Abstract
Background
Deafness, autosomal recessive 77 (DFNB77) is a rare non-syndromic hearing loss (NSHL) worldwide, which is caused by deleterious variants within lipoxygenase homology domains 1 (LOXHD1). Here we identified that a novel missense variant of LOXHD1 was associated with NSHL in a Chinese family under consanguineous marriage.
Case presentation
A 28-year-old woman suffered a bilateral profound NSHL. Impedance audiometry, temporal bone computerized tomography (TBCT) scans and magnetic resonance imaging-inner ear hydrography (MRI-IEH) did not find any obvious abnormality of middle or inner ear. Routine genetic detection did not find pathogenic variants in common HL-associated genes. Therefore, we performed a whole-exome sequencing (WES) in this family. By trio-WES, co-segregation validation and bioinformatics analysis, we revealed that a novel homozygous variant in this patient, LOXHD1: c.5948C > T (p.S1983F), might be the pathogenic factor. Her parents (heterozygotes) and brother (wild-type) were asymptomatic.
Conclusions
We successfully identified a novel variant of LOXHD1 associated with a rare NSHL from a Chinese family. Our finds highlight the effectiveness of trio-WES for molecular diagnosis of rare NHSL, and expand the genotypic spectrum of DFNB77.
Electronic supplementary material
The online version of this article (10.1186/s12881-019-0758-2) contains supplementary material, which is available to authorized users.
Keywords: Deafness, autosomal recessive 77 (DFNB77); Non-syndromic hearing loss (NSHL); Lipoxygenase homology domains 1 (LOXHD1); Genetic variant; Whole-exome sequencing (WES)
Background
Hearing loss (HL) is the most common sensory deficit that affects 466 million people in the world (Available at http://www.who.int/pbd/deafness/estimates/en/). At least 60% of HL cases are accounted for genetic causes [1, 2]. Non-syndromic HL (NSHL) is the predominant type (~ 80%) of hereditary HL [3]. Nowadays, more than 100 genes have been related to NSHL (Available at https://hereditaryhearingloss.org/). However, except for several genes, many genes are insufficiently described due to low mutated frequencies, thus handicapping evidence-based genetic counseling on HL patients. Currently, the introduction of whole-exome sequencing (WES) makes it possible to screen all potential disease-causing genes for hereditary HL [4–6]. Benefiting from this technology, many HL patients could have molecular diagnosis when conventional methods identify no pathogenic variants within common HL-associated genes, thus helping to determine novel and more detailed genotype-phenotype correlations.
Deafness, autosomal recessive 77 (DFNB77, MIM # 613079) is a typical example of rare NSHL, which is caused by deleterious variants within lipoxygenase homology domains 1 (LOXHD1) located at chromosome 18q21.1 (MIM #613072) [7]. LOXHD1 is a highly conserved stereociliary protein consisting of 15 polycystin-1/lipoxygenase/alpha-toxin (PLAT) domains, which facilitates proteins interacting with the plasma membrane [8]. Loxhd1 in mice is mainly expressed in hair cell stereocilia and plays a crucial role in maintaining normal function of cochlear hair cells [7]. Mutations within LOXHD1 are rare that only 33 pedigrees have been reported worldwide, harboring less than 50 different HL-causing variants to date [2, 7, 9–22]. Specially, these variants are extremely rare in East Asian population and only reported once in China [2, 14]. According to the HGMD database (http://www.hgmd.cf.ac.uk/docs/login.html, professional 2018.3 version), there are 47 missenses/nonsenses, 5 splicing variants, 5 small deletions, 1 small insertion, 1 small indel and 1 gross deletion identified in the LOXHD1 gene. In these variants, 47 variants are associated with hearing loss and 16 variants are associated with late-onset Fuchs corneal dystrophy (FCD, MIM #136800). More studies are necessary to uncover potential genotype-phenotype correlations between LOXHD1 variants and HL.
