Abstract
Serine palmitoyltransferase (SPT), an endoplasmic reticulum-localized membrane enzyme composed of a catalytic LCB1/LCB2 heterodimer and a small activating subunit (Tsc3 in yeast; ssSPTs in mammals), is negatively regulated by the evolutionarily conserved family of proteins known as the ORMs. In yeast, SPT, the ORMs, and the PI4P phosphatase Sac1, copurify in the “SPOTs” complex. However, neither the mechanism of ORM inhibition of SPT nor details of the interactions of the ORMs and Sac1 with SPT are known. Here we report that the first transmembrane domain (TMD1) of Lcb1 is required for ORM binding to SPT. Loss of binding is not due to altered membrane topology of Lcb1 since replacing TMD1 with a heterologous TMD restores membrane topology but not ORM binding. TMD1 deletion also eliminates ORM-dependent formation of SPT oligomers as assessed by co-immunoprecipitation assays and in vivo imaging. Expression of ORMs lacking derepressive phosphorylation sites results in constitutive SPT oligomerization, while phosphomimetic ORMs fail to induce oligomerization under any conditions. Significantly, when LCB1–RFP and LCB1ΔTMD1–GFP were coexpressed, more LCB1ΔTMD1–GFP was in the peripheral ER, suggesting ORM regulation is partially accomplished by SPT redistribution. Tsc3 deletion does not abolish ORM inhibition of SPT, indicating the ORMs do not simply prevent activation by Tsc3. Binding of Sac1 to SPT requires Tsc3, but not the ORMs, and Sac1 does not influence ORM-mediated oligomerization of SPT. Finally, yeast mutants lacking ORM regulation of SPT require the LCB-P lyase Dpl1 to maintain long-chain bases at sublethal levels.
Keywords: serine palmitoyltransferase, ORMs, sphingolipids, Lcb1, Tsc3, Dpl1
INTRODUCTION
The sphingolipids are a complex family of molecules with crucial structural and signaling roles and perturbations in their synthesis, trafficking, and degradation underlie many human diseases (1). Thus, understanding the mechanisms responsible for maintaining homeostasis of the sphingolipids is an important goal. Serine palmitoyltransferase (SPT) catalyzes the committed and rate-limiting step in sphingolipid biosynthesis. In prokaryotes, SPT is a soluble homodimer that structural studies have shown contains two catalytic sites, each at the subunit interface (2). In eukaryotes, the enzyme is composed of three subunits, two of which, LCB1 and LCB2, are evolutionarily conserved and form a catalytically competent ER-localized heterodimer with a single active site at the dimer interface (3). The third subunit is a much less conserved small protein that increases the catalytic activity of the enzyme nearly 100-fold. While in yeast there is a single isoform of this protein (Tsc3) (4), in higher eukaryotes there are two, and sometimes three, highly related isoforms (ssSPTs) that also specify acyl-CoA chain preference (5).
While much progress has been made in elucidating the structural organization of SPT, determining the mechanism(s) that regulate its activity has proven more difficult. However, the Weissman (6) and Chang (7) laboratories provided major insights into how SPT is regulated by showing that the yeast ORM proteins negatively regulate SPT in response to sphingolipid availability. This negative regulation was shown to be relieved by phosphorylation of the N-terminal domains of the ORM proteins (6). In addition, the ORM proteins were shown to reside in a complex (SPOTS) with SPT that also contains Sac1 (a PI4P lipid phosphatase), and repressed SPT appeared to be localized to the peripheral ER, while derepressed SPT was largely perinuclear (6). Subsequently, it has been determined that phosphorylation of the N-terminal domain is catalyzed by Ypk1, which is regulated by TORC2 (8-10), and by Npr1, which is regulated by TORC1 (11).
In higher eukaryotic cells the ORM orthologues, the ORMDLs (of which there are three), also regulate SPT activity (6,12-14). However, they lack the N-terminal domain present in the yeast ORMs, and therefore do not appear to be regulated by phosphorylation. Rather, since silencing the ORMDLs increases long-chain base (LCB) synthesis, simple mass action may account for their regulation of SPT. This is consistent with our previous report that increasing SPT activity results in a compensatory increase in ORMDL expression that is required to maintain sphingolipid homeostasis (12).
