Abstract
Protein tyrosine phosphatase receptor type J (PTPRJ, DEP1) is a tumour suppressor gene that negatively regulates such processes as angiogenesis, cell proliferation and migration and is one of the genes important for tumour development. Similar to other phosphatase genes, PTPRJ is also described as an oncogene. Among various genetic changes characteristic for this gene, single nucleotide polymorphisms (SNPs) constituting benign genetic variants that can modulate its function have been described. We focused on Gln276Pro and Arg326Gln missense polymorphisms and performed a meta-analysis using data from 2930 and 852 patients for Gln276Pro and Arg326Gln respectively in different cancers. A meta-analysis was performed based on five articles accessed via the PubMed and Research Gate databases. Our meta-analysis revealed that for Arg326Gln, the presence of the Arg (C) allele was associated with lower risk of some cancers, the strongest association was observed for colorectal cancer patients, and there was no association between Gln276Pro (G>T) polymorphism and cancer risk. The polymorphisms Arg326Gln and Gln276Pro of the PTPRJ gene are not associated with an increased risk of cancer except for the Arg326Gln polymorphism in colorectal cancer. Large-scale studies should be performed to verify the impact of this SNP on individual susceptibility to colorectal cancer for given individuals.
Keywords: PTPRJ, SNP, Cancer, Meta-analysis
Introduction
Protein tyrosine phosphatase receptor type J (PTPRJ, DEP1) is described as a tumour suppressor gene that negatively regulates angiogenesis, cell proliferation and migration and therefore is involved in tumour progression in some human cancers (Aya-Bonilla et al. 2013; Bilotta et al. 2016; Fournier et al. 2016; Zhang et al. 2017). The proven impact of PTPRJ on cellular biology results from its role in dephosphorylation of some kinases such as PDGFR, EGFR, VEGFR2, HGFR, PI3K (p85), MAPK (ERK1/2), FLT3 and RET and also proteins involved in cell adhesion such as c-Src, p120-catenin, VE-cadherin, THBS1 and ZO-1 (Aya-Bonilla et al. 2013; Fournier et al. 2016; Qiao et al. 2016; Steinman et al. 2016). Although PTPRJ is mainly classified as a tumour suppressor gene, it has also been described as an oncogene. PTPRJ loss of heterozygosity (LOH) has been reported in several cancers such as colon, breast, thyroid, meningioma, non-Hodgkin lymphoma and cervical carcinoma (Ruivenkamp et al. 2003; Iuliano et al. 2010; Aya-Bonilla et al. 2013; Yan et al. 2015). Moreover, germline epigenetic silencing of this gene in early-onset familiar colorectal cancer (Venkatachalam et al. 2010) as well as the loss of PTPRJ expression in association with an advanced tumour stage and poor differentiation in oesophageal squamous cell carcinoma was described (Qiao et al. 2016). In contrast, the role of PTPRJ in VEGF-dependent Src activation, and therefore capillary formation and permeability, has been demonstrated in both a mouse model and human breast cancer (Spring et al. 2015; Fournier et al. 2016). Also a soluble short isoform, sPTPRJ, secreted into endothelial and tumour cells was shown to down-regulate endothelial adhesion molecules and to promote angiogenesis in glioblastoma (Bilotta et al. 2016). It was also reported that PTPRJ agonist nonapeptide led to reduction of cell proliferation and promotion of apoptosis in cancer cell lines and also inhibited tube formation in an in vitro experiment (Ortuso et al. 2013). These findings confirmed the hypothesis that protein tyrosine phosphatases via dephosphorylation play a dual role in carcinogenesis (as tumour suppressors and oncogenes) as they are crucial regulators of the activity of a variety of proteins (Julien et al. 2011; Zhao et al. 2015).
As has been demonstrated, PTPRJ is expressed in various types of cells (e.g. epithelial, haematopoietic and endothelial cells and many cancer cell lines), and thus its effect on different cancer types seems to be indisputable (Fournier et al. 2016).