Here, we examined a Chinese family by trio-WES analyses and identified a novel missense variant, c.5948C > T (p.S1983F) within LOXHD1 gene. This variant results in a bilateral NSHL.
Case presentation
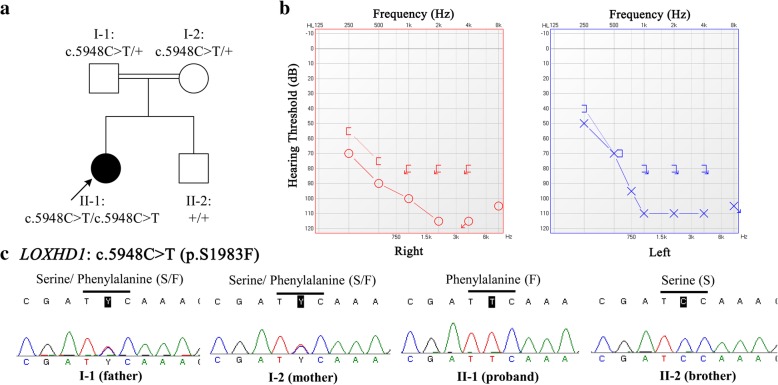
The pedigree was shown in Fig. 1a. The proband (II-1) was a 28-year-old woman, who suffered a profound HL without any syndromic phenotype. She demonstrated a bilateral hearing loss at all frequencies and predominantly at middle to high frequencies, based on pure tone audiometry (PTA) test. The pure tone averages of 500 Hz, 1000 Hz and 2000 Hz were 97 dB HL in her both ears (see Fig. 1b). Impedance audiometry exhibited a typical A-type tympanogram for each ear. Temporal bone computerized tomography (TBCT) scans and magnetic resonance imaging-inner ear hydrography (MRI-IEH) did not find any obvious abnormality of middle or inner ear. Other associated symptoms were also not observed in the proband (II-1), including vestibular disorders (dizziness, vertigo, etc.), optic problems (blurred or distorted vision, eye pain, etc.), mal-development and intellectual disability. According to information provided by the family, II-1 had congenital HL but did not find obvious progression these years. No hearing or associated symptoms were found in the proband’s parents (I-1 and I-2) or brother (II-2). Her parents had consanguineous marriage. No deafness history was found in the last three generations of their family.
Fig. 1.
Pedigree, audiological evaluation and Sanger sequencing validation. a. Pedigree of this Chinese family under consanguineous marriage. The proband was indicated by arrows. “+” indicates wild type. b. Pure-tone audiometry evaluation of this proband. c. LOXHD1: c.5948C > T variants were validated by Sanger sequencing
To identify the genetic cause of NSHL in this proband, we routinely applied a Sanger sequencing of four common HL-associated genes, including gap junction protein beta-2 (GJB2), gap junction protein beta-3 (GJB3), solute carrier family 26 member 4 (SLC26A4) and mitochondrially encoded 12S RNA (MT-RNR1). DNA preparation, PCR conditions and Sanger sequencing process were described previously [23]. The coding regions of GJB2 and GJB3, hotspot region (exon7–8 and exon19) of SLC26A4, and the full-length region of MT-RNR1 were carefully screened, only a homozygous variant, m.827A > G within MT-RNR1, was identified. However, previous studies reported conflicting interpretations of pathogenicity for this variant [24–26], which was insufficient to result in hearing impairment.