Since the catalytic heterodimers of yeast and mammalian SPT are activated by a small third subunit, it is possible that ORM repression is the result of antagonism of the activity of the activating subunits. Alternatively, the ORMs could directly inhibit the catalytic heterodimer. Because phosphorylation of wild-type yeast ORMs is regulated by sphingolipid availability, and ORM phosphorylation (and derepression) would therefore be expected to increase in the absence of the activating small subunit, these possibilities are difficult to distinguish. However, mutation of the YPK phosphorylation sites of the yeast ORMs results in proteins that are constitutive repressors of SPT activity (6). In the present study, we have used these mutants to show that the ORMs bind to and directly inhibit the catalytic heterodimer.
Regardless of the mechanism by which the ORMs regulate SPT activity, binding to SPT is required. Our topological analysis of heterotrimeric SPT showed that: i) LCB1 has three transmembrane domains with its N-terminus in the ER lumen and its C-terminus in the cytosol (15); ii) LCB2 has two transmembrane domains with both the N- and C-termini in the cytosol (unpublished); and iii) the small activating subunit has a single transmembrane domain with its N-terminus in the cytosol (16). In addition, Han et al. have shown that Lcb1 is sufficient to bind the yeast ORMs (7). Furthermore, the N-terminal domain, including TMD1, of Lcb1 can be deleted without compromising membrane targeting or enzymatic activity (15). Thus, TMD1 could be a regulatory domain important for interaction with the ORMs, integral ER membrane proteins containing four membrane spanning domains (17).
Based on the interaction between epitope-tagged hLCB2b (SPTLC3) and hLCB2a (SPTLC2), blue-native gel electrophoresis and size exclusion chromatography of solubilized SPT, Hornemann et al. proposed that mammalian SPT is an octamer of catalytic heterodimers (18). In addition, Breslow et al. proposed that the yeast SPOTS complex also exists in higher order structures whose assembly is regulated by sphingolipid availability (6). Both the yeast and mammalian ORM proteins have themselves been proposed to exist in higher order complexes (6,19), but it is unclear whether the formation of SPT multimers is dependent upon ORM oligomerization. It is also unclear what role, if any, ORM oligomerization plays in the regulation of SPT activity. In the present study, we have defined a region of LCB1 required for ORM binding and used mutants in which this region has been deleted to address these questions. Our results suggest that SPT multimerization is ORM-dependent, and that, like many other enzymes, its activity is regulated by its multimeric state.
MATERIALS AND METHODS
Reagents
Tunicamycin, stearylamine, sucrose monolaurate (SML), Triton X-100 (TX100), and myriocin were purchased from Sigma. Digitonin (DIG) and n-dodecyl-β-d-maltoside (DDM) were purchased from Anatrace. Endoglycosidase H, restriction enzymes, DNA polymerases and DNA modifying enzymes were from New England Biolabs. QuikChange mutagenesis kits were from Agilent. PHS, and DHS were from Avanti Polar Lipids and 3-ketosphinganine was from Matreya. The Phos-tag™ reagent was purchased from Wako Chemicals. [3]H-serine (30 Ci/mmol) was obtained from PerkinElmer. Anti-FLAG beads were from Sigma-Aldrich and GFP-Trap®_A beads were from Chromotek. Yeast and bacterial media were obtained from BD Difco. 3-amino-1,2,4-triazole (3-AT) was from MP Biomedicals, LLC. AccQ-Tag, used to derivatize long-chain bases for detection, was from Waters.
Antibodies
The rabbit polyclonal anti-Lcb1 and anti-Lcb2 antibodies were generated using recombinant yeast proteins prepared from E. coli. as previously described (4). Rabbit anti-hLCB1 antibodies were from BD Transduction Laboratories. Rabbit polyclonal anti-Tsc3, anti-Orm1, anti-Orm2 and anti-Elo3 antibodies were made using peptides as immunogens. For anti-Tsc3, two pooled peptides, MTQHKSSMVYIC, residues 1-11, and CIPTTKEAKRRNGKSE, residues 11-25, (cysteines added for coupling) were used. The peptides were synthesized and the antibodies were generated by Covance. The antibodies were validated using microsomes prepared from a yeast tsc3Δ mutant and from yeast expressing epitope-tagged Tsc3 proteins. For anti-Orm1, a single peptide (SASSSIKTTEPVKDHRC, residues 32-47) was used; for anti-Orm2, two pooled peptides, SQSNKISTPVTDHRC, residues 29-42, and SHVEQETFEDENDQC, residues 51-64, were used; and for anti-Elo3, a single peptide KKTVKKESEVSGSVASGS, residues 311-328, was used. The Orm1, Orm2 and Elo3 peptides and antibodies were produced by New England Peptide, Inc. Antibodies were affinity purified using CNBr-activated Sepharose 4B beads (GE Healthcare Life Sciences) to which the immunizing peptides were covalently coupled according to the manufacturer’s instructions. Antibodies were validated using yeast knockout mutants and yeast expressing epitope-tagged variants of the Orm1, Orm2, or Elo3 proteins. HRP-conjugated goat anti-rabbit and anti-mouse secondary antibodies from Bio-Rad were used for detection. HA or FLAG-tagged proteins were detected using commercially available HRP-conjugated anti-HA (Covance) or anti-FLAG (GenScript) antibodies.