Single nucleotide polymorphisms (SNPs) are benign genetic variants that can modulate expression, folding, activity, binding affinity and other protein functions and therefore are intensively studied in many different diseases (Katsonis et al. 2014). The Gln276Pro (rs1566734) and Arg326Gln (rs1503185) polymorphisms in PTPRJ are missense SNPs that were previously genotyped in colorectal, thyroid, lung, head and neck, oesophageal and breast cancers (Iuliano et al. 2004, 2010; Toland et al. 2008; Mita et al. 2010; Wei et al. 2013; Shangkuan et al. 2017). Both polymorphisms are located in the extracellular region of PTPRJ and are involved in fibronectin-like type III domain formation (Ruivenkamp et al., 2002).
In our study, we investigated whether Gln276Pro and Arg326Gln missense polymorphisms are risk factors in various cancers. Because a meta-analysis is a proper tool for evaluating the association between gene polymorphisms and cancer risk, we analysed data collected from all five studies published to date focusing on these SNPs (n = 2930 and n = 852 for Gln276Pro and Arg326Gln respectively) in different cancers.
Materials and methods
Meta-analysis—selection of studies
We searched PubMed and ResearchGate databases to compare our investigation with research on other populations by collecting articles published until December 2017 using the following terms: “PTPRJ[All Fields] OR “DEP-1[All Fields] AND (“neoplasms“[MeSH Terms] OR “neoplasms“[All Fields] OR “cancer”[All Fields])”. Reference lists and conference reports were included in the analysis. Finally, 82 publications which were focused on comparison between cancer patients and a healthy control group were found. Review papers, meta-analyses and papers describing other PTPRJ gene polymorphisms were excluded. After applying the above restriction, five papers that compared the frequency of Arg326Gln and Gln276Pro of the PTPRJ gene in cancer patients and healthy controls were included in our final meta-analysis. Odds ratios (OR), together with 95% confidence intervals (CIs), were used to assess the strength of associations between polymorphisms of PTPRJ and the risk of cancer. The search strategy is reported according to the PRISMA (http://www.prisma-statement.org/) reporting guidelines.
Meta-analysis—statistical analysis
The number of alleles was calculated according to the following formula: NA = n × q, where n is the total number of genotypes in the group, and q is the probability of the allele (R and Q for both polymorphisms) (Łaczmański et al. 2015).
The fixed-effects model and the DerSimonian-Laird random-effects model (with weights based on the inverse variance) were used to calculate summary odds ratios (ORs), and both within- and between-study variations were considered. Variants of this model were considered both within our study and for other studies. The significance level was set at 5% and random-effects analysis was selected. All statistical analyses were performed using Statistica ver. 10 software (StatSoft, USA) with the add-on Medical Package.
Results
Five articles describing the association between PTPRJ Arg326Gln and Gln276Pro polymorphisms in six different cancers (breast, colorectal, oesophagus, head and neck, lung and thyroid) were included in the analysis. In total, for the Arg326Gln polymorphism, 852 cases and 1071 controls while, for the Gln276Pro polymorphism, 2930 cases and 3408 controls were included in the pooled analyses (Tables 1, 2).
Table 1.
Characteristics of studies included in the meta-analysis of the SNP at C>T, Arg326Gln (rs1503185) of PTPRJ
| Study | Area | Race | Type of cancer | No. of cases | No. of controls | Genotypes in case group n (%) | Genotypes in controls n (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Arg326Gln (R326Q) (rs1503185) | Arg326Gln (R326Q) (rs1503185) | ||||||||||
| RR | RQ | RR | RQ | ||||||||
| Iuliano et al. 2004 | Italy | Caucasian | TA TC |
22 66 |
54 54 |
11 45 |
10 17 |
1 4 |
32 32 |
20 20 |
2 2 |
| Mita et al. 2010 | Japan | Asian | LAD LSQ HNSCC CRC ESCC |
108 59 92 113 193 |
819 819 819 819 819 |
66 36 58 51 106 |
34 17 27 45 77 |
8 6 7 17 10 |
510 510 510 510 510 |
272 272 272 272 272 |
37 37 37 37 37 |
| Wei et al. 2013 | China | Japan | BC | 199 | 198 | 104 | 75 | 20 | 87 | 92 | 19 |
TA thyroid adenoma, TC thyroid carcinoma, CRC colorectal cancer, LAD lung adenocarcinoma, LSQ lung squamous cell carcinoma, HNSCC head and neck squamous cell carcinoma, ESCC oesophageal cancer, BC breast cancer
Table 2.