Therefore, we further performed a WES analysis for the trio (I-1, I-2 and II-1) by using the Illumina HiSeq platforms. Details of the process of WES analysis are shown in Additional file 1: Supplementary materials. The target mean depths in the trio were all greater than 128X and > 97.8% of the target regions were covered by at least 20X. More than 77 thousands of variants were annotated for each person, and we summarize these results in Additional file 1: Table S1. Two analyses were applied in the trio data. One was de novo variants analysis, but we found no deleterious HL-associated variant. The other was shared variants analysis. A promising variant within LOXHD1 (c. c.5948C > T; p.S1983F) was identified after rigorous filters (see Additional file 1: Tables S1 and S2). It was co-segregated and validated in this family by Sanger sequencing (see Fig. 1c). The primer sequences (5′ → 3′) were: forward-p, ATCGTGGTGCTTTTAACCTGC; reverse-p, GGGTGCTTGCACAGGATTG. Although homogeneous MT-RNR1: m.827A > G was identified in the proband, but her asymptomatic brother and mother also carrier this variant, implying that MT-RNR1: m.827A > G contributed little to the pathogenesis of the proband (Additional file 1: Table S3). LOXHD1: c.5948C > T was a missense variant, which was not found in all public databases (dbSNP, 1000 Genomes, ExAC and gnomAD), and predicted as damaging by multiple bioinformatics tools (SIFT, Polyphen2, and Mutation Taster, etc.). Evolution analysis also indicated that this variant was located at the well conserved region (Additional file 1: Table S2). Nowadays there have been 47 variants within LOXHD1 associated with hearing impairment according to HGMD database, but c.5948C > T (p.S1983F) was not reported previously.
Discussion and conclusions
NSHL is a complex disorder with highly genetic and clinical heterogeneity. Routinely, hotspot regions of four common HL-associated genes, such as GJB2, GJB3, SLC26A4 and MT-RNR1, are recommended to be initially detected for molecular diagnosis for NSHL. If results are negative, gene-panel sequencing or WES are applied for further detection. Specially, a trio-WES is quite suitable for those rare NSHL. DFNB77 is a rare NSHL with autosomal recessive inheritance, caused by homozygous mutations within LOXHD1 gene, firstly described in 2009 [7]. In the past ten years, about 60 variants within this gene were identified in NSHL cases. They showed different auditory characteristics, varying from stable to progressive and from mild to profound HL. The limited variant spectrum of LOXHD1 strongly requires more studies to fill in gaps in the genotype-phenotype correlations of DFNB77. In this study, we used a trio-WES to successfully identify a novel homozygous variant, c.5948C > T (p.S1983F), within LOXHD1 gene in a Chinese family. To the best of our knowledge, this is the second pedigree report of LOXHD1-related NSHL in China.
LOXHD1 encodes an important protein consisting of 15 PLAT domains, which mediates protein interactions to maintain normal hair cell function [7]. Deleterious variants within LOXHD1 could lead to various severities and various types of NSHL including progressive and non-progressive congenital HL [2, 7, 9–22]. Table 1 summarizes the published genotype-phenotype correlations of DFNB77 confirmed by segregation analysis. HL-associated variants within LOXHD1 could occur in various races. Homozygotes (c.71delT/c.71delT, c.1588G > T/c.1588G > T, c.4212 + 1G > A/c.4212 + 1G > A, etc.) appeared to show a trend toward severe or profound HL, and compound heterozygotes showed different HL phenotypes. No overlapping genotype was reported by these studies performing segregation analysis. The quite limited information hindered to explore more genotype-phenotype correlations, requiring more studies to uncover variant spectrum of LOXHD1 and related HL phenotype. Here, we identified a novel missense variant, LOXHD1: c.5948C > T, was associated with non-progressive NSHL in a family under consanguineous marriage. The proband carried homozygous c.5948C > T, her parents carried heterozygous c.5948C > T, and her brother did not carry this variant, which was compatible with the autosomal recessive inheritance of DFNB77. Comprehensive analyses, including family history, trio-WES, co-segregation validation, rarity in control population, and bioinformatics prediction, strongly support that LOXHD1: c.5948C > T could be a pathogenic factor. It makes effect on all the transcript isoforms of LOXHD1 gene: NM_144612:exon38:c.5948C > T: p.S1983F, NM_001145473:exon7:c.851C > T:p.S284F, NM_001173129:exon7:c.851C > T:p.S284F, NM_001308013:exon19:c.2513C > T:p.S838F, and NM_001145472:exon21:c.2801C > T:p.S934F. Variants within LOXHD1 are quite rare and recently, Hu et al. reported a first affected Chinese pedigree with progressive NSHL [2]. Compared to the compound heterozygotes (c.1751C > T/c.5815G > A) found by Hu et al., we identified a novel homozygote, c.5948C > T/ c.5948C > T, was associated with non-progressive NSHL. In addition, c.5948C > T is located in the 14th PLAT domain of LOXHD1 protein, which harbors the most published variants to date, compared with other PLAT domains [20]. Another five published variants (c.5869G > T, c.5885C > T, c.5934C > T, c.5944C > T and c.6162_6164delCCT) from different races are also concentrated in here [14, 18, 20], indicating that the 14th PLAT domain could be a hotspot mutated region of LOXHD1.