Cells and Growth Conditions
The yeast strains used in this study are listed in Table 1. Yeast cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or in synthetic complete medium with 2% glucose (SD) according to standard procedures (20). CHO LyB cells were grown in Ham’s F-12 medium (Invitrogen) supplemented with 10% fetal bovine serum (Biofluids, Inc.), 100 U/ml penicillin and 100 μg/ml streptomycin. CHO LyB cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Table 1.
Strains used in this study
| Strains | Genotype | Source |
| TDYG4110 | MATa his3Δ1 leu2Δ0 met15Δ 0 lys2Δ0 ura3Δ0 lcb1:: KanMX4 | Open biosystems |
| ACX164-1C | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Orm1::NatRorm2::HIS3 | Chang lab |
| TDYK1085 | MATα his3Δ1 leu2Δ0 ura3Δ0 orm1:: NatRorm2:: HIS3 lcb1:: KanMX4 | Dunn lab |
| TDYG4a | MATa his3Δ1 leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 | Dunn lab |
| TDYD513 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lcb1::LCB1ΔTMD1 | Dunn lab |
| TDYD529 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Orm1::NatRorm2::HIS3, lcb1 ::LCB1ΔTMD1 | Dunn lab |
| TDYD514 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lcb1:LCB1TMDlFVT | Dunn lab |
| TDYG12115 | MATα his3Δ1 leu2Δ0 ura3Δ0 Orm1::NatRorm2::HIS3, lcb1:: LCB1TMD1FVT | Dunn lab |
| TDYG1197 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ypk1:: KanMX4 | Open biosystems |
| TDYG1199 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ypk2:: KanMX4 | Open biosystems |
| TDYG12040 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ypk1:: KanMX4 ypk2:: KanMX4 | Dunn lab |
| TDYG12100 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ypk1:: KanMX4 ypk2:: KanMX4 +pRS315LCB1ΔTMD1 | Dunn lab |
| TDYG12073 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ypk1:: KanMX4 ypk2 :: KanMX4 Orm1::NatR | Dunn lab |
| TDYG12083 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ypk1:: KanMX4 ypk2:: KanMX4 orm2:: HIS3 | Dunn lab |
| TDYG8040 | MATa, trp1-901 leu2-3,112 his3-200 lys2-;801 ade2 ura3Δ0 LYS2:: lexA-HIS3 ura3:: lexA-lacZ | Dunn lab |
| TDYG9113 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trp1Δ lcb1:: KanMX4 tsc3::NatR | Dunn lab |
| TDYG1191 | MATa his3Δ1 leu2Δ0 met15Δ60 ura3Δ0 sacl:: KanMX4 | Open biosystems |
| TDY14038 | MATa his3Δ1 leu2Δ0 lysΔ2Δ0 ura3Δ0 lcb1:: KanMX4 sac1:: KanMX4 | Dunn lab |
| TDYG14027 | MATa his3Δ1 leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 lcb1:: KanMX4 sac1:: KanMX4 tsc3::NatR | Dunn lab |
| TDYG6028 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 lcb2:: KanMX4 | Open biosystems |
| TDYG6032 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 lcb2:: KanMX4 | Open biosystems |
| TDYG6041 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 lcb1:: KanMX4 lcb2:: KanMX4 | Dunn lab |
| TDYG1211 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lcb2:: KanMX4 sacl:: KanMX4 | Dunn lab |
| TDYG1152 | MATΔ his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Orm1::NatRorm2::HIS3 lcb2:: KanMX4 | Dunn lab |
| TDYG4b | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Dunn lab |
| TDYG4071 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tsc3::NatR | Dunn lab |
| TDYK1145 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Orm1::NatRtsc3::URA | Dunn lab |
| TDYK1147 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 orm2::HIS3 tsc3::URA | Dunn lab |
| TDYG1130 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Orm1::NatRorm2::HIS3 tsc3::URAFOA | Dunn lab |