leftacteristics of studies included in the meta-analysis of the SNP at G>T, Gln276Pro (rs1566734) of PTPRJ
| Study | Area | Race | Type of cancer | No. of cases | No. of controls | Genotypes in case group n (%) | Genotypes in controls n (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gln276Pro (Q276P) (rs1566734) | Gln276Pro (Q276P) (rs1566734) | ||||||||||
| QP | PP | QP | PP | ||||||||
| Iuliano et al. 2004 | Italy | Caucasian | TA | 22 | 126 | 11 | 10 | 1 | 32 | 20 | 74 |
| TC | 66 | 54 | 45 | 17 | 4 | 32 | 20 | 2 | |||
| Puijenbroek et al. 2005 | The Netherlands | Caucasian | CRC | 222 | 156 | 149 | 64 | 9 | 103 | 47 | 6 |
| Toland et al. 2008 | Israel | Mostly of Ashkenazi Jewish ancestry | CRC | 1897 | 1954 | 1047 | 701 | 149 | 1094 | 717 | 143 |
| Mita et al. 2010 | Japan | Asian | LAD LSQ HNSCC CRC ESCC |
108 59 92 113 193 |
814 814 814 814 814 |
66 36 58 51 106 |
34 17 27 45 77 |
8 6 7 17 10 |
499 499 499 499 499 |
281 281 281 281 281 |
34 34 34 34 34 |
| Iuliano et al. 2010 | Finland France Italy |
Caucasian | TC | 156 | 299 | 111 | 36 | 9 | 197 | 91 | 11 |
TA thyroid adenoma, TC thyroid carcinoma, CRC colorectal cancer, LAD lung adenocarcinoma, LSQ lung squamous cell carcinoma, HNSCC head and neck squamous cell carcinoma, ESCC oesophageal cancer, BC breast cancer
Arg326Gln
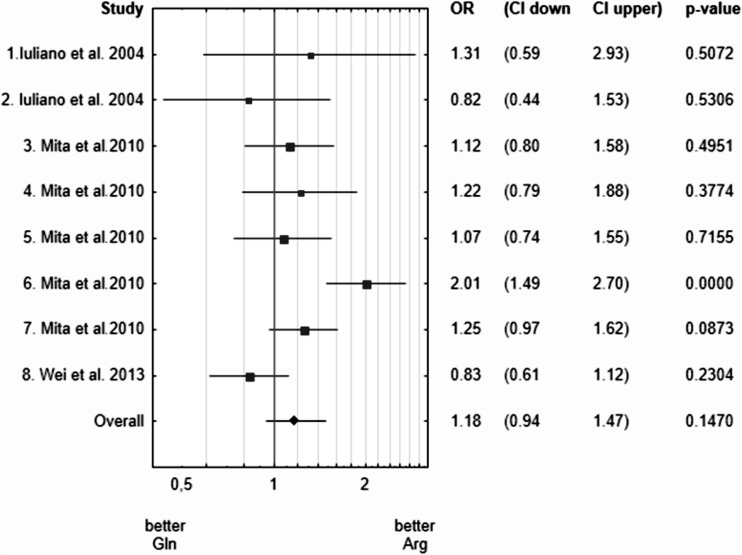
The presence of the Arg (C) allele is associated with lower risk of different cancers. The strongest association was observed for colorectal cancer patients (Fig. 1).
Fig. 1.
