anal. Chem 77 (2005) 6729–6736. [DOI] [PubMed] [Google Scholar]
- [31].Boccard J, Rutledge DN, A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion, Anal. Chim. Acta 769 (2013) 30–39. [DOI] [PubMed] [Google Scholar]
- [32].Gika HG, Theodoridis GA, Earll M, Snyder RW, Sumner SJ, Wilson ID, Does the mass spectrometer define the marker? A comparison of global metabolite profiling data generated simultaneously via UPLC-MS on two different mass spectrometers, Anal. Chem 82 (2010) 8226–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Casale M, Casolino C, Oliveri P, Forina M, The potential of coupling information using three analytical techniques for identifying the geographical origin of Liguria extra virgin olive oil, Food Chemistry 118 (2010) 163–170. [Google Scholar]
- [34].Richards SE, Dumas ME, Fonville JM, Ebbels TMD, Holmes E, Nocholson JK, Intra- and inter-omic fusion of metabolic profiling data in a systems biology framework, Chemometr. Intell. Lab. System 104 (2010) 121–131. [Google Scholar]
- [35].Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, Rainville P, Stumpf CL, Nicholson JK, Statistical heterospectroscopy, an approach to the integrated analysis of NMR and UPLC-MS data sets: application in metabonomic toxicology studies, Anal. Chem 78 (2006) 363–371. [DOI] [PubMed] [Google Scholar]
- [36].Blanchet L, Smolinska A, Attali A, Stoop MP, Ampt KA, van Aken H, Suidgeest E, Tuinstra T, Wijmenga SS, Luider T, Buydens LM, Fusion of metabolomics and proteomics data for biomarkers discovery: case study on the experimental autoimmune encephalomyelitis, BMC Bioinformatics 12 (2011) 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lowenstein JE, Cohen AI, Dry mass, lipid content and protein content of the intact and zona–free mouse ovum, J. Embryol. Exp. Morphol 12 (1964) 113–121. [PubMed] [Google Scholar]
- [38].Ferguson EM, Leese HJB, Triglyceride content of bovine oocytes and early embryos, J. Reprod. Fertil 116 (1999) 373–378. [DOI] [PubMed] [Google Scholar]
- [39].Coull GD, Speake BK, Staines ME, Broadbent PJ, McEvoy TG, Lipid and fatty acid composition of zona-intact sheep oocytes, Theriogenology 49 (1998) 179. [Google Scholar]
- [40].Prather RS, Lorson M, Ross JW, Whyte JJ, Walters E, Genetically engineered porcine models for human diseases, Annu. Rev. Anim. Biosci 1 (2013) 203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang W, Yi K, Yan H, Zhou X, Advances on in vitro production and cryopreservation of porcine embryos, Anim. Reprod. Sci 132 (2012) 115–22. [DOI] [PubMed] [Google Scholar]
- [42].Kwak SS, Yoon JD, Cheong SA, Jeon Y, Lee E, Hyun SH, The new system of shorter porcine oocyte in vitro maturation (18 hours) using >/=8 mm follicles derived from cumulus-oocyte complexes, Theriogenology 81 (2014) 291–301. [DOI] [PubMed] [Google Scholar]
- [43].Lee K, Wang C, Chaille JM, Machaty Z, Effect of resveratrol on the development of porcine embryos produced in vitro, J. Reprod. Dev 56 (2010) 330–335. [DOI] [PubMed] [Google Scholar]
- [44].Jackson AU, Shum T, Sokol E, Dill A, Cooks RG, Enhanced detection of olefins using ambient ionization mass spectrometry: Ag+ adducts of biologically relevant alkenes, Anal. Bioanal. Chem 399 (2011) 367–376. [DOI] [PubMed] [Google Scholar]
- [45].Fahy E, Sud M, Cotter D, Subramaniam S, LIPID MAPS online tools for lipid research, Nucleic. Acids Res. 35 (2007) W606–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G, METLIN: a metabolite mass spectral database, Ther. Drug Monit 27 (2005) 747–751. [DOI] [PubMed] [Google Scholar]
- [47].Oliveri P, Casolino MC, Forina M, Chemometric brains for artificial tongues, Adv. Food Nutr. Res 61 (2010) 57–117. [DOI] [PubMed] [Google Scholar]
- [48].Fearn T The effect of spectral pre-treatments on interpretation. NIR news (2009) 20: 16–17. [Google Scholar]
- [49].Jolliffe IT. (2002) Principal component analysis. New York: Springer-Verlag. 519 p. [Google Scholar]
- [50].Bro R, Age K. Smildea, Principal component analysis, Anal. Methods 6 (2014) 2812–2831. [Google Scholar]
- [51].Pirro V, Eberlin LS, Oliveri P, Cooks RG, Interactive hyperspectral approach for exploring and interpreting DESI-MS images of cancerous and normal tissue sections, Analyst 137 (2012) 2374–2380. [DOI] [PubMed] [Google Scholar]
- [52].Forshed J, Idborg H, Jacobsson SP, Evaluation of different techniques for data fusion of LC/MS and 1H-NMR, Chemometr. Intel. Lab. Syst 85 (2007) 102–109. [Google Scholar]
- [53].McEvoy TG, Coull GD, Broadbent PJ, Hutchinson JS, Speake BK, Fatty acid composition of lipids in immature cattle, porcine and sheep oocytes with intact zona pellucid, J. Reprod. Fertil 118 (2000) 163–170. [PubMed] [Google Scholar]
- [54].Prates EG, Alves SP, Marques CC, Baptista MC, Horta AE, Bessa RJ, Pereira RM, Fatty acid composition of porcine cumulus oocyte complexes (COC) during maturation: effect of the lipid modulators trans-10, cis-12 conjugated linoleic acid (t10,c12 CLA) and forskolin, In vitro Cell. Dev. Biol. Anim 49 (2013) 335–345. [DOI] [PubMed] [Google Scholar]
- [55].Homa S, Racow C, McCaughy R, Lipid analysis of immature porcine oocytes. J. Reprod. Fertil 77 (1986) 425–434. [DOI] [PubMed] [Google Scholar]
- [56].Colonna R, Tacone C, Malgaroli A, Eusebi F, Mangia F. Gamete Res, Effects of protein kinase C stimulation and free Ca2+ rise in mammalian egg activation, Gamete Res. 24 (1989) 171–183. [DOI] [PubMed] [Google Scholar]
- [57].Ma JY, Li M, Luo YB, Song S, Tian D, Yang J, Zhang B, Hou Y, Schatten H, Liu Z, Sun QY, Maternal factors required for oocyte developmental competence in mice: transcriptome analysis of non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) oocytes, Cell Cycle 12 (2013) 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen W, Zhou H, Liu S, Fhaner CJ, Gross BC, Lydic TA, Reid GE, Altered lipid metabolism in residual white adipose tissues of Bscl2 deficient mice, PLOS One (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sturmey RG, Leese HJ, Energy metabolism in porcine oocytes and early embryos, Reproduction 126 (2003) 197–204. [DOI] [PubMed] [Google Scholar]
- [60].Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG, Desorption electrospray ionization mass spectrometry: imaging drugs and metabolites in tissues, Proc. Natl. Acad. Sci. USA 105 (2008) 18120–18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Eberlin LS, Liu X, Ferreira CR, Santagata S, Agar NY, Cooks RG, Desorption electrospray ionization then MALDI mass spectrometry imaging of lipid and protein distributions in single tissue sections. Anal. Chem 83 (2011) 8366–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR, Mass spectrometry imaging under ambient conditions, Mass. Spectrom. Rev 32 (2013) 218–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.