| TDYG1150 | MATα his3Δ1 leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 orm2::HIS3 lcb2:: KanMX4 | Dunn lab |
| TDYK1166 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 dpl1:: KanMX4 | Open biosystems |
| TDYK1544 | MATα his3Δ1 leu2Δ0 ura3Δ0 Orm1::NatRorm2::HIS3 dpl1:: KanMX4 | Dunn lab |
| TDYK1135 | MATα his3Δ1 leu2Δ0 ura3Δ0 Orm1::NatRorm2::HIS3 dpl1:: KanMX4 tsc3::URA3 | Dunn lab |
Plasmids
Plasmids were constructed using standard procedures and relevant features are listed in Table 2. For the construction of yeast Lcb1TMD1FVT1, a plasmid containing Lcb1 with codons 50 to 85 of Lcb1 replaced with an NheI site (15) was linearized with Nhe1 and co-transformed with a PCR-generated fragment containing TMD1 of FVT1 (codons 2-26) flanked with 50 base pairs upstream and 50 base pairs downstream of TMD1 of Lcb1. After allowing for in vivo recombination, the plasmid (pTDG-3, Table 2) was extracted, amplified by passage through E. coli, and the construct was confirmed by DNA sequencing. To replace TMD1 of hLCB1 with TMD1 from yeast Lcb1, the N-terminal domain (codons 1 to 40) of hLCB1 was replaced with the yeast N-terminal domain (codons 1-85) by a similar in vivo recombination strategy to generate pTDG-24 (Table 2). The pAL2-URA plasmid was constructed by replacing the GAL1 and GAL10 promoters of pESC-URA with the yeast LCB2 and ADH2 promoters respectively. A fragment encoding E. coli biotin ligase (birA) was PCR-amplified for insertion immediately before the stop codon of Lcb1 in a yeast expression vector and the R118G mutation (21) was introduced into BirA by QuikChange mutagenesis.
Table 2.
Plasmids used in this study
| Names | Plasmids | Description |
|---|---|---|
| pTDG-1 | pRS315 | LCB1, CEN LEU |
| pTDG-2 | pRS315 | LCB1ΔTMD1, CEN LEU |
| pTDG-3 | pRS315 | LCB1TMD1FVT CEN LEU |
| pTDG-4 | PRS315 | LCB1-Cub-LexA-VP16, CEN LEU |
| pTDG-5 | pRS315 | LCB1ΔTMD1- Cub-LexA-VP16, CEN LEU |
| pTDG-6 | pRS315 | LcB1-3xFLAG CEN LEU |
| pTDG-7 | pRS315 | LcB1ΔTMDl-3x FLAG, CEN LEU |
| pTDG-8 | pRS315 | LCB1TMD1FVt-3x FLAG, CEN LEU |
| pTDG-9 | pRS315 | LCB1 GC@4, CEN LEU |
| pTDG-10 | pRS315 | LCB1TMD1FVT GC@4, CEN LEU |
| pTDG-11 | pRS315 | LCB1-GFP, CEN LEU |
| pTDG-12 | pRS315 | LCB1-RFP, CEN LEU |
| pTDG-13 | pRS315 | LCB1ΔTMD1-GFP, CEN LEU |
| pTDG-14 | PRS316 | LCB2-GFP@82, CEN URA |
| pTDG-14-1 | PRS315 | LCB2-GFP@82, CEN LEU |
| pTDG-15 | pRS316 | LCB2-Nub, CEN URA |
| pTDG-15 | pRS316 | ORM2 CEN URA |
| pTDG-17 | pRS316 | ORM2-3A, S46A S47A S48A CEN URA |
| pTDG-18 | pRS316 | ORM2-3D, S46D S47D S48D CEN URA |
| pTDG-19 | pRS316 | Nub-Orm1, CEN URA |
| pTDG-20 | pRS316 | RFP-ORM2, CEN URA |
| pTDG-21 | pRS316 | Nub-ORM2, CEN URA |
| pTDG-22 | pRS316 | 3x FLAG-ORM2, CEN URA |
| pTDG-23 | pRS316 | 3xHA-Sac1, CEN URA |
| pTDG-24 | pRS315 | hLCB1TMD1yeast CEN URA |
| pTDG-25 | pRS313 | hLcb1, CEN HIS3 |
| pTDG-27 | pRS316 | hLCB2a, CEN URA |
| pTDG-28 | pPR3N | Nub-HA-ssSPTa BS TRP |
| pTDG-29 | pESC-URA | ORM1 (GAL10) / ORM2 (GAL1) 2μ URA |
| pTDG-30 | pESC-URA | ORM1-3A (GAL10) /ORM2-3A (GAL1) 2μ URA |
| pTDG-31 | pESC-URA | ORM1-3D (GAL10) /ORM2-3D (GAL1) 2μ URA |
| pTDG-32 | pAL2-URA | ORM2 (pLCB2) 2μ URA |
| pTDG-33 | pAL2-URA | ORM2-3A (pLCB2) 2μ URA |
| pTDG-34 | pAL2-URA | ORM2-3D (pLCB2) 2μ URA 1 |
| pTDG-35 | pRS315 | Lcb1-BirA CEN LEU |
| pTDS-1 | pCMV6-XL5 | hLCB1C133W |
| pTDS-2 | pCMV6-XL5 | hLCB1c133W ΔTMD1 |