A Forest plot for meta-analysis of association between Arg326Gln PTPRJ polymorphism and cancer risk. 1, Thyroid adenomas; 2, Thyroid carcinomas; 3, LAD; 4, LSQ; 5, HNSCC; 6, CRC; 7, ESCC; 8, BC
Gln276Pro
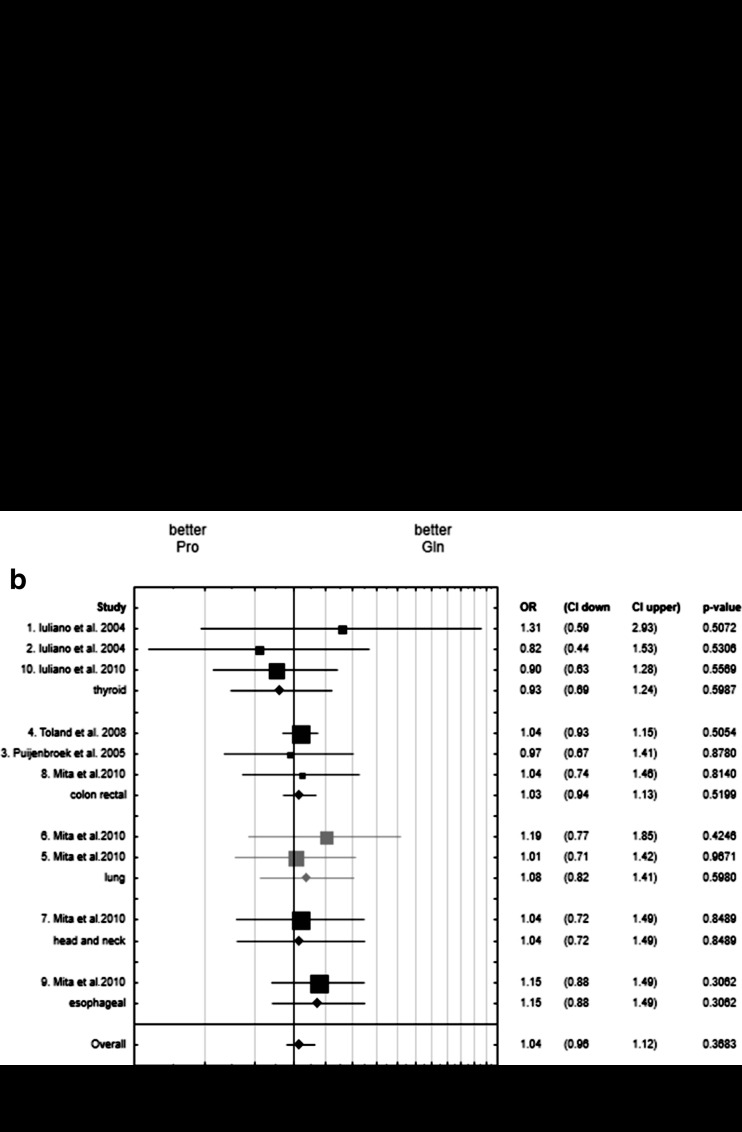
There was no association between Gln276Pro (G>T) polymorphism and cancer risk (Fig. 2).
Fig. 2.
a Forest plot for meta-analysis of association between Gln276Pro PTPRJ polymorphism and cancer risk. 1, Thyroid adenomas; 2, Thyroid carcinomas; 3, CRC; 4, CRC; 5, LAD; 6, LSQ; 7, HNSCC; 8, CRC; 9, ESCC; 10, Thyroid carcinomas. b Forest plot for meta-analysis of association between Gln276Pro PTPRJ polymorphism and cancer risk in different sub-groups of cancers
Discussion
The dual role of protein phosphatases makes them one of the most important gene groups (together with kinase genes) in cancer development and progression. Many studies on protein tyrosine phosphatase alterations at epigenetic and genetic levels confirm that loss of their function is a characteristic feature for cancer cells. Beside genetic alterations such as loss of heterozygosity, point mutations and aberrant methylation, also single nucleotide polymorphisms in protein tyrosine phosphatase genes are suspected to be responsible for variable risk of cancers in patients.
Due to the fact that data from many studies on different types of cancers are nowadays easily available, the meta-analysis of a huge number of results is possible. The strength of meta-analysis lies in its ability to collect together all published data from different laboratories for different ethnic groups of patients. In our meta-analysis, to our best knowledge, we analysed all published data for PTPRJ Arg326Gln and Gln276Pro polymorphisms in different cancers (breast, colorectal, oesophagus, head and neck, lung and thyroid cancer).
Our meta-analysis revealed that:
-
For Arg326Gln, the presence of the Arg (C) allele was associated with lower risk of some cancers and the strongest association was observed for colorectal cancer patients; however, the data for CRC were obtained only from one study (Mita et al. 2010).
The association for Arg326Gln we revealed was strongly influenced by data for 113 patients with CRC from Mita et al. 2010. When these data were excluded from the analysis there was no association of Arg326Gln and cancer risk (p value = 0.2959).
There was no association between Gln276Pro (G>T) polymorphism and cancer risk.
Cancer development and progression are complex processes, and, except for major impact genes, there is usually no simple answer as to which moderate and especially minor impact genes are important for them. Genetic polymorphisms (mainly SNPs) have been widely studied for many years, but although many experiments show the statistical significance of these variables, their effect is always dependent on other genetic and environmental factors and cannot be precisely defined. A meta-analysis of all available data can show which genetic changes are important and which may be excluded from further studies.
Both polymorphisms we studied are missense SNPs, and the changed amino acids are located in the extracellular region of PTPRJ involved in fibronectin-like type III domain formation. Because the role of PTPRJ in cancers has been widely described and many studies confirm its role in this process, we may suppose that the location of Gln276Pro (rs1566734) and Arg326Gln (rs1503185) polymorphisms in PTPRJ or amino acid changes and what they underlie are not important for the role of PTPRJ in carcinogenesis. Nevertheless, the association of the presence of the Arg (C) allele for the Arg326Gln polymorphism with lower risk especially for colorectal cancer seems to be an interesting subject for further studies to finally confirm or not the results of Mita et al. (2010).
Some limitations of our meta-analysis should be pointed out, such as the low number of studies included in the analysis (only five publications) with only 852 cases and 1071 controls for Arg326Gln and 2930 cases and 3408 controls for the Gln276Pro polymorphism. The ethnicity of patients was also different, for the Arg326Gln polymorphism the majority of patients were Asians, while for Gln276Pro the biggest group had Ashkenazi Jewish ancestry, and the second in number was Asian. Despite these considerations, our results were surprisingly consistent for all analysed cancer sub-types. Another limitation of our paper is that the meta-analysis included one study with 5 analyses of different cancer patients groups for which there was only a single control group. However, the control group consisted of more than 800 healthy individuals (Mita et al. 2010). In our opinion, one group of healthy individuals as a control group for 5 different cancer groups is permissible, because the segregation of polymorphic alleles in healthy individuals should be in Hardy-Weinberg equilibrium.
Conclusion
The polymorphisms Arg326Gln and Gln276Pro of the PTPRJ gene are not associated with an increased risk of cancer except for the Arg326Gln polymorphism in colorectal cancer. Thus, further large-scale studies should be performed to verify the impact of this SNP on individual susceptibility to colorectal cancer for given individuals.
Acknowledgements
The authors are grateful to Pawel Karpinski and Lukasz Laczmanski for their support in preparing the manuscript.
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aya-Bonilla C, Green MR, Camilleri E et al (2013) High-resolution loss of heterozygosity screening implicates PTPRJ as a potential tumor suppressor gene that affects susceptibility to non-Hodgkin’s lymphoma. Genes Chromosom Cancer. 10.1002/gcc.22044 [DOI] [PubMed]
- Bilotta A, Dattilo V, D’Agostino S et al (2016) A novel splice variant of the protein tyrosine phosphatase PTPRJ that encodes for a soluble protein involved in angiogenesis. Oncotarget. 10.18632/oncotarget.14350 [DOI] [PMC free article] [PubMed]