Table 1.
Genotype-phenotype correlation of DFNB77 confirmed by segregation analysis
| Genotype | Ethnicity | Severity of HL | Progression of HL | Reference |
|---|---|---|---|---|
| c.71delT/c.71delT | Turkish | Severe or profound | NA | [13] |
| c.442A > T/c.4217C > T | NA | NA | NA | [19] |
| c.1588G > T/c.1588G > T | Qatary | Severe to profound | Progressive | [12] |
| c.1618dup/c.1730 T > G | Dutch | Moderate to severe | Stable to progressive | [20] |
| c.1751C > T/c.5815G > A | Chinese | Severe | Progressive | [2] |
| c.1828G > T/c.2641G > A | Dutch | Mild | Stable | [20] |
| c.1904 T > C/c.4678 T > C | Dutch | Mild | Stable to progressive | [20] |
| c.2008C > T/c.2008C > T | Iranian | Mild to profound | Progressive | [7] |
| c.2696G > C/c.3834G > C | Dutch | Moderate | Stable | [20] |
| c.2696G > C/c.5934C > T | Dutch | Mild | NA | [20] |
| c.2863G > T/c.2863G > T | Turkish | NA | NA | [10] |
| c.3061C > T/c.5885C > T | Indian | Severe | Stable | [20] |
| c.3061 + 1G > A/c.6353G > A | Dutch | Moderate | NA | [20] |
| c.3076G > T/c.4375 + 1G > T | Japanese | Profound | Stable | [17] |
| c.3169C > T/c.6353G > A | Dutch | Severe | Stable | [20] |
| c.3371G > A/c.3979 T > A | Cameroonian | Profound | NA | [15] |
| c.3748 + 1G > C/c.6353G > A | Dutch | Moderate to severe | Stable to progressive | [20] |
| c.4212 + 1G > A/c.4212 + 1G > A | Japanese | Profound | Stable | [14] |
| c.4212 + 1G > A/c.5674G > T | Japanese | Mild to profound | Progressive | [16] |
| c.4480C > T/c.4480C > T | Turkish | NA | NA | [10] |
| c.4480C > T/c.5869G > T | Japanese | Moderate to severe | Stable | [14] |
| c.4623C > G/c.5545G > A | Czech | Severe | NA | [22] |
| c.4714C > T/c.4714C > T | Ashkenazi Jewish | Severe to profound | NA | [9] |
| c.5894dupG/c.5894dupG | Arab | Profound | NA | [21] |
| c.5948C > T/c.5948C > T | Chinese | Profound | Stable | This study |
Abbreviation: NA not available
Variants within LOXHD1 were also linked to late-onset FCD, a genetic degenerative disease of corneal endothelium towards blindness. In 2012, Riazuddin, et al. first reported a heterozygous damaging variant within LOXHD1 in a multiplex family with dominant-inherited late-onset FCD [27]. However, subsequent studies failed to provide a strong association between LOXHD1 variants and FCD [20, 28–30]. Specially, results from a Chinese multi-generational FCD pedigree demonstrated that no pathogenic variants were identified in LOXHD1 [28]. In line with these previous studies, our work also did not observed any symptoms of FCD in the proband and her blood relatives within three generations. However, a limitation of our study is that the identified missense variant is lacking in animal models or in the verification of other HL patients. More functional and population studies are required to further verify our results.
In summary, we demonstrated that a novel missense variant, LOXHD1: c.5948C > T, was associated with non-progressive NSHL in a Chinese family under consanguineous marriage. Our work highlights the effectiveness of trio-WES for molecular diagnosis of rare NHSL and expands the variant spectrum of LOXHD1 in hearing impairment.
Additional file
Supplementary Materials and Tables. (a) The process of whole-exome sequencing (WES) analysis. (b) Table S1. Filtering process of WES analysis in our study. (c) Table S2. Candidate gene and variant identified by trio-WES. (d) Table S3. Variants validated by Sanger sequencing. (DOC 59 kb)
Acknowledgements
We would like to thank the patient and her family for their participation in this study. We also would like to thank the editors and reviewers of our manuscript for their constructive comments that improved its quality.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81271072) in the study design, sample collection and experimental analysis.
Availability of data and materials
Data are available from the corresponding author on reasonable request.
Abbreviations
- DFNB77
Deafness, autosomal recessive 77
- FCD
Fuchs corneal dystrophy
- GJB2
Gap junction protein beta-2
- GJB3
Gap junction protein beta-3
- HL
Hearing loss
- LOXHD1
Lipoxygenase homology domains 1 l
- MT-RNR1
Mitochondrially encoded 12S RNA
- NSHL
Non-syndromic hearing loss
- SLC26A4
Solute carrier family 26 member 4
- WES
Whole-exome sequencing
Authors’ contributions
YL and AL conceived the study and revised this paper. TW and DL conducted experiments. NS analyzed and interpreted the data, and was a major contributor in writing this paper. YL and AL cared the patient and performed medical examinations. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. Written informed consent were obtained from all subjects.
Consent for publication
All subjects have given their written informed consents for publication of the medical information.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Na Shen, Email: shenna0920@163.com.
Ting Wang, Email: wtingwy@163.com.
Delei Li, Email: 284464659@qq.com.
Aiguo Liu, Email: aiguoliu309@163.com.
Yanjun Lu, Email: junyanlu_2000@163.com.
References
- 1.Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354(20):2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Hu S, Sun F, Zhang J, Tang Y, Qiu J, Wang Z, Zhang L. Genetic etiology study of ten Chinese families with nonsyndromic hearing loss. Neural plasticity. 2018;2018:4920980. doi: 10.1155/2018/4920980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 4.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 5.Zazo Seco C, Wesdorp M, Feenstra I, Pfundt R, Hehir-Kwa JY, Lelieveld SH, Castelein S, Gilissen C, de Wijs IJ, Admiraal RJ, et al. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in the Netherlands. European journal of human genetics : EJHG. 2017;25(3):308–314. doi: 10.1038/ejhg.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bademci G, Foster J, 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, Menendez I, Diaz-Horta O, Shirkavand A, Zeinali S, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med. 2016;18(4):364–371. doi: 10.1038/gim.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grillet N, Schwander M, Hildebrand MS, Sczaniecka A, Kolatkar A, Velasco J, Webster JA, Kahrizi K, Najmabadi H, Kimberling WJ, et al. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet. 2009;85(3):328–337. doi: 10.1016/j.ajhg.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateman A, Sandford R. The PLAT domain: a new piece in the PKD1 puzzle. Curr Biol :CB. 1999;9(16):R588–R590. doi: 10.1016/S0960-9822(99)80380-7. [DOI] [PubMed] [Google Scholar]
- 9.Edvardson S, Jalas C, Shaag A, Zenvirt S, Landau C, Lerer I, Elpeleg O. A deleterious mutation in the LOXHD1 gene causes autosomal recessive hearing loss in Ashkenazi Jews. Am J Med Genet A. 2011;155A(s):1170–1172. doi: 10.1002/ajmg.a.33972. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Horta O, Duman D, Foster J, 2nd, Sirmaci A, Gonzalez M, Mahdieh N, Fotouhi N, Bonyadi M, Cengiz FB, Menendez I, et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One. 2012;7(11):e50628. doi: 10.1371/journal.pone.0050628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eppsteiner RW, Shearer AE, Hildebrand MS, Deluca AP, Ji H, Dunn CC, Black-Ziegelbein EA, Casavant TL, Braun TA, Scheetz TE, et al. Prediction of cochlear implant performance by genetic mutation: the spiral ganglion hypothesis. Hear Res. 2012;292(1–2):51–58. doi: 10.1016/j.heares.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vozzi D, Morgan A, Vuckovic D, D'Eustacchio A, Abdulhadi K, Rubinato E, Badii R, Gasparini P, Girotto G. Hereditary hearing loss: a 96 gene targeted sequencing protocol reveals novel alleles in a series of Italian and Qatari patients. Gene. 2014;542(2):209–216. doi: 10.1016/j.gene.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Atik T, Onay H, Aykut A, Bademci G, Kirazli T, Tekin M, Ozkinay F. Comprehensive analysis of deafness genes in families with autosomal recessive nonsyndromic hearing loss. PLoS One. 2015;10(11):e0142154. doi: 10.1371/journal.pone.0142154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori K, Moteki H, Kobayashi Y, Azaiez H, Booth KT, Nishio SY, Sato H, Smith RJ, Usami S. Mutations in LOXHD1 gene cause various types and severities of hearing loss. Ann Otol Rhinol Laryngol. 2015;124(Suppl 1):135S–141S. doi: 10.1177/0003489415574067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebeko K, Sloan-Heggen CM, Noubiap JJ, Dandara C, Kolbe DL, Ephraim SS, Booth KT, Azaiez H, Santos-Cortez RL, Leal SM, et al. Targeted genomic enrichment and massively parallel sequencing identifies novel nonsyndromic hearing impairment pathogenic variants in Cameroonian families. Clin Genet. 2016;90(3):288–290. doi: 10.1111/cge.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minami SB, Mutai H, Namba K, Sakamoto H, Matsunaga T. Clinical characteristics of a Japanese family with hearing loss accompanied by compound heterozygous mutations in LOXHD1. Auris Nasus Larynx. 2016;43(6):609–613. doi: 10.1016/j.anl.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Sakuma N, Moteki H, Takahashi M, Nishio SY, Arai Y, Yamashita Y, Oridate N, Usami S. An effective screening strategy for deafness in combination with a next-generation sequencing platform: a consecutive analysis. J Hum Genet. 2016;61(3):253–261. doi: 10.1038/jhg.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, Ephraim SS, Shibata SB, Booth KT, Campbell CA, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135(4):441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, Walkiewicz M, Bi W, Xiao R, Ding Y, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376(1):21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wesdorp M, Schreur V, Beynon AJ, Oostrik J, van de Kamp JM, Elting MW, van den Boogaard MH, Feenstra I, Admiraal RJC, Kunst HPM, et al. Further audiovestibular characterization of DFNB77, caused by deleterious variants in LOXHD1, and investigation into the involvement of Fuchs corneal dystrophy. Clin Genet. 2018;94(2):221–231. doi: 10.1111/cge.13368. [DOI] [PubMed] [Google Scholar]
- 21.Danial-Farran N, Brownstein Z, Gulsuner S, Tammer L, Khayat M, Aleme O, Chervinsky E, Zoubi OA, Walsh T, Ast G, et al. Genetics of hearing loss in the Arab population of northern Israel. Eur J Hum Genet. 2018. [DOI] [PMC free article] [PubMed]
- 22.Plevova P, Paprskarova M, Tvrda P, Turska P, Slavkovsky R, Mrazkova E. STRC deletion is a frequent cause of slight to moderate congenital hearing impairment in the Czech Republic. Otol Neurotol. 2017;38(10):e393–e400. doi: 10.1097/MAO.0000000000001571. [DOI] [PubMed] [Google Scholar]
- 23.Shen N, Peng J, Wang X, Zhu Y, Liu W, Liu A, Lu Y. Association between the p.V37I variant of GJB2 and hearing loss: a pedigree and meta-analysis. Oncotarget. 2017;8(28):46681–46690. doi: 10.18632/oncotarget.17325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing G, Chen Z, Wei Q, Tian H, Li X, Zhou A, Bu X, Cao X. Maternally inherited non-syndromic hearing loss associated with mitochondrial 12S rRNA A827G mutation in a Chinese family. Biochem Biophys Res Commun. 2006;344(4):1253–1257. doi: 10.1016/j.bbrc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Xing G, Chen Z, Wei Q, Tian H, Li X, Zhou A, Bu X, Cao X. Mitochondrial 12S rRNA A827G mutation is involved in the genetic susceptibility to aminoglycoside ototoxicity. Biochem Biophys Res Commun. 2006;346(4):1131–1135. doi: 10.1016/j.bbrc.2006.05.208. [DOI] [PubMed] [Google Scholar]
- 26.Chaig MR, Zernotti ME, Soria NW, Romero OF, Romero MF, Gerez NM. A mutation in mitochondrial 12S rRNA, A827G, in Argentinean family with hearing loss after aminoglycoside treatment. Biochem Biophys Res Commun. 2008;368(3):631–636. doi: 10.1016/j.bbrc.2008.01.143. [DOI] [PubMed] [Google Scholar]
- 27.Riazuddin SA, Parker DS, McGlumphy EJ, Oh EC, Iliff BW, Schmedt T, Jurkunas U, Schleif R, Katsanis N, Gottsch JD. Mutations in LOXHD1, a recessive-deafness locus, cause dominant late-onset Fuchs corneal dystrophy. Am J Hum Genet. 2012;90(3):533–539. doi: 10.1016/j.ajhg.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang H, Zhang W, Yan XM, Wang LP, Dong H, Shou T, Lei H, Guo Q. Analysis of SLC4A11, ZEB1, LOXHD1, COL8A2 and TCF4 gene sequences in a multi-generational family with late-onset Fuchs corneal dystrophy. Int J Mol Med. 2016;37(6):1487–1500. doi: 10.3892/ijmm.2016.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skorodumova LO, Belodedova AV, Antonova OP, Sharova EI, Akopian TA, Selezneva OV, Kostryukova ES, Malyugin BE. CTG18.1 expansion is the best classifier of late-onset Fuchs' corneal dystrophy among 10 biomarkers in a cohort from the European part of Russia. Invest Ophthalmol Vis Sci. 2018;59(11):4748–4754. doi: 10.1167/iovs.18-24590. [DOI] [PubMed] [Google Scholar]
- 30.Rao BS, Ansar S, Arokiasamy T, Sudhir RR, Umashankar V, Rajagopal R, Soumittra N. Analysis of candidate genes ZEB1 and LOXHD1 in late-onset Fuchs' endothelial corneal dystrophy in an Indian cohort. Ophthalmic Genet. 2018;39(4):443–449. doi: 10.1080/13816810.2018.1474367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Tables. (a) The process of whole-exome sequencing (WES) analysis. (b) Table S1. Filtering process of WES analysis in our study. (c) Table S2. Candidate gene and variant identified by trio-WES. (d) Table S3. Variants validated by Sanger sequencing. (DOC 59 kb)
Data Availability Statement
Data are available from the corresponding author on reasonable request.