| pTDS-3 | pCMV6-XL4 | hLCB2a |
| pTDS-4 | pcDNA3.1 (+) | NubG-HA-ORMDL3, pCMV |
| pTDS-5 | pcDNA3.1 (+) | 3x FLAG-ORMDL3, pCMV |
| pTDS-6 | pcDNA3.1 (+) | NubG-HA-ssSPTa, pCMV |
Yeast microsomal membrane preparation
Cells in exponential growth were pelleted at 5000 x g, and washed with water and with TEGM buffer (0.05 M Tris (pH 7.5), 1 mM EGTA, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml aprotinin). Cell pellets were resuspended in TEGM buffer at 50 OD cells/ 1 ml and glass beads (0.5 mm diameter zirconia beads (BioSpec)) were added to the meniscus. Cells were disrupted by vortexing 4 x 1 min, with 1 min cooling on ice between vortexing. The lysate was transferred to a fresh tube along with several washings of the beads and the sample was centrifuged at 8000 x g for 10 min. The supernatant was transferred to a new tube and spun at 100,000 x g for 30 min. The resulting pellet was resuspended by Dounce homogenization in at least 10 volumes of TEGM buffer and repelleted at 100,000 x g. The final membrane pellet was resuspended in TEGM containing 33% glycerol and stored at −80°C. Protein concentration was determined by Bio-Rad dye reagent using IgG as standard.
CHO-LyB microsomal membrane preparation
CHO-LyB cells were harvested by scraping, washed with PBSE (phosphate-buffered saline containing 1 mM EGTA and 1 mM EDTA) and transferred into 15 ml conical tubes. Following centrifugation at 1000 × g for 5 min, the cell pellets were resuspended in TEGM buffer. Following cell lysis by sonication (three 6 sec pulses at a setting of 5), the lysate was centrifuged at 8000 × g for 10 min. Microsomes were recovered from the supernatant by centrifugation at 100,000 × g for 30 min, and the microsomal pellet was washed and resuspended in 100 μl TEGM buffer containing 15% glycerol.
SPT assay
The reaction was initiated by adding 200 μg of microsomal membrane to a reaction cocktail (final volume 300 μl) containing 50 mM HEPES (pH, 8.1), 50 μM pyridoxal-5’-phosphate, 2.5 mM serine, 20 μCi [3]H-serine, 100 μM palmitoyl-CoA and 20 μM BSA. After 10 min at 37 °C, NH4OH to 0.25 M was added, followed by the addition of 2 ml of chloroform:methanol (1:2), and the sample was vortexed. Long chain bases (LCBs) were extracted by adding 1 ml chloroform and 2 ml of 0.5 M NH4OH, vortexing and brief centrifugation. The upper aqueous layer was aspirated off and the lower layer was washed with 2 ml of 60 mM KCl and centrifuged. The washing was repeated and 1 ml of the sample was dried under nitrogen and counted in a Packard Tri-Carb scintillation counter.
Extraction and quantification of LCBs by HPLC
Total LCBs were isolated from 5 OD600 of exponentially growing yeast cells by HCl methanolysis. Briefly, washed cells were resuspended in 1 ml 1 N methanolic HCl (Supelco, Bellefonte, PA) and the tubes were placed in boiling water for 30 min. After cooling on ice, 1 ml of 0.9% NaCl and 2 ml hexane:diethyl ether (1:1) were added and following centrifugation the upper phase was removed and the LCBs were extracted from the lower phase by adding 0.25 ml 10 N NaOH, vortexing and then adding 2 ml hexane and vortexing. Following centrifugation, 1.5 ml of the upper phase was dried under N2, resuspended in 80 μl of methanol: 190 mM triethylamine (2:0.3, v/v) and 20 μl of AccQ reagent (Waters, Milford, MA) was added and allowed to react for 60 min. Derivatized LCBs from 1.0 OD600 of yeast cells were analyzed by HPLC as described below.