- Fournier P, Dussault S, Fusco A et al (2016) Tyrosine phosphatase PTPRJ/DEP-1 is an essential promoter of vascular permeability, angiogenesis, and tumor progression. Cancer Res. 10.1158/0008-5472.CAN-16-1071 [DOI] [PubMed]
- Iuliano R, Le PI, Cristofaro C, et al. The tyrosine phosphatase PTPRJ/DEP-1 genotype affects thyroid carcinogenesis. Oncogene. 2004;23:8432–8438. doi: 10.1038/sj.onc.1207766. [DOI] [PubMed] [Google Scholar]
- Iuliano R, Palmieri D, He H et al (2010) Role of PTPRJ genotype in papillary thyroid carcinoma risk. Endocr Relat Cancer. 10.1677/ERC-10-0143 [DOI] [PMC free article] [PubMed]
- Julien SG, Dubé N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. 2011;11:35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- Katsonis P, Koire A, Wilson SJ, et al. Single nucleotide variations: biological impact and theoretical interpretation. Protein Sci. 2014;23:1650–1666. doi: 10.1002/pro.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łaczmański Ł, Jakubik M, Bednarek-Tupikowska G, et al. Vitamin D receptor gene polymorphisms in Alzheimer’s disease patients. Exp Gerontol. 2015;69:142–147. doi: 10.1016/j.exger.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Mita Y, Yasuda Y, Sakai A et al (2010) Missense polymorphisms of PTPRJ and PTPN13 genes affect susceptibility to a variety of human cancers. J Cancer Res Clin Oncol. 10.1007/s00432-009-0656-7 [DOI] [PubMed]
- Ortuso F, Paduano F, Carotenuto A, et al. Discovery of PTPRJ agonist peptides that effectively inhibit in vitro cancer cell proliferation and tube formation. ACS Chem Biol. 2013;8:1497–1506. doi: 10.1021/cb3007192. [DOI] [PubMed] [Google Scholar]
- Qiao D, Li M, Pu J, et al. Loss of protein tyrosine phosphatase receptor J expression predicts an aggressive clinical course in patients with esophageal squamous cell carcinoma. Pathol Oncol Res. 2016;22:541–547. doi: 10.1007/s12253-015-0036-3. [DOI] [PubMed] [Google Scholar]
- Ruivenkamp C, Hermsen M, Postma C et al (2003) LOH of PTPRJ occurs early in colorectal cancer and is associated with chromosomal loss of 18q12-21. Oncogene. 10.1038/sj.onc.1206246 [DOI] [PubMed]
- Shangkuan W-C, Lin H-C, Chang Y-T, et al. Risk analysis of colorectal cancer incidence by gene expression analysis. PeerJ. 2017;5:e3003. doi: 10.7717/peerj.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring K, Fournier P, Lapointe L, et al. The protein tyrosine phosphatase DEP-1/PTPRJ promotes breast cancer cell invasion and metastasis. Oncogene. 2015;34:5536–5547. doi: 10.1038/onc.2015.9. [DOI] [PubMed] [Google Scholar]
- Steinman KJ, Spence SJ, Ramocki MB, et al. 16p11.2 deletion and duplication: characterizing neurologic phenotypes in a large clinically ascertained cohort. Am J Med Genet Part A. 2016;170:2943–2955. doi: 10.1002/ajmg.a.37820. [DOI] [PubMed] [Google Scholar]
- Toland AE, Rozek LS, Presswala S et al (2008) PTPRJ haplotypes and colorectal cancer risk. Cancer Epidemiol Biomark Prev. 10.1158/1055-9965.EPI-08-0513 [DOI] [PubMed]
- Venkatachalam R, Ligtenberg M, Hoogerbrugge N et al (2010) Germline epigenetic silencing of the tumor suppressor gene PTPRJ in early-onset familial colorectal cancer. Gastroenterology. 10.1053/j.gastro.2010.08.063 [DOI] [PubMed]
- Wei W, Jiang M, Luo L, et al (2013) Colorectal cancer susceptibility variants alter risk of breast cancer in a Chinese Han population. funpecrp.com.br Genet Mol Res Genet Mol Res 12:6268–6274. 10.4238/2013.December.4.14 [DOI] [PubMed]
- Yan CM, Zhao YL, Cai HY et al (2015) Blockage of PTPRJ promotes cell growth and resistance to 5-FU through activation of JAK1/STAT3 in the cervical carcinoma cell line C33A. Oncol Rep. 10.3892/or.2015.3769 [DOI] [PubMed]
- Zhang XF, Tu R, Li K et al (2017) Tumor suppressor PTPRJ is a target of miR-155 in colorectal cancer. J Cell Biochem. 10.1002/jcb.25995 [DOI] [PubMed]
- Zhao S, Sedwick D, Wang Z. Genetic alterations of protein tyrosine phosphatases in human cancers. Oncogene. 2015;34:3885–3894. doi: 10.1038/onc.2014.326. [DOI] [PMC free article] [PubMed] [Google Scholar]