Abstract
Background
Neuroblastoma is the most common extracranial solid tumor in children. This cancer has a low frequency of TP53 mutations and its downstream pathway is usually intact. This study assessed the efficacy of the p53 activator, PRIMA-1MET, in inducing neuroblastoma cell death.
Methods
CellTiter 2.0 was used to study susceptibility and specificity of NB cell lines to PRIMA-1MET. Real-time PCR and western blot were used to assess the most common p53 transactivation targets. Induction of p53 and Noxa, and inhibition of Cas3/7, were used to assess impact on cell death after PRIMA-1MET treatment. Flow cytometry was used to analyze cell cycle phase and induction of apoptosis, reactive oxygen species, and the collapse of mitochondrial membrane potential.
Results
Neuroblastoma cell lines were at least four times more susceptible to PRIMA-1MET than were primary fibroblasts and keratinocyte cell lines. PRIMA-1MET induced cell death rapidly and in all cell cycle phases. Although PRIMA-1MET activated p53 transactivation activity, p53’s role is likely limited because its main targets remained unaffected, whereas pan-caspase inhibitor demonstrated no ability to prevent cell death. PRIMA-1MET induced oxidative stress and modulated the methionine/cysteine/glutathione axis. Variations of MYCN and p53 modulated intracellular levels of GSH and resulted in increased/decreased sensitivity of PRIMA-1MET. PRIMA-1MET inhibited thioredoxin reductase, but the effect of PRIMA-1MET was not altered by thioredoxin inhibition.
Conclusions
PRIMA-1MET could be a promising new agent to treat neuroblastoma because it demonstrated good anti-tumor action. Although p53 is involved in PRIMA-1MET-mediated cell death, our results suggest that direct interaction with p53 has a limited role in neuroblastoma but rather acts through modulation of GSH levels.
Electronic supplementary material
The online version of this article (10.1186/s13046-019-1066-6) contains supplementary material, which is available to authorized users.
Keywords: Neuroblastoma, PRIMA-1MET, p53, MYCN, Glutathione
Background
Neuroblastoma (NB) is the most common extracranial solid tumor in children. Current event-free survival rates range from 75% to more than 85% for low- and very low-risk groups, to less than 50% for high-risk patients despite dose-intensive therapy [1, 2]. NB progression to an advanced stage and poorer overall survival is characterized by specific molecular events, the most common of which are MYCN amplification (MNA) [2, 3] and 11q deletion [4]. NB show a low rate of point mutations, and predominant events leading to tumor progression are chromosomal rearrangements due to apparent chromosomal instabilities [5–8]. Fifty percent of all human cancers contain mutation in the tumor suppressor gene TP53, and some estimations contend that almost all cancers evolve a way to circumvent the p53 pathway [9]. NB has been suggested as a good candidate for reactivation of the p53 pathway because it has a low frequency of mutations in TP53 [10, 11]. The downstream pathway is usually intact, with most of the mutations appearing to be in the upstream MDM2-p14(ARF)-p53 network [12]. Nutlin-3 and its cis-imidazoline analogues activate p53 by inhibiting p53-MDM2 interaction. Preclinical investigation on NB cell lines was encouraging, demonstrating good responses in vitro [11, 13]. In vivo studies in mice suggest that MDM2 inhibitors could be well-tolerated [14]. Clinical trials in liposarcoma patients using Nutlin-3 analogues did not prove effective, however, and revealed an association with severe thrombocytopenia and neutropenia [15]. In addition, resistance can readily develop in cancer cells exposed to selection pressure by selecting cells with TP53 mutation, which dramatically reduces the efficacy of Nutlin-3 [16].
A new group of molecules that are able to directly activate mutated p53 was recently developed [17, 18]. The most promising, PRIMA-1MET, is currently being investigated in several early-stage adult clinical trials (NCT02098343, NCT02999893, NCT03072043, NCT03588078, NCT03745716, NTC03391050, NTC03268382 and NTC00900614). In vivo, PRIMA-1MET is converted into the active compound methylene quinuclidinone (MQ), which reacts with the thiol group of cysteine in proteins. Studies by Lambert et al demonstrated that PRIMA-1MET binds to p53, thus restoring p53 function by refolding the protein in its native structure [18]. In vitro cells and in vivo mouse studies on various cell lines suggest good efficacy of PRIMA-1MET on adenocarcinoma and non-small cell lung cancer [19, 20], colorectal cancer [21], glioblastoma [22], multiple myeloma [23, 24], acute myeloid leukemia [25], breast cancer [26], and ovarian cancer [27] cell lines. Interestingly, depending on the cancer type, PRIMA-1MET induced death was not always p53 dependent. Different off-target effects involving ROS toxicity or autophagy were reported (recently reviewed by Perdrix et al [28]).
This study aimed to evaluate the efficacy of PRIMA-1MET in NB cell lines and to explore the roles of p53, MYCN, glutathione (GSH) and thioredoxin (TXN) systems in PRIMA-1MET efficacy and cellular response to PRIMA-1MET.
Methods
Cell lines and chemicals
The NB cell lines CHP212, LAN6, NBL-S, NGP, SK-N-DZ and SK-N-SH were provided by Dr. E. Attiyeh and Prof. J. Maris (Children’s Hospital of Philadelphia, Philadelphia, USA). The CLB-GA NB cell line was provided by Dr. V. Combaret (Centre de Ressources Biologiques du Centre Léon Bérard, Lyon, France). BE-(2)C, LA1–55 N, and SK-N-DZ were purchased from ATCC (USA). All NB cell lines were maintained in a standard NB medium composed of DMEM supplemented with 10% FBS, 1% antibiotic/antimycotic solution, and 1% L-glutamine. All NB cell lines passed identity and mycoplasma testing performed independently by Microsynth AG (Switzerland). Human normal primary keratinocytes and fibroblasts (LGC, Germany) were maintained in a dermal cell basal medium supplemented with keratinocyte growth kit and low serum fibroblast basal medium, respectively, prepared according to the manufacturer’s recommendations (LGC, Germany). LCL (lymphoblastoid cell lines, LGC, Germany) were maintained in RPMI 1640 supplemented with 10% FBS and 1% antibiotic/antimycotic solution according to manufacturer’s recommendations. The following compounds were used: PRIMA-1MET (50 mM in H2O, Abcam, UK), pan-caspase inhibitor Z-VAD-FMK (25 mM in H2O, LubioScience, Switzerland), etoposide (42,4 mM in DMSO), glutathione (100 mM in H2O), JQ-1 (1 mM in H2O), L-cysteine (50 mM in H2O), L-homocysteine (100 mM in H2O), L-methionine (12 g/L in H2O), N-acetyl-cysteine (100 mM in H2O), orlistat (20 mM in DMSO) (Sigma-Aldrich, Germany), buthionine sulfoximine (BSO, 14.4 mM in H2O, TRC, Canada). A stably expressing p53 LA1–55 N cell line was obtained by transfection of pcDNA3, containing the p53 gene, using X-tremeGENE HP DNA Transfection Reagent (Roche, Switzerland) and subsequent selection with G418 (Promega, USA) according to the manufacturer’s recommendations. Concentrations of PRIMA-1MET were adjusted for each cell line to obtain precise 50% inhibitory concentration (IC50) values. Vehicle control was used in all experiments.
Cell survival assay
CellTiter 2.0 (Promega, USA) was used according to the manufacturer’s recommendations to measure the number of live cells. 100 μL of CellTiter 2.0 was added to each well containing 7500 cells treated with PRIMA-1MET for 24 h in 100 μL of medium. Plates were incubated for 2 min on a horizontal shaker at medium speed and at room temperature. Chemiluminescence was measured using Victor3 (Thermo Fisher Scientific, USA) for 1 s, after shaking at medium speed and without the use of filters. Real-time cell viability after treatment with PRIMA-1MET or etoposide and inhibition of caspase by pan-caspase inhibitor was followed using RealTime-Glo MT Cell Viability Assay (G9711, Promega, Madison, USA) according to the manufacturer’s recommendations. Chemiluminescence was measured using SpectraMax iD3 (Molecular Devices, USA) every 10 min for 22 h at 37 °C.
Flow cytometry
CyAn ADP flow cytometer (Beckman Coulter, USA) was used with the following kits according to manufacturers’ recommendations: FITC Annexin V Apoptosis Detection Kit I (BD Pharmingen, USA), CellROX Oxidative Stress Reagent (Molecular Probes, USA), Autophagy detection kit (Abcam, UK) and mitochondrial JC-1 dye (Invitrogen, USA). Cell cycle, induction of caspase 3 and 7 (Cas3/7), and cellular membrane stability were investigated simultaneously using the FORTESSA flow cytometer system (BD Biosciences, USA). Results were analyzed using Kaluza software (Beckman Coulter, USA).
In-cell Western blots and Western blots
The intracellular concentration of proteins was measured using CellTag 700 Stain and In-Cell Western (ICW) Assay Kit 1 (LI-COR, USA) according to the manufacturer’s recommendations, 6 h after exposure to 60 μM PRIMA-1MET. The following primary mouse antibodies were used for protein labelling. Abcam (UK): Anti-ATM (ab-78), Anti-TXNRD1 (ab16847), Anti-Bax (ab77566) and Anti-Actinβ (ab6276). Cell Signaling Technology (USA): Phospho-p53 (Ser-15) (16G8) (9286), Phospho-ATM (Ser1981) (4526). Santa Cruz (USA): Anti-p53 (sc-126), Anti-p21 (sc-53,870). Antibody specificity was assessed using WB as described in our previous publication [29]. Etoposide, a known inducer of the ATM/p53 pathway [29], was used as a positive control.