Free LCBs were extracted from 5 OD600 of exponentially growing cells. Cells were pelleted, washed with water and resuspended in 200 μl 1 N NH4OH. Zirconia beads (0.5 mm, BioSpec) were added to the meniscus and the pellets were vortexed for 90 sec. LCBs were extracted by the addition of 2 ml chloroform:methanol (1:2) containing 0.25 mM C17-sphingosine standard (Avanti). The extract was vortexed an additional 90 sec and transferred to a new tube. The beads were washed with an additional 1 ml chloroform:methanol (1:2) without standards, and the combined extracts were briefly centrifuged to remove beads and debris. Chloroform (1.5 ml) was added to the extract, which was then partitioned into phases by the addition of 3 ml 0.5 N NH4OH. After centrifugation, the upper aqueous layer was aspirated off and the lower organic phase was washed 2 times with 60 mM KCl. The final cleared organic phase was dried and resuspended in 100 μl methanol:triethylamine (20:3). Eighty μl of the final extract was transferred to an HPLC vial and 20 μl AccQ reagent (Waters) was added to derive the LCBs. After 60 min, the reaction was subjected to base hydrolysis using 0.1 N KOH. The equivalent of 3.2 OD600 was then separated by HPLC as described below.
HPLC analysis of AccQ-derivatized LCBs
AccQ-derivatized LCB samples were separated on a reverse phase 0.46 × 25 cm C18 column (4 μm) (GraceVydac) using an HP 1100 liquid chromatograph coupled to an Agilent 1100 series fluorescence detector. The LCBs were resolved using solvent A (acetonitrile:methanol: water:acetic acid:triethylamine (480:320:165:30:7, v/v)) for 60 min at a flow rate of 1.5 ml/min. The AccQ-derivatized LCB peaks were identified using fluorescence excitation at 244 nm and 398 nm emission. Between runs, the column was washed using solvent B (acetonitrile:methanol, 60:40, v/v) at 1 ml/min for 6 min before returning to solvent A for 8 min at 1.5 ml/min.
Quantification of LCBs from the orm1Δorm2Δdpl1Δ mutant by mass spectrometry
Cells were cultured overnight in YPD media containing 90 ng/ml myriocin to an OD600nm of ~0.5, washed with water and transferred to YPD media at 0.1 OD600/ml. After collection of all time points, LCBs were extracted from frozen cells by the addition of 1 ml water: ethanol: diethyl ether: pyridine: NH4OH (15:15:5:1:0.018) containing 4 μl of 25 μM sphingolipid standards (Avanti, LM-6002) at 65 °C for 20 min. The extract was briefly centrifuged, the supernatant was collected, and the pellet was extracted once more with 1 ml extraction buffer without internal standards. The combined supernatants were subjected to base hydrolysis with 0.1 N KOH in methanol for 60 min at 37 °C, neutralized with 0.1 M acetic acid in methanol and dried under N2. The dried residue was suspended in 100 μl (80:20) LC/MS running buffers, sonicated, briefly centrifuged, and the liquid was transferred to LC/MS vials for analysis. LC buffers were: A, tetrahydrofuran: methanol: 5 mM ammonium formate (30:20:50); B, tetrahydrofuran: methanol: 5 mM ammonium formate (70:20:10). Both buffers contained 0.1% formic acid. LCBs were separated on a 75 mm C18 Bio-Spec column at 750 μl/min using a binary gradient as follows: 20% buffer B for 1 min, ramp to 50% buffer B in 1 min and hold for 3 min. Ramp to 100% buffer B in 1 min and hold for 6 min before returning to 20% in 1 min and hold for 2 min. LCBs were detected in MRM mode essentially as described by Merrill et. al (22) and quantified using authentic standards. Each sample was injected a minimum of two times at various volumes (2-20 μl) to ensure linearity.
Analysis of Protein-Protein Interactions
Split-ubiquitin 2-hybrid assays were performed as described previously (5). The plasmids expressing the Nub- and Cub-tagged proteins are as in Table 2. Orm1 and Orm2 were Nub-tagged at their N-termini by insertion of a NubG-HA cassette immediately following the start codon. Lcