Abstract
How cells build their internal structures remains one of the central mysteries in cell biology. If the cell nucleus is anything to go by, stochastic assembly and self-organization seem to be key.
As anyone knows who has ever put together one of those home-assembly bookshelves, most man-made structures can only be built by following a defined sequence of steps. It is difficult to set aside this preconceived notion of linear assembly when thinking about how cellular structures emerge. Yet a landmark paper by Kaiser, Intine and Dundr1, published in Science, makes a powerful case for the formation of biological structures by apparently random pathways and self-organization.
The mammalian cell nucleus is a prototypical, highly organized biological structure2. Not only does it hold most of an individual’s genetic information, but it is also the site of many essential processes, such as reading, copying and repairing the genome. To coordinate these events — and presumably to make them more efficient — the nucleus is highly compartmentalized and contains numerous nuclear bodies that have distinct functions2.
Kaiser and colleagues1 set out to investigate the formation of a prominent nuclear compartment called the Cajal body, a roughly spherical structure of 0.5–1 micrometres diameter. There are typically several Cajal bodies in every nucleus, often near clusters of genes encoding histone proteins or small nuclear RNAs (snRNAs)3. Their precise function is unclear, but the fact that they contain factors involved in small-RNA processing suggests they might be involved in modifying nuclear RNAs and in recycling RNA-processing factors3. Like all other nuclear compartments, Cajal bodies lack a membrane, and are highly dynamic yet stable steady-state structures, exchanging their proteins rapidly and continuously with the surrounding nuclear space2.
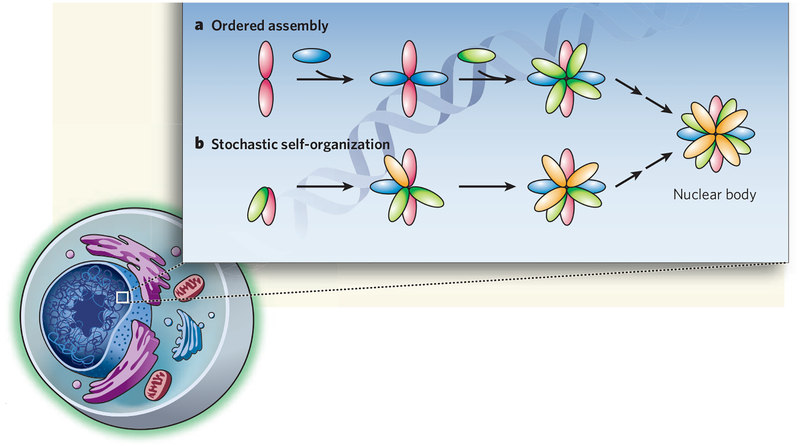
How Cajal bodies, and indeed all other nuclear bodies, are formed and maintained without membrane boundaries has been a puzzle. Two fundamentally different models have been considered4. One idea — akin to the bookshelf-assembly strategy — is that Cajal-body proteins bind to scaffold proteins in an orderly manner to gradually build up the structure. Indeed, scaffold function has been ascribed to two prominent Cajal-body marker proteins, coilin and SMN (Fig. 1a). Alternatively, it has been suggested that nuclear bodies form through self-organization, whereby their components simply aggregate in a stochastic, largely random way (Fig. 1b).
Figure 1 |. Formation of nuclear structures.
a, Nuclear bodies may assemble by the association of their constituents in a defined, sequential way, where the association of a factor with the growing body depends on the presence of others already there. b, Alternatively, they could form by random assembly of components through self-organization. Kaiser and colleagues’ data1 strongly support the self-organization model.
To distinguish between these two models, Kaiser et al.1 first asked a fundamental question: can Cajal bodies form de novo? The authors used a bacterial tethering system to immobilize individual Cajal-body proteins at an engineered random site in the genome of mammalian cells. Not entirely unexpectedly, coilin and SMN were each sufficient to form an apparently normal, functional Cajal body. Surprisingly, however, when the authors tethered minor Cajal-body components to the same genomic site, just about any Cajal-body component could initiate the formation of this structure, including all RNA-processing factors. In fact, minor components were more efficient at forming Cajal bodies than coilin and SMN.
These findings lead to two main conclusions. First, the formation of Cajal bodies does not require a specific gene locus, and could potentially occur anywhere in the genome. Second, this process does not require a strict hierarchical pathway, as any Cajal-body protein can initiate the formation of the entire structure.
These observations do not agree well with a linear-assembly model and point to self-organization as a driving force in the assembly. Several other features of Cajal bodies also favour a self-organization model. First, Kaiser and colleagues1 show that the recruitment of any native protein to a newly formed Cajal body occurs with similar kinetics, suggesting largely random assembly. Second, previous work has shown that coilin and SMN can self-assemble5. This tendency of the two proteins might be crucial for the assembly process, as Kaiser et al. show that Cajal bodies form less efficiently in the absence of either coilin or SMN, hinting that these marker proteins serve to stabilize transient interactions among other Cajal-body components. Third, Cajal-body proteins are highly dynamic, and move rapidly between this structure and its surroundings — a requirement for a self-organization process. But perhaps most telling is the authors’ finding1 that, despite the ready availability of building blocks, the size of the structure formed de novo is limited, a classic hallmark of self-organization.
Are these results relevant to other nuclear structures? Probably. For one thing, Kaiser and colleagues report that tethering of PML — a marker protein of another nuclear body — leads to the formation of a PML body. Furthermore, much of what we know about nuclear bodies suggests shared principles of organization. Like the Cajal body, all known nuclear bodies represent dynamic steady-state systems that undergo rapid exchange of components2. Moreover, other nuclear bodies also seem to form at specific nuclear sites. Whereas Cajal bodies preferentially associate with clustered histone and snRNA genes6, the nucleolus — the most prominent nuclear substructure — forms around sites of ribosomal-RNA transcription, and compartments enriched in factors mediating pre-messenger-RNA splicing form near the sites where RNA is transcribed. The likeliest possibility, therefore, is that nuclear bodies assemble at sites of high gene activity through the accumulation of freely diffusing proteins, which leads to the nucleation of a distinct body. In that sense, Kaiser and colleagues’ tethering experiments may simply recapitulate the immobilization these factors normally experience at their sites of action.
Having said that, the main caveat of this study1 is uncertainty about its physiological relevance. It is not known whether de novo assembly of Cajal bodies occurs naturally, or, if it does, whether it is a frequent occurrence. In fact, Dundr and colleagues previously observed that, when activated, snRNA gene loci can be recruited to pre-existing Cajal bodies7. So it is essential now to establish whether de novo assembly or recruitment of pre-assembled Cajal bodies is the preferred means of pairing up these structures with their target genes.
Regardless of this, the present work1 represents a major step forward. Not only does it provide a conceptual framework for probing nuclear-body formation further, but it also gives us a potential means of succeeding in one of the remaining quests of modern cell biology — finding out how cellular structures form in vivo.
References
- 1.Kaiser TE, Intine RV & Dundr M Science doi: 10.1126/science.1165216 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Misteli T Cell 128, 787–800 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Handwerger KE & Gall J Trends Cell Biol. 16, 19–26 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Misteli TJ Cell Biol. 155, 181–185 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebert MD & Matera AG Mol. Biol. Cell 11, 4159–4171 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shopland LS et al. Mol. Biol. Cell 12, 565–576 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dundr M et al. J. Cell Biol. 179, 1095–1103 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]