Real-time PCR and gene expression
Real-time PCR was used to measure the expression of p21, 14–3-3 (YWHAQ), GADD45, PUMA (BBC3), Bax, MDM2, Noxa (PMAIP1), CXCR4, and NMYC. GAPDH, EEF1A1, and TBP were used as a normalization control (Additional file 2: Table S1). PrimeScript Reverse Transcriptase enzyme (Takara Bio Inc., Japan) and 2x Power SYBR Green Master Mix (Applied Biosystems, USA) was used for generation of cDNA and real-time PCR according to the manufacturer’s recommendations. PCR was performed on an SDS 7900HT instrument (Applied Biosystems, USA). Raw Ct values obtained with SDS 2.2 (Applied Biosystems, USA) were imported in Excel and normalization factor and fold changes were calculated using the GeNorm method [30].
Luciferase reporter assay
pGL10.4 (Promega, USA) was used to study the impact of PRIMA-1MET on the Noxa promoter region. Three plasmids containing NOXA promoter region from + 198 to − 157 were designed. In two of the plasmids, additional mutations were inserted in p53 binding site and CREB binding site. All plasmids were prepared by GenScript (USA). The pRL-SV40 Renilla luciferase control reporter vector (Promega, USA) was used as a normalization control at a 1:10 weight ratio. Plasmids were transfected into SK-N-SH cells using X-tremeGENE HP DNA Transfection Reagent (Roche, Switzerland) according to manufacturer’s recommendations. The Dual-Luciferase Reporter Assay System (Promega, USA) and Victor3 (Thermo Fisher Scientific, USA) were used to measure luciferase activity (5 s shaking at medium speed prior to 1 s measurement time without the use of filters).
Cysteine, glutathione and S-adenozyl-homocysteine concentration, glutathione transferase activity and thioredoxin reductase activity assay
The concentration of total intracellular glutathione (GSH-Glo Glutathione Assay, Promega, USA), cysteine (Cysteine Assay Kit (Fluorometric), Abcam, UK), S-adenozyl-homocysteine (S-Adenosylhomocysteine (SAH) ELISA Combo Kit, Cell Biolabs, USA), glutathione transferase activity (Glutathione S-Transferase (GST) Assay Kit, Sigma Aldrich, USA) and the activity of thioredoxin reductase (Thioredoxin Reductase (TrxR) Assay Kit, Abcam, UK) was measured according to the manufacturer’s recommendations. The Victor3 reader and Versamax microplate reader (Molecular Devices, USA) were used to measure luminescence and absorbance (412 nm), respectively.
Whole-exome sequencing and bioinformatics
The Illumina HiSeq 2500 sequencer (Illumina, USA) was used for whole exome sequencing according to manufacturer’s recommendations. Illumina paired-end reads were aligned to the human UCSC hg19 reference sequence using BWA software (version 0.7.10). Single-nucleotide variants were called with GATK (version 3.4.46) and annotated with Annovar (version 2016 Feb 01) and SnpEff (version 4.3). Ingenuity variant analysis (QIAgen, Germany) was used to predict the number of cancer driver mutations and to search for ALK and TP53 variants. Affymetrix expression data from GSE3960 were normalized using Robust Multichip Average (RMA) and was used to investigate the association between MNA and expression of genes in the TXN and GSH pathways.
Statistics
ANOVA p-values were used to determine significant differences in expression levels between MNA and non-MNA samples. A t-test was used to determine association of MNA and difference in expression of GCLM versus GCLC genes. A two-way t-test was used to calculate statistical significance for cell-based experiments. Robust nonlinear fit of dose vs. response method (GraphPad Prism 7, USA) was used to calculate the IC50 from relative cell survival, as determined by using CellTiter2.0. The coefficient of drug interaction (CDI) was calculated according to the following formula: CDI = AB/(AxB), where AB represents the relative cell viability for the combination of drugs and A and B represent the relative viability for each compound alone. CDI less than 0.8 was considered indicative of synergy. CDI between 0.8 and 1.2 as additive, while CDI more than 1.2 indicated antagonism [31]. Activity of glutathione S - transferase was calculated using GraphPad Prism 7. If not otherwise noted, measurements were taken in at least triplicate in each experiment, and at least two independent experiments were performed.
Results
Genetic characteristics of the eight neuroblastoma cell lines used in this study
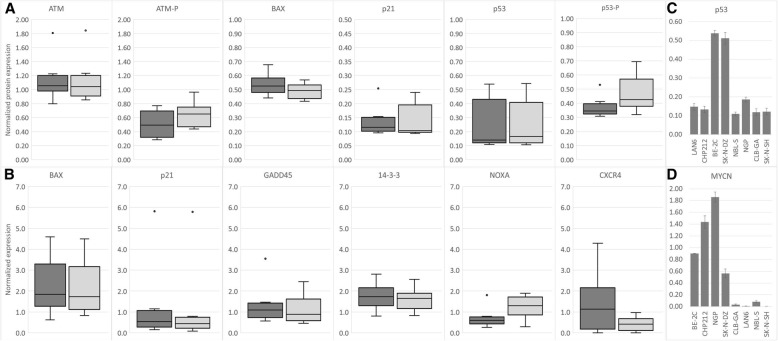
The eight NB cell lines used in this study were analyzed for their genetic features (Table 1). MYCN amplification and 11q-deletion were detected in four cell lines each. These two features overlap in the cell lines NGP and SK-N-DZ, which were found to have both MNA and 11q-deletion. MNA and 11q-deletion were assessed using real-time PCR (Fig. 3d) and loss of heterozygosity, respectively. The results concur with previous publications, ours and others’ [29, 32, 33]. Mutations in cancer driver genes were detected with whole exome sequencing. The NB cell lines contained an average of 783 cancer-driving variants in an average of 488 different genes (Additional file 1). Three of the cell lines (CLB-GA, LAN6, and SK-N-SH) carry ALK variants; the activating mutations F1174 L and R1275Q have been demonstrated to be oncogenic in an NB mouse model [34]. The NGP cell line exhibits high expression of MDM2 as assessed by real-time PCR. TP53 mutations were found in three NB cell lines (BE-2C, NGP, and SK-N-DZ). Mutation R110L, found in SK-N-DZ, has been reported in patients with Li-Fraumeni syndrome and results in high expression and abnormal localization of p53, as well as impairment of proper p53 oligomerization [35]. Our results confirm this observation, as SK-N-DZ exhibited high baseline p53 expression (Fig. 3c). The TP53 mutations found in the NGP cell line, A159D (COSM11496) and C141W (COSM44204), are both deleterious mutations leading to amino acid substitution. No loss of heterozygosity at the TP53 locus was detected in NGP. This last result suggests that the NGP cell line has no gross deletions at this locus, but it is unknown whether both alleles are inactivated. BE-2C carries the previously described homozygous C135F mutation and has a loss of chromosome 17 at the site of TP53 [36]. Interestingly, in the three cell lines carrying one or more TP53 variants, MYCN was found to be amplified (Table 1).
Table 1.
Characteristics of the neuroblastoma cell lines used in this study
| Cell line | MYCN a | 11qb | TP53 status and p53 expressionc | MDM2 expressiond | ALK status c | Number of cancer driver variants (genes) | Reference |
|---|---|---|---|---|---|---|---|
| BE-2C | amp | no del | C135Fe High expression |
– | wt | 908 (629) | [29, 50, 51] |
| CHP212 | amp | no del | wt | – | wt | 619 (354) | [29, 50] |
| CLB-GA | wt | no del | wt | – | R1275Q (rs113994087) | 715 (394) | [29, 32] |
| LAN6 | wt | del | wt | – | D1091N (rs864309584) | 754 (444) | [11, 29, 51] |
| NBL-S | wt | del | wt | – | wt | 755 (434) | [29, 50, 51] |
| NGP | amp | del | A159Df C141Wf |
High expression | wt | 1120 (810) | [29, 50] |
| SK-N-DZ | amp | del | R110Le (rs11540654) High expression |
– | wt | 733 (453) | [29, 50] |
| SK-N-SH | wt | no del | wt | – | F1174 L (rs863225281) | 660 (388) | [29, 50] |
amp amplified, del deletion, no del no deletion, wt wild type
aResults obtained from the literature and confirmed by real-time PCR expression of MYCN
bResults verified with analysis of heterozygosity and/or Fish analysis
cResults obtained by whole-exome sequencing and confirmed by the literature
dResults obtained by real-time PCR and confirmed by the literature
eHomozygous mutation/variant
fHeterozygous mutation/variant
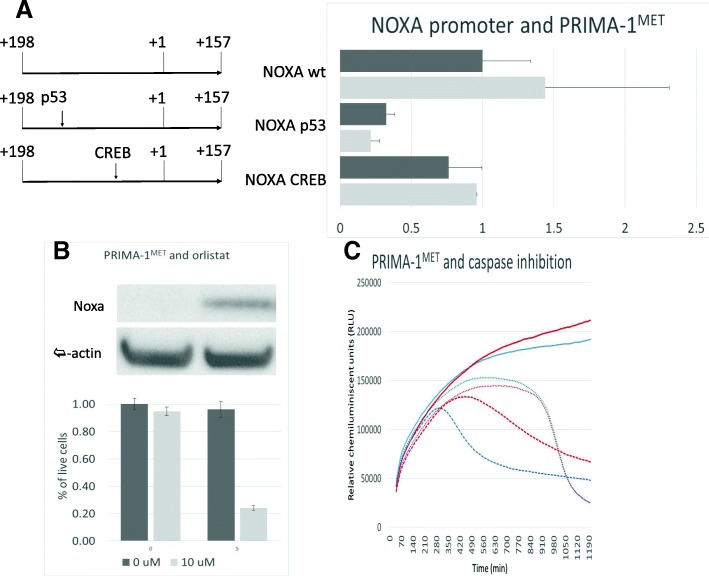
Fig. 3.
Protein and gene expression after treatment with PRIMA-1MET. a: Results of protein expression measured by ICW demonstrate that 6 h of 60 μM PRIMA-1MET does not induce phosphorylation of ATM or p53 in NB cell lines (dark grey: non-treated vehicle control, light grey: PRIMA-1MET, dots: outliers). There is no significant upregulation of p21 or Bax due to PRIMA-1MET treatment. b: Results of gene expression measured by real-time PCR demonstrate that major p53 target genes involved in cell cycle arrest (14–3-3, Gadd45, p21) remain unchanged (dark grey: non-treated vehicle control, light grey: PRIMA-1MET, dots: outliers). Bax is involved in signaling apoptosis and did not show any changes in levels as measured by ICW (A) or real-time PCR (B). Noxa was consistently and significantly upregulated and showed on average a 2-fold increase. CXCR4, a marker of aggressive NB, was downregulated after PRIMA-1MET treatment. c: BE-2C and SK-N-DZ (both MNA cell lines) showed accumulation of TP53 mRNA. d: BE-2C, CHP212, NGP and SK-N-DZ have high expression of MYCN, which is in accordance with the previously-described presence of MNA in these cell lines
PRIMA-1MET is a potent inducer of neuroblastoma cell death
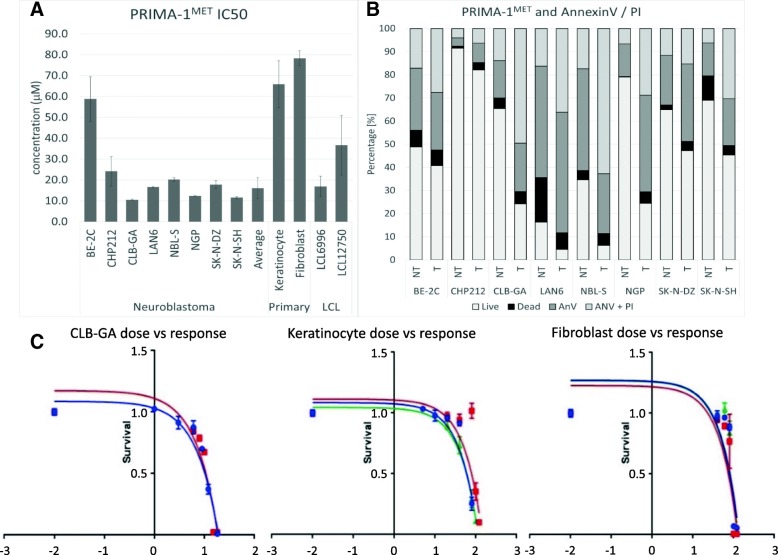
To investigate the ability of PRIMA-1MET to induce NB cell death, eight NB cell lines were tested to determine IC50 values (Fig. 1a). Seven of the eight cell lines exhibited cell death at a low PRIMA-1MET concentration (mean 16.1 + − 5.02 μM); BE-2C was resistant to high PRIMA-1MET concentrations (58.8 + − 10.72 μM). Next, the cell lines were compared for IC50 and presence of 11q-deletion and MNA, the two most prominent molecular events leading to highly aggressive clinical behavior. No significant difference in IC50 was observed in the subgroupings by these features. However, the two most resistant cell lines (BE-2C and CHP212) have MNA, while the two most sensitive (CLB-GA and SK-N-SH) have normal MYCN expression (Fig. 1a). NGP and SK-N-DZ, carrying heterozygous A159D and C141W, and homozygous R110L, respectively, do not differ in IC50 in comparison to the average NB cell line, suggesting these mutations do not affect PRIMA-1MET activity. The BE-2C cell line contains a homozygous substitution of cysteine 135 for phenylalanine, suggesting that C135F could be involved in BE-2C’s increased resistance to PRIMA-1MET. To understand whether p53 was associated with IC50 the LA1–55 N cell line, which lacks expression of p53, was transfected with pcDNA3-p53 plasmid. Stably expressing p53 LA1–55 N cells (Fig. 6b) had 1.39-fold higher IC50 values than control LA1–55 N cells transfected with empty pcDNA3 plasmid. IC50 was also investigated in four non-NB cell lines (2 primary and 2 lymphoblast) in order to understand the specificity of PRIMA-1MET to tumor cell lines. Primary keratinocytes and primary fibroblasts were 4.1 times and 4.9 times more resistant, respectively, than the average of the seven sensitive NB cell lines (Fig. 1a). Two immortalized peripheral lymphoblast cell lines (LCL) exhibited significantly lower PRIMA-1MET resistance than both primary cell lines (Fig. 1a). LCL6996 did not demonstrate any significant difference in resistance compared to the average of the NB cells, while LCL12750 was 2.17 times more resistant than LCL6996 (Fig. 1a). This result suggests that Epstein-Barr immortalization, as well as normal population polymorphisms, could affect resistance to PRIMA-1MET. Analyzing the dose-response relationship demonstrated that PRIMA-1MET exhibits rapid killing and a narrow activation interval (Fig. 1c). Once the threshold concentration is exceeded, complete cell death is observed within 15 h (Additional file 2: Data for Figure S1).
Fig. 1.
PRIMA-1MET IC50 values. a: PRIMA-1MET IC50 values. Eight NB cell lines were tested for PRIMA-1MET IC50 values: BE-2C (58.8 + − 10.72 μM), CHP212 (24.2 + − 7.02 μM), CLB-GA (10.5 + − 0.34 μM), LAN6 (16.5 μM + − 0.48), NBL-S (20.1 + − 0.80 μM), NGP (12.3 + − 0.27 μM), SK-N-DZ (17.8 + − 1.95 μM) and SK-N-SH (11.6 + − 0.55 μM). The average IC50 (excluding resistant BE-2C) for NB cell lines was 16.1 + − 4.99 μM. The PRIMA-1MET IC50 values for the non-tumor cells were: 65.8 + − 11.2 μM (keratinocytes), 78.3 + − 3.69 μM (fibroblasts), 16.9 + − 4.96 μM (LCL6996) and 36.6 + − 14.32 μM (LCL12750). No significant impact of MNA or 11q status on IC50 was observed. b: Graph shows increase in dead (black), early apoptotic (dark grey), and late apoptotic (light grey) cells and decrease in live cells (white) 16 h after PRIMA-1MET treatment at IC50 concentration. Treatment with PRIMA-1MET results in significant overall increase in the three populations (dead, early, and late apoptosis) combined and in AnnexinV-positive cells (early and late apoptosis), demonstrating activation of apoptosis after PRIMA-1MET. NT – non-treated vehicle control, T – treatment with PRIMA-1MET. c: The graph shows a non-linear regression fitting for IC50 calculation (BE-2C, keratinocytes, and fibroblasts). The steepness of the curve illustrates PRIMA-1MET’s narrow activation window
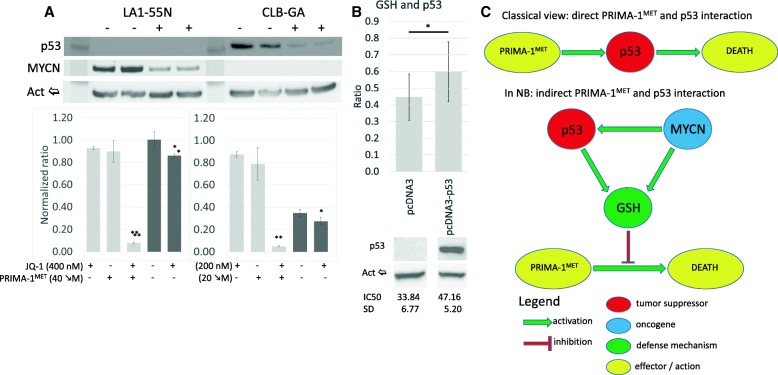
Fig. 6.
PRIMA-1MET, p53 and MYCN. a: Inhibition of MYCN and p53 expression by JQ-1. LA1–55 N (p53-null) cells showed a significant decrease in MYCN, whereas CLB-GA (non-MNA) cells demonstrated almost complete suppression of p53 expression after 16 h treatment. - – Non-treatment control, + − 16 h, 200 nM JQ-1. PRIMA-1MET and JQ-1 demonstrated synergism in both cell lines (CDILA1–55N = 0.1, CDICLB-GA = 0.07). Treatment with JQ-1 resulted in a significant decrease in the concentration of GSH in LA1–55 N (1.163-fold, p < 0.01) and CLB-GA (1.26-fold, p = 0.028). Light grey – JQ-1/PRIMA-1MET co-treatment, dark grey – GSH concentration after JQ-1 treatment, *p < 0.05, **p < 0.01. b: Concentrations of GSH and IC50 in LA1–55 cells with and without p53 gene expression. Results demonstrated 1.34-fold (p = 0.029) increase in total cellular GSH and 1.39-fold (p = 0.007) IC50 increase in cells expressing p53. Upper – concentration of GSH normalized to the number of cells using CellTiter2.0. Middle – WB for p53. Bottom – IC50 values for cells with and without p53. * - p < 0.05. c: New proposed model of interaction between PRIMA-1MET, p53, and MYCN
PRIMA-1MET induces accumulation of cells in the sub-G1 phase and triggers apoptosis irrespective of cell cycle phase through induction of caspase 3 and 7
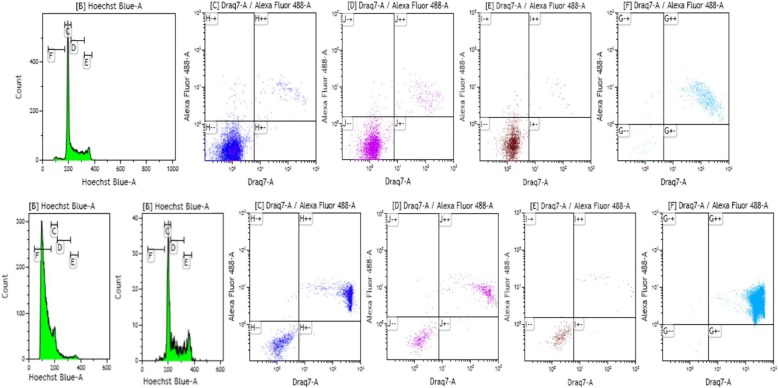
To understand how PRIMA-1MET affects NB cell proliferation, cell cycle analysis was performed on the seven sensitive NB cell lines (CHP212, CLB-GA, LAN6, NBL-S, NGP, SK-N-DZ and SK-N-SH). A 20 μM concentration of PRIMA-1MET was used to induce partial cell death, allowing us to analyze the cell cycle after a 24-h exposure. The results show that PRIMA-1MET induces the accumulation of cells in the sub-G1 phase (Fig. 2 and Additional file 2: Data for Figure S2). Simultaneous measurement of Cas3/7 activity and membrane integrity showed that the majority of cells in sub-G1 had significantly increased Cas3/7 activity and compromised membranes (Fig. 2). This phenotype is typical for apoptotic cells; necrotic cells usually do not express high levels of Cas3/7. Results on the cells that survived PRIMA-1MET treatment demonstrate that the cell cycle does not change substantially (Fig. 2, bottom row). Analyzing the Cas3/7 activity and membrane integrity of cells in the G1, S, and G2/M phases revealed that cells undergo apoptosis in all three phases of the cell cycle (Fig. 2 and Additional file 2: Data for Figure S2).
Fig. 2.
Cell cycle analysis of the NGP neuroblastoma cell line. Top row: non-treated NGP cell line. Bottom row: treated (20 μM PRIMA-1MET) NGP cell line. Gates: Green: cell cycle (20 μM Hoechst) c: G1 phase (dark blue); d: S phase (purple); e: G2/M phase (brown); f: sub-G1 phase (light blue). Cells in gates C, D, E, and F are distributed according to DRAQ7 and Cas3/7 (AlexaFluor 488) intensity. −/− live cells with stable membrane; −/+ live cells with triggered Cas3/7; +/+ dead cells with triggered Cas3/7 and nonfunctional membrane; +/− dead cells without activated Cas3/7 but with nonfunctional membrane. Results demonstrate that cells treated with PRIMA-1MET die in all phases of the cell cycle and that PRIMA-1MET activates apoptosis with similar intensity to non-treated control cells undergoing apoptosis
To corroborate this observation, we tested for early stage markers of apoptosis, AnnexinV/PI and collapse of mitochondrial membrane potential. The results showed a significant increase in AnnexinV positive cells, indicating early and late apoptosis after PRIMA-1MET treatment (Fig. 1b). This is supported by results using JC-1 labeling that demonstrate previously described “ballooning” of mitochondria [37] which leads to their subsequent collapse and induction of apoptosis (Additional file 2: Data for Figure S2).
Overall results demonstrate that PRIMA-1MET efficiently kills tumor cells irrespective of the cell cycle phase by triggering apoptosis, as measured by increased Cas3/7 activity and increased levels of AnnexinV. This result is not surprising, considering that most cells die within 15 h of sufficient exposure to PRIMA-1MET.
PRIMA-1MET increases Noxa and decreases CXCR4
Studies performed to date demonstrate that PRIMA-1MET acts by covalently binding to wild-type or mutant p53 and restoring its function, driving cancer cells to apoptosis [18, 38]. To further explore this process, an ICW assay and WB were used to investigate phosphorylation of p53 and ATM and induction of the downstream targets Bax and p21. Concentrations higher than IC50 were used in all subsequent experiments because they reflect better in vivo conditions. Mice tolerated PRIMA-1MET concentrations of up to 100 mg/kg (500 mmol/kg) without apparent side effects [17] while in clinical trials peak concentrations up to 82.6 mg/L (415 μmol/L) demonstrated mild but reversible side effects [39]. Results show that 60 μM PRIMA-1MET does not induce phosphorylation of either ATM or p53 (Fig. 3a). Likewise, no change was detected in the basal levels of ATM, p53, p21, or Bax (Fig. 3a). The only exception to these results was the NGP cell line, in which PRIMA-1MET treatment induced phosphorylation of ATM and p53, as well as overexpression of p21 (data not shown). Investigating the induction of reactive oxidative species (ROS) after PRIMA-1MET treatment in the NGP cell line revealed an increased presence of ROS (Additional file 2: Supplementary data 7), which could explain the induction of ATM, p53, and p21 due to DNA damage. WB was performed on BE-2C and SK-N-DZ to control for the accuracy of the ICW method. Results using WB demonstrate complete concordance of protein expression, as there was no difference after PRIMA-1MET treatment. Etoposide, a positive control, induced expression of all tested proteins (Additional file 2: Data for Figure S3).
Even though we did not observe a change in p53 phosphorylation in our experiments, it has been demonstrated that treatment with PRIMA-1MET is able to restore the p53 transactivation function and signal apoptosis without p53 phosphorylation in the p53-null Saos-2 cell line [17, 18]. It was also reported that, after exposure to PRIMA-1MET, Bax could be triggered in p53WT and Noxa in mutant p53R175H cell lines [18]. In our experiments, we observed no change in expression of Bax as measured by ICW and real-time PCR (Fig. 3a and b). Because Bax works through translocation and destabilization of mitochondria, increased expression might not be a prerequisite for its action. Analysis of Bax by immunofluorescence did not demonstrate any change in its localization (data not shown), suggesting that, in NB cell lines, the Bax pathway is not involved in PRIMA-1MET-induced apoptosis. PUMA, GADD45, 14–3-3, and Noxa were the other p53 transcriptional targets investigated using real-time PCR. Results demonstrate that Noxa is consistently upregulated after PRIMA-1MET treatment, while GADD45 and 14–3-3 show no change (Fig. 3b). PUMA expression was below reliable detection range in the eight NB cell lines we tested (data not shown). Lastly, CXCR4, a marker of NB aggressiveness and metastasis, and a target for p53 transactivation, was consistently downregulated after PRIMA-1MET treatment, suggesting a potential benefit of PRIMA-1MET in inhibiting tumor spread (Fig. 3B).
Noxa expression correlates with IC50 and is upregulated through p53
Saha et al recently reported that, in multiple myeloma cells, Noxa is involved in apoptosis triggered by PRIMA-1MET [23]. In our NB cell lines, a significant correlation (r2 = 0.59, p = 0.026) was demonstrated between IC50 and PRIMA-1MET-induced Noxa expression, suggesting that together with mitochondrial dysfunction (Additional file 2: Data for Figure S2), Noxa could play a role in PRIMA-1MET-induced NB apoptosis. Unchanged Noxa expression in the resistant cell line BE-2C, which carries a TP53 C135 mutation, further supports the involvement of p53. However, other classical p53 targets did not show any induction after PRIMA-1MET, thus necessitating investigation of the relationship between p53 and Noxa after PRIMA-1MET treatment. Experiments using the SK-N-SH cell line transfected with plasmids containing mutations in p53 or CREB binding segments of the Noxa promoter demonstrate 3.1-fold (p < 0.0001) or 1.3-fold (p < 0.026) decrease in activity when the p53 or CREB site is mutated, respectively (Fig. 4a). When transfected SK-N-SH cells were exposed to PRIMA-1MET, we observed significant induction of luciferase in cells with wt (1.44-fold, p = 0.03) and CREB-mutated (1.26-fold, p = 0.025) plasmids, but no significant difference in cells with plasmids containing a p53 binding site mutation (Fig. 4a). To control for p53 action on this particular site, cells with wt and p53-mutated plasmids were treated with etoposide, a known inducer of p53. Similarly, significant luciferase induction was observed in SK-N-SH cells containing wt plasmid (1.86-fold, p = 0.007) and no change was observed in SK-N-SH cells bearing plasmid with p53 binding site mutation (0.92-fold, p = 0.07) (Additional file 2: Data for Figure S4A). Although Noxa gene expression was clearly up-regulated after PRIMA-1MET treatment, we were unable to demonstrate an increase in Noxa protein expression in all eight of the NB cell lines tested, either before or after PRIMA-1MET treatment, because Noxa protein levels were not high enough for detection using WB. Next, Orlistat, a known Noxa stabilizer was used to increase Noxa protein expression. While no significant difference was detected in cell survival after treatment with orlistat alone, a significant decrease in live cells was observed when PRIMA-1MET was added (Fig. 4b). To address whether inhibition of general apoptosis, of which Noxa is an essential part, is enough to inhibit death induced by PRIMA-1MET, pan-caspase inhibitor was used. Our results demonstrate a partial but significant rescue of SK-N-SH cells from apoptosis induced by etoposide, used as positive control, but not from death induced by PRIMA-1MET (Fig. 4c).
Fig. 4.
PRIMA-1MET and Noxa. a: Results of luciferase reporter assay. A significant increase in luciferase activity after PRIMA-1MET treatment in Noxa wt and Noxa CREB but not Noxa p53 pGL10.4 plasmids. Dark grey: non-treated vehicle control. Light grey: PRIMA-1MET treatment (24 h, 15 μM). NOXA wt: pGL10.4 plasmid with normal Noxa promoter sequence. Noxa p53: pGL10.4 plasmid with Noxa promoter sequence containing mutation at the p53 binding site. Noxa CREB: pGL10.4 plasmid with Noxa promoter sequence containing mutation at the CREB binding site. b: Sensitization of SK-N-SH cells by pretreatment with orlistat. Dark grey: non-treated vehicle control. Light grey: PRIMA-1MET treatment (24 h, 10 μM). Left columns: orlistat non-treated vehicle control. Right columns: orlistat treatment (5 μM, 6 h prior to PRIMA-1MET treatment). c: Impact of pan-caspase inhibitor on the growth curve of SK-N-SH cells. Red lines: cells treated with pan-caspase inhibitor (22 h, 50 μM). Blue lines: Non-treated vehicle controls. Full line: non-treated vehicle control for PRIMA-1MET. Dotted line: PRIMA-1MET treated cell line (22 h, 20 μM). Dashed line: etoposide treatment (22 h, 30 μM)
PRIMA-1MET, GSH pathway and p53
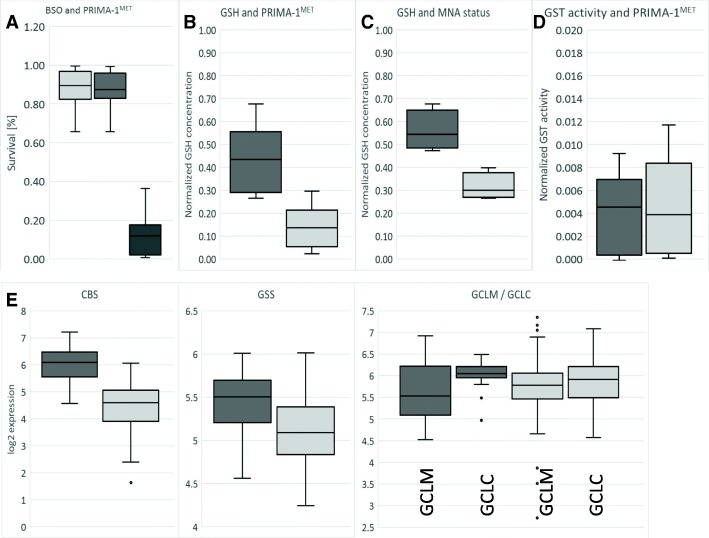
GSH plays a central role in cellular redox control and in the xenobiotic neutralizing system. It has been reported that inhibiting GSH with BSO sensitizes lung cancer cells to PRIMA-1MET [18] by decreasing the intracellular concentration of GSH. To investigate if the same mechanism works in NB, a combinatorial treatment of BSO and PRIMA-1MET was tested. BSO treatment alone has a limited short-term effect on NB cell survival. After 30 h of BSO treatment, only SK-N-DZ and NGP had less than 80% survival. All the NB cells, including p53-null LA1–55 N, treated with 100 μM of BSO 6 h prior to 24 h treatment with PRIMA-1MET were highly sensitized to PRIMA-1MET (Fig. 5a and Additional file 2: Data for Figure S5A). To understand how the intracellular concentration of GSH is affected by PRIMA-1MET, it was measured 8 h after exposure to PRIMA-1MET. A significant decrease (3.1-fold on average) of GSH concentration was observed in all eight of the NB cell lines (Fig. 5b), including in p53-null LA1–55 N (Additional file 2: Data for Figure S5A). Because this result suggested an alteration of the methionine/cysteine/GSH axis, we investigated whether supplementation of cysteine-related metabolites (GSH, N-acetyl-cysteine, cysteine, homocysteine, and methionine) was able to reverse the action of PRIMA-1MET (Additional file 2: Data for Figure S5B). The results of co-treatments demonstrated that 400 μM of either N-acetyl-cysteine, cysteine, homocysteine, or GSH were able to reverse the effect of PRIMA-1MET completely, but 400 μM of methionine could not do so in any of the six different NB cells, including p53-null LA1–55 N (Additional file 2: Data for Figure S5B). A complete reversal effect was possible, even after prolonged exposure to PRIMA-1MET, immediately before cell death, as indicated by profound morphological changes and complete reversal of autophagy (Additional file 2: Data for Figure S5D). To understand where possible inhibition of the methionine/cysteine/GSH axis might occur, we measured concentrations of the pathway’s main metabolites (S-adenozyl-methionine (SAH) and cysteine) in the cells, 1 h and 11 h after PRIMA-1MET exposure (Additional file 2: Data for Figure S5C). Results indicated that treatment using PRIMA-1MET significantly reduced SAH concentrations (0.71-fold and 0.66-fold at 1 h and 11 h, respectively) but increased cysteine concentrations (1.27-fold at 11 h), whereas the cysteine/SAH ratio had increased by 1.56-fold at 1 h to 1.92-fold at 11 h (Additional file 2: Data for Figure S5C). These results demonstrated that GSH was an important defense mechanism against PRIMA-1MET that was modulated by PRIMA-1MET.
Fig. 5.
PRIMA-1MET and glutathione. a: Concurrent BSO and PRIMA-1MET treatment. All cell lines were treated with fixed 100 μM BSO for 6 h prior to PRIMA-1MET treatment at a concentration of less than IC90 on average. The Y-axis indicates the survival ratio. A significant synergistic effect (CDI = 0.16) due to BSO treatment was detected in all cell lines, indicating that the depletion of GSH significantly influences the sensitivity of the cells to PRIMA-1MET. Light grey: survival at 100 μM BSO, dark grey: survival at IC90 concentration of PRIMA-1MET, black: BSO and PRIMA-1MET. b: Concentration of GSH decreases 3.1-fold (p < 0.001) in NB cell lines treated with 60 μM PRIMA-1MET for 6 h (light grey) in comparison to non-treated (dark grey). c: Concentration of GSH is 1.8-fold (p < 0.05) lower in MNA NB (dark grey) in comparison to non-MNA NB (light grey). d: No significant difference in GST activity due to PRIMA-1MET (light grey) in comparison to non-treated control (dark grey) NB cell lines. e: Higher expression of CBS (2.97-fold, p < 0.001) and GSS (1.25-fold, p < 0.001), and significant change in GCLC/GCLM ratio (1.3-fold, p = 0.037) in MNA NB samples (dark grey) in comparison to non-MNA NB samples (light grey). Dots: outliers
P53 and MNA status affects concentration of GSH and modulates resistance of neuroblastoma cells against PRIMA-1MET
Because MNA is one of the main genomic events in NB, we investigated MNA’s impact on GSH. Before PRIMA-1MET treatment, GSH concentration correlates with MNA, but not with IC50 or 11q-deletion. MNA cells have on average 1.8 times (p < 0.05) more GSH (Fig. 5c). The decrease in GSH concentration due to PRIMA-1MET was not significantly associated with MNA, 11q-deletion, or IC50 (data not shown). To investigate how GSH levels differ between MNA and non-MNA NB, correlation between MYCN status and gene expression of the main enzymes involved in GSH synthesis was analyzed using the Affymetrix expression array dataset consisting of 101 NB primary tumors (GEO accession: GSE3960). The results demonstrate that expression of CBS and GSS, genes involved in cystathionine and glutathione synthesis, are 2.97 times (p < 0.001) and 1.25 times (p < 0.001) higher, respectively, in MNA than in non-MNA tumors (Fig. 5e). GCLC and GCLM, which form a heterodimeric enzyme, are responsible for the synthesis of γ-glutamyl-cysteine. In MNA NB tumors, expression of GCLC (the catalytic subunit) is 1.3 times higher (p < 0.05) than GCLM expression, but there was no difference in non-MNA NB tumors (Fig. 5e). The other components of GSH metabolism (GSTM1, GSTM2, GSTM3, GSTA1, GSTT1) were also investigated in order to understand the potential influence of MNA on GSH, but no significant differences were detected between MNA and non-MNA NB tumors (data not shown). Finally, to explore the interaction between PRIMA-1MET and the glutathione-S-transferase (GST) system, we measured overall GST activity in the eight NB cell lines. No significant change was observed after PRIMA-1MET treatment (Fig. 5d). MNA’s involvement in the resistance of neuroblastoma cells was explored using JQ-1, a known inhibitor of MYCN expression [40]. Results using JQ-1 demonstrated a simultaneous decrease in MYCN expression and GSH levels (1.16-fold, p < 0.01) and sensitized p53-null LA1–55 N cells to higher doses of PRIMA-1MET (Fig. 6a). In contrast, expressing p53 in LA1–55 N resulted in a significant 1.34-fold increase in GSH and a 1.39-fold (p = 0.007) increase in IC50 (Fig. 6b). Because MYCN is a known transcription factor of p53 and JQ-1 is known to reduce p53 [40], we investigated p53 expression in CLB-GA (non-MNA, normal p53) cells after exposure to 200 nM JQ-1. JQ-1-treated CLB-GA cells demonstrated a marked decrease in p53 expression, no change in MYCN levels, a significant decrease in GSH levels (1.26-fold, p = 0.028), and increased susceptibility to higher doses of PRIMA-1MET (Fig. 6a). Cumulatively, these results demonstrated that MYCN and p53 are implicated in the survival of cells exposed to PRIMA-1MET via the modulation of GSH levels.
PRIMA-1MET inhibits thioredoxin reductase activity in neuroblastoma cell lines
Another system potentially implicated in resistance to PRIMA-1MET is thioredoxin (TXN). Recently, Peng et al demonstrated that, in lung adenocarcinoma, osteosarcoma, and Burkitt lymphoma cells, PRIMA-1MET can also target thioredoxin reductase 1 (TXNRD1) by binding to its active site, causing a decrease in reductase activity. Interestingly, it was demonstrated that even when inhibited by PRIMA-1MET, TXNRD1 is still functional as a pro-oxidant NADPH oxidase. This kind of modulation of TXNRD1 can cause an increase in oxidative stress and cell death, but implications for NB cell survival are questionable [41]. To understand how PRIMA-1MET might influence the TXN system in NB cells, we first used ICW to measure intracellular TXN levels. The results demonstrate that PRIMA-1MET treatment induces no change in total TXN levels in NB cell lines (Additional file 2: Supplementary data 6C). Comparing TXN levels to MNA and 11q status of the cells showed that high levels of TXN are significantly associated with the presence of MNA (Additional file 2: Supplementary data 6B) but not with 11q-deletion. MNA was correlated with 1.2-fold (p < 0.02) and 1.22-fold (p = 0.02) upregulation in respective expression of TXN and TXN2 in NB tumoral samples. However, no difference in TXNRD1 expression was observed between MNA and non-MNA NB tumors (Additional file 2: Supplementary data 6E). Because TXN reduction capacity is dependent on the activity of TXNRD1, we measured reductase activity in NB cells. Results demonstrate a significant drop in TXNRD1 activity in all eight NB cell lines (Additional file 2: Supplementary data 6D). No correlation was observed between the reduction rate of TXNRD1 activity and MNA, 11q status, or IC50. BE-2C and SK-N-DZ, two cell lines with aberrant p53 accumulation, demonstrated the smallest inhibition of TXNRD1 activity, and cell lines with TP53 mutations showed the highest TXNRD1 activity (data not shown). Finally, to address the possible role of the TXN system in resistance to PRIMA-1MET, cells were treated concurrently with PX-12 (a known TXN inhibitor) and PRIMA-1MET, and survival was analyzed. The results showed an additive effect of PX-12 and PRIMA-1MET, but no synergy was observed in any of the eight NB cell lines tested (data not shown). PX-12 alone at 5 μM exhibited the ability to inhibit growth in all NB cell lines, particularly those with MNA (0.54-fold growth compared to 0.82-fold growth in non-MNA lines, p = 0.016) (Additional file 2: Supplementary data 6A). This finding could be explained by the higher basal level of TXN in MNA cells (Additional file 2: Supplementary data 6B). An additive effect observed with PX-12 and PRIMA-1MET suggests that PRIMA-1MET action is independent of TXN status. On the other hand, PRIMA-1MET was found to interact with the TXN system through inhibition of TXNRD1, suggesting that drugs interacting with the TXN system might synergize or antagonize with PRIMA-1MET.
PRIMA-1MET and induction of oxidative stress in neuroblastoma cell lines
Because PRIMA-1MET interacts with the GSH and TXN systems, we hypothesized that it could induce oxidative stress in NB. The eight NB cell lines were treated with 60 μM and 120 μM concentrations of PRIMA-1MET for 6 h to understand how PRIMA-1MET could affect the generation of reactive oxygen species in NBs. Higher concentrations were used because they better reflect the physiological reality. Flow cytometry results demonstrate that PRIMA-1MET induced oxidative stress in three cell lines (LAN6, NGP, SK-N-DZ) at 60 μM and in two additional cell lines (BE-2C, CH212) at 120 μM (Additional file 2: Supplementary data 7). Induction of oxidative stress in NGP by PRIMA-1MET could explain ATM- and p53-phosphorylation and p21 overexpression because ROS could lead to an increase in DNA damage. Partially supporting results obtained in other cancer cell lines [18, 41, 42], our data demonstrate that PRIMA-1MET has the ability to induce oxidative stress in NB cell lines. However, this effect is specific to concentration and cell line. Acquired or germline genetic variations might play an important role in the development of oxidative stress due to PRIMA-1MET treatment.
Discussion
In this study, we have explored the efficacy of PRIMA-1MET in eight NB cell lines and investigated the possible molecular pathways involved in its action and resistance. The data demonstrate that PRIMA-1MET is effective and particularly active in seven of the eight NB cell lines tested. PRIMA-1MET exhibited a narrow activation range, and killed NB cells within 15 h of exposure to IC90 concentration. Unlike initial reports about PRIMA-1MET [18], we demonstrated that the main effect of p53 in NB cells occurs at the level of GSH and not through direct activation of the main p53 targets (Fig. 6). MYCN, which is an important factor in the development of NB, also activates the GSH pathway, resulting in increased levels of GSH and resistance to PRIMA-1MET.
PRIMA-1MET is known to restore p53 function after conversion to MQ, a Michael acceptor, which can bind covalently to cysteine and lead to p53 refolding to its native state and inducing cell death [18]. It was therefore surprising that BE-2C, which carries the TP53 C135F homozygous mutation, was highly resistant. On the other hand, stable expression of p53 in LA1–55 N cells, which otherwise lack p53 expression, increased LA1–55 N resistance to PRIMA-1MET. To address these discrepancies, we comprehensively analyzed current evidence about PRIMA-1MET in the context of NB cells. We confirmed that PRIMA-1MET is a potent inducer of apoptosis by analyzing several key markers of apoptosis, namely, induction of AnnexinV, end-stage Cas3/7, perforation of cellular membranes and damage to mitochondria. Our results demonstrate that PRIMA-1MET does not affect the cell cycle, as evidenced by cellular DNA content and low p21 levels. Furthermore, it can induce apoptosis in NB cell lines irrespective of cell cycle phase. This property of PRIMA-1MET could be particularly beneficial since chemotherapeutics such as cisplatin and etoposide usually induce DNA damage followed by cell cycle arrest [43], which leads to the development of resistance and cancer cell survival [7]. It is known that p21, the primary mediator of p53 and a potent cyclin-dependent kinase inhibitor, plays a dual role: it promotes cell cycle arrest during the G1 phase and inhibits proliferation, but it can also promote cell survival by inhibiting apoptosis and giving cells time to repair damaged DNA [44, 45].
Next, the p53 transactivation function was analyzed in detail. Results show no changes in p53, phosphorylated p53, ATM, phosphorylated ATM, or Bax expression levels after PRIMA-1MET treatment. Furthermore, the absence of ATM- and p53-phosphorylation is significant because it demonstrates indirectly that PRIMA-1MET is probably not a DNA-damaging agent, as the ATM/p53 pathway itself is not triggered. Testing for p53 transactivation function showed no induction of many of its major targets, including GADD45, 14–3-3, p21, and Bax. The only exception is Noxa, which is upregulated 2-fold on average. Despite indirect evidence such as Noxa gene expression and IC50 correlation, mitochondrial membrane potential collapse and activation of apoptotic cascade, these results still raised doubts about the direct relationship between PRIMA-1MET, p53 and Noxa in NB cell lines. Using luciferase reporter assays, we demonstrated that Noxa upregulation is due to p53, confirming p53 transactivation after PRIMA-1MET treatment. Further investigation of the role of Noxa, using WB and pan-caspase inhibitor, demonstrated that Noxa was not present in NB cells, whereas the inhibition of apoptosis was insufficient to rescue cells from death, thus questioning Noxa’s involvement in PRIMA-1MET-mediated death. Nevertheless, Noxa may still prove useful, either as a drug target for Orlistat, which sensitizes cells to PRIMA-1MET, or as a marker of PRIMA-1MET potency.
Next, we investigated the role of cellular redox pathways in PRIMA-1MET activity, because PRIMA-1MET is cleared through GSH [18] and influences TXNRD1 reductase activity [41]. Our results demonstrated significant synergy between PRIMA-1MET and BSO, providing strong evidence of GSH but not GST involvement in clearing PRIMA-1MET in NB. Further investigations of the methionine/cysteine/GSH axis demonstrated that metabolites carrying free SH-groups were involved in the inhibition of PRIMA-1MET, which leads to a fast reversal of morphology and autophagy. However, the same 400 μM concentration of methionine was unable to rescue the cells. Analysis of intracellular concentrations of cysteine and SAH demonstrated that most of the inhibition of GSH-synthesis by PRIMA-1MET likely happens after cysteine, as its concentration is higher even though the concentration of SAH decreases. Finally, investigating GSH concentration in the context of p53 expression demonstrated an increase of GSH in p53-competent cells, which is in accordance with a recent report by Tarangelo et al [46]. These results suggest that indirect interaction between p53 and PRIMA-1MET, through the modulation of GSH, is more relevant than the direct p53 transactivation activity observed in the context of NB (Fig. 6c). Likewise, our results showed that MNA was associated with higher levels of GSH via the increased expression of enzymes associated with GSH synthesis. Further experiments on p53-null LA1–55 N confirmed MYCNs’ independent role in the reduction of GSH levels and increased sensitivity to PRIMA-1MET.
In exploring the involvement of the TXN system in PRIMA-1MET efficacy, we found that TXN does not appear to be important for PRIMA-1MET action but that PRIMA-1MET is able to induce TXNRD1 inhibition. It is interesting that PX-12 on its own exhibits a significant effect on NB cell viability, particularly in NB cell lines without MNA. This is probably due to lower gene and subsequent TXN protein expression in comparison to MNA NB. Interestingly, in contrast to previous studies [18, 41, 42], our results suggest that induction of ROS by PRIMA-1MET is frequent but not a universal event and instead depends on molecular context. Interestingly, despite the induction of ROS inactivity of ATM/p53 pathway suggests indirectly that, in NB cell lines, PRIMA-1MET is not involved in DNA damage.
Finally, we found that CXCR4, which has been reported to promote NB tumor growth, metastasis and resistance to therapeutics [47, 48] and was therefore chosen in this study as a marker of aggressiveness, was found to be downregulated after PRIMA-1MET exposure. While it was demonstrated that p53 negatively regulates CXCR4 in breast cancer [49], to our knowledge this modulation has never been reported in NB. Nevertheless, our result suggests that downregulation of CXCR4 could participate in NB growth inhibition induced by PRIMA-1MET.
Conclusions
PRIMA-1MET was identified as a potent and specific compound that inhibits growth in NB cell lines and kills cells rapidly by activation of apoptosis, autophagy, and oxidative stress. Although PRIMA-1MET triggers p53 transactivation activity [18], its main targets remain inactive and inhibition of apoptosis is ineffective, suggesting an indirect interaction between PRIMA-1MET and p53 in cases of NB. Importantly, PRIMA-1MET is deactivated by free SH-groups, but it also causes a deep decrease in GSH concentration, through both direct binding and the inhibition of GSH synthesis. Finally, by showing that p53 and MYCN are involved in the modulation of resistance to PRIMA-1MET through the modulation of intracellular GSH levels, we established a new model of the interaction between p53 and PRIMA-1MET. In the proposed model based on the presented results, p53 and MYCN induce PRIMA-1MET resistance in an indirect manner, via GSH modulation, rather than via direct binding between PRIMA-1MET and p53, which is the currently held view in many other cancer models (Fig. 6c).
Additional files
Whole exome sequencing of neuroblastoma cells. (XLSX 439 kb)
Supplementary information for figures. (PPTX 6460 kb)
Acknowledgements
We thank Dr V. Combaret (Centre des Ressources Biologiques du Centre Léon Berard) and Dr E. Attiyeh for providing the cell lines. Many thanks to Mary Boudal-Khoshbeen for her excellent technical assistance, to the iGE3 genomics platform of the University of Geneva, in particular to Natacha Civic and Didier Collet for their contributions with WES and real-time PCR, and to Donna Furlani and to Darren Hart for English editing.
Funding
This work was supported by the CANSEARCH Foundation, Le Printemps des Enfants, Fondation pour la lutte contre le cancer et pour des recherches médico-biologiques, Fondation Simone et Gustave Prévot, and the Giulio D’Angio Endowed Chair.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Abbreviations
- BSO
Buthionine sulfoximine
- GSH
Glutathione,
- IC50
50% inhibitory concentration
- ICW
In-cell western blot
- LCL
Lymphoblastoid cell lines
- MNA
MYCN amplification
- MQ
Methylene quinuclidinone
- NB
Neuroblastoma
- r2
Correlation coefficient
- SAH
S-adenosylhomocysteine
- TXN
Thioredoxin
- TXNRD1
Thioredoxin reductase 1
Authors’ contributions
VM, FGP: conceived the study and prepared the first draft. VM, SJM, LL, DM: executed cell line experiments. KSR, JMM: collected clinical NB samples, analyzed transcriptomes, evaluated statistics. VM, SJM, KSR, JMM, MA, FGP: prepared figures and Tables. VM, FGP, JMM, KSR, SJM, MA: wrote, discussed and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vid Mlakar, Email: vid.mlakar@unige.ch.
Simona Jurkovic Mlakar, Email: simona.mlakar@unige.ch.
Laurence Lesne, Email: laurence.lesne@unige.ch.
Denis Marino, Email: denis.marino@unige.ch.
Komal S. Rathi, rathik@email.chop.edu
John M. Maris, maris@email.chop.edu
Marc Ansari, Email: marc.ansari@hcuge.ch.
Fabienne Gumy-Pause, Phone: +41 22 372 47 31, Email: fabienne.gumypause@hcuge.ch.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, Weiss WA. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 3.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3:a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mlakar V, Jurkovic Mlakar S, Lopez G, Maris JM, Ansari M, Gumy-Pause F. 11q deletion in neuroblastoma: a review of biological and clinical implications. Mol Cancer. 2017;16:114. doi: 10.1186/s12943-017-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasorsa VA, Formicola D, Pignataro P, Cimmino F, Calabrese FM, Mora J, Esposito MR, Pantile M, Zanon C, De Mariano M, et al. Exome and deep sequencing of clinically aggressive neuroblastoma reveal somatic mutations that affect key pathways involved in cancer progression. Oncotarget. 2016;7:21840–21852. doi: 10.18632/oncotarget.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 7.Padovan-Merhar OM, Raman P, Ostrovnaya I, Kalletla K, Rubnitz KR, Sanford EM, Ali SM, Miller VA, Mosse YP, Granger MP, et al. Enrichment of targetable mutations in the relapsed neuroblastoma genome. PLoS Genet. 2016;12:e1006501. doi: 10.1371/journal.pgen.1006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wasylishen AR, Lozano G. Attenuating the p53 pathway in human cancers: many means to the same end. Cold Spring Harb Perspect Med. 2016;6. [DOI] [PMC free article] [PubMed]
- 10.Barone G, Tweddle DA, Shohet JM, Chesler L, Moreno L, Pearson AD, Van Maerken T. MDM2-p53 interaction in paediatric solid tumours: preclinical rationale, biomarkers and resistance. Curr Drug Targets. 2014;15:114–123. doi: 10.2174/13894501113149990194. [DOI] [PubMed] [Google Scholar]
- 11.Van Maerken T, Speleman F, Vermeulen J, Lambertz I, De Clercq S, De Smet E, Yigit N, Coppens V, Philippe J, De Paepe A, et al. Small-molecule MDM2 antagonists as a new therapy concept for neuroblastoma. Cancer Res. 2006;66:9646–9655. doi: 10.1158/0008-5472.CAN-06-0792. [DOI] [PubMed] [Google Scholar]
- 12.Van Maerken T, Rihani A, Dreidax D, De Clercq S, Yigit N, Marine JC, Westermann F, De Paepe A, Vandesompele J, Speleman F. Functional analysis of the p53 pathway in neuroblastoma cells using the small-molecule MDM2 antagonist nutlin-3. Mol Cancer Ther. 2011;10:983–993. doi: 10.1158/1535-7163.MCT-10-1090. [DOI] [PubMed] [Google Scholar]
- 13.Barbieri E, Mehta P, Chen Z, Zhang L, Slack A, Berg S, Shohet JM. MDM2 inhibition sensitizes neuroblastoma to chemotherapy-induced apoptotic cell death. Mol Cancer Ther. 2006;5:2358–2365. doi: 10.1158/1535-7163.MCT-06-0305. [DOI] [PubMed] [Google Scholar]
- 14.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 15.Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012;13:1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 16.Aziz MH, Shen H, Maki CG. Acquisition of p53 mutations in response to the non-genotoxic p53 activator Nutlin-3. Oncogene. 2011;30:4678–4686. doi: 10.1038/onc.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 18.Lambert JM, Gorzov P, Veprintsev DB, Soderqvist M, Segerback D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Zandi R, Selivanova G, Christensen CL, Gerds TA, Willumsen BM, Poulsen HS. PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin Cancer Res. 2011;17:2830–2841. doi: 10.1158/1078-0432.CCR-10-3168. [DOI] [PubMed] [Google Scholar]
- 20.Bykov VJ, Zache N, Stridh H, Westman J, Bergman J, Selivanova G, Wiman KG. PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene. 2005;24:3484–3491. doi: 10.1038/sj.onc.1208419. [DOI] [PubMed] [Google Scholar]
- 21.Li XL, Zhou J, Chan ZL, Chooi JY, Chen ZR, Chng WJ. PRIMA-1met (APR-246) inhibits growth of colorectal cancer cells with different p53 status through distinct mechanisms. Oncotarget. 2015;6:36689–36699. doi: 10.18632/oncotarget.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patyka M, Sharifi Z, Petrecca K, Mansure J, Jean-Claude B, Sabri S. Sensitivity to PRIMA-1MET is associated with decreased MGMT in human glioblastoma cells and glioblastoma stem cells irrespective of p53 status. Oncotarget. 2016;7:60245–60269. doi: 10.18632/oncotarget.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha MN, Jiang H, Yang Y, Reece D, Chang H. PRIMA-1Met/APR-246 displays high antitumor activity in multiple myeloma by induction of p73 and Noxa. Mol Cancer Ther. 2013;12:2331–2341. doi: 10.1158/1535-7163.MCT-12-1166. [DOI] [PubMed] [Google Scholar]
- 24.Tessoulin B, Descamps G, Moreau P, Maiga S, Lode L, Godon C, Marionneau-Lambot S, Oullier T, Le Gouill S, Amiot M, Pellat-Deceunynck C. PRIMA-1Met induces myeloma cell death independent of p53 by impairing the GSH/ROS balance. Blood. 2014;124:1626–1636. doi: 10.1182/blood-2014-01-548800. [DOI] [PubMed] [Google Scholar]
- 25.Ali D, Mohammad DK, Mujahed H, Jonson-Videsater K, Nore B, Paul C, Lehmann S. Anti-leukaemic effects induced by APR-246 are dependent on induction of oxidative stress and the NFE2L2/HMOX1 axis that can be targeted by PI3K and mTOR inhibitors in acute myeloid leukaemia cells. Br J Haematol. 2016;174:117–126. doi: 10.1111/bjh.14036. [DOI] [PubMed] [Google Scholar]
- 26.Synnott NC, Murray A, McGowan PM, Kiely M, Kiely PA, O'Donovan N, O'Connor DP, Gallagher WM, Crown J, Duffy MJ. Mutant p53: a novel target for the treatment of patients with triple-negative breast cancer? Int J Cancer. 2017;140:234–246. doi: 10.1002/ijc.30425. [DOI] [PubMed] [Google Scholar]
- 27.Mohell N, Alfredsson J, Fransson A, Uustalu M, Bystrom S, Gullbo J, Hallberg A, Bykov VJ, Bjorklund U, Wiman KG. APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis. 2015;6:e1794. doi: 10.1038/cddis.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perdrix A, Najem A, Saussez S, Awada A, Journe F, Ghanem G, Krayem M. Prima-1 and PRIMA-1(Met) (APR-246): From Mutant/Wild Type p53 Reactivation to Unexpected Mechanisms Underlying Their Potent Anti-Tumor Effect in Combinatorial Therapies. Cancers (Basel). 2017;9(12):E172. 10.3390/cancers9120172. [DOI] [PMC free article] [PubMed]
- 29.Mandriota SJ, Valentijn LJ, Lesne L, Betts DR, Marino D, Boudal-Khoshbeen M, London WB, Rougemont AL, Attiyeh EF, Maris JM, et al. Ataxia-telangiectasia mutated (ATM) silencing promotes neuroblastoma progression through a MYCN independent mechanism. Oncotarget. 2015;6:18558–18576. doi: 10.18632/oncotarget.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3:e00149. doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiele CJ. Neuroblastoma cell lines. Lancester, UK: Kluwer academic publisher; 1998. [Google Scholar]
- 33.Harenza JL, Diamond MA, Adams RN, Song MM, Davidson HL, Hart LS, Dent MH, Fortina P, Reynolds CP, Maris JM. Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data. 2017;4:170033. doi: 10.1038/sdata.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montavon G, Jauquier N, Coulon A, Peuchmaur M, Flahaut M, Bourloud KB, Yan P, Delattre O, Sommer L, Joseph JM, et al. Wild-type ALK and activating ALK-R1275Q and ALK-F1174L mutations upregulate Myc and initiate tumor formation in murine neural crest progenitor cells. Oncotarget. 2014;5:4452–4466. doi: 10.18632/oncotarget.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine JC, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 36.Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61:8–13. [PubMed] [Google Scholar]
- 37.Mancini M, Anderson BO, Caldwell E, Sedghinasab M, Paty PB, Hockenbery DM. Mitochondrial proliferation and paradoxical membrane depolarization during terminal differentiation and apoptosis in a human colon carcinoma cell line. J Cell Biol. 1997;138:449–469. doi: 10.1083/jcb.138.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert JM, Moshfegh A, Hainaut P, Wiman KG, Bykov VJ. Mutant p53 reactivation by PRIMA-1MET induces multiple signaling pathways converging on apoptosis. Oncogene. 2010;29:1329–1338. doi: 10.1038/onc.2009.425. [DOI] [PubMed] [Google Scholar]
- 39.Lehmann S, Bykov VJ, Ali D, Andren O, Cherif H, Tidefelt U, Uggla B, Yachnin J, Juliusson G, Moshfegh A, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30:3633–3639. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- 40.Shahbazi J, Scarlett CJ, Norris MD, Liu B, Haber M, Tee AE, Carrier A, Biankin AV, London WB, Marshall GM, et al. Histone deacetylase 2 and N-Myc reduce p53 protein phosphorylation at serine 46 by repressing gene transcription of tumor protein 53-induced nuclear protein 1. Oncotarget. 2014;5:4257–4268. doi: 10.18632/oncotarget.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng X, Zhang MQ, Conserva F, Hosny G, Selivanova G, Bykov VJ, Arner ES, Wiman KG. APR-246/PRIMA-1MET inhibits thioredoxin reductase 1 and converts the enzyme to a dedicated NADPH oxidase. Cell Death Dis. 2013;4:e881. doi: 10.1038/cddis.2013.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grellety T, Laroche-Clary A, Chaire V, Lagarde P, Chibon F, Neuville A, Italiano A. PRIMA-1(MET) induces death in soft-tissue sarcomas cell independent of p53. BMC Cancer. 2015;15:684. doi: 10.1186/s12885-015-1667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piacentini M, Fesus L, Melino G. Multiple cell cycle access to the apoptotic death programme in human neuroblastoma cells. FEBS Lett. 1993;320:150–154. doi: 10.1016/0014-5793(93)80081-5. [DOI] [PubMed] [Google Scholar]
- 44.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 45.McKenzie PP, Guichard SM, Middlemas DS, Ashmun RA, Danks MK, Harris LC. Wild-type p53 can induce p21 and apoptosis in neuroblastoma cells but the DNA damage-induced G1 checkpoint function is attenuated. Clin Cancer Res. 1999;5:4199–4207. [PubMed] [Google Scholar]
- 46.Tarangelo A, Magtanong L, Bieging-Rolett KT, Li Y, Ye J, Attardi LD, Dixon SJ. p53 suppresses metabolic stress-induced Ferroptosis in Cancer cells. Cell Rep. 2018;22:569–575. doi: 10.1016/j.celrep.2017.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein S, Abraham M, Bulvik B, Dery E, Weiss ID, Barashi N, Abramovitch R, Wald H, Harel Y, Olam D, et al. CXCR4 promotes neuroblastoma growth and therapeutic resistance through miR-15a/16-1-mediated ERK and BCL2/cyclin D1 pathways. Cancer Res. 2018;78:1471–1483. doi: 10.1158/0008-5472.CAN-17-0454. [DOI] [PubMed] [Google Scholar]
- 48.Muhlethaler-Mottet A, Liberman J, Ascencao K, Flahaut M, Balmas Bourloud K, Yan P, Jauquier N, Gross N, Joseph JM. The CXCR4/CXCR7/CXCL12 Axis is involved in a secondary but complex control of neuroblastoma metastatic cell homing. PLoS One. 2015;10:e0125616. doi: 10.1371/journal.pone.0125616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehta SA, Christopherson KW, Bhat-Nakshatri P, Goulet RJ, Jr, Broxmeyer HE, Kopelovich L, Nakshatri H. Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene. 2007;26:3329–3337. doi: 10.1038/sj.onc.1210120. [DOI] [PubMed] [Google Scholar]
- 50.Hart LS, Rader J, Raman P, Batra V, Russell MR, Tsang M, Gagliardi M, Chen L, Martinez D, Li Y. Preclinical therapeutic synergy of MEK1/2 and CDK4/6 inhibition in neuroblastoma. Clin Cancer Res. 2017;23. [DOI] [PubMed]
- 51.Krytska K, Ryles HT, Sano R, Raman P, Infarinato NR, Hansel TD, Makena MR, Song MM, Reynolds CP, Mosse YP. Crizotinib synergizes with chemotherapy in preclinical models of neuroblastoma. Clin Cancer Res. 2016;22:948–960. doi: 10.1158/1078-0432.CCR-15-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole exome sequencing of neuroblastoma cells. (XLSX 439 kb)
Supplementary information for figures. (PPTX 6460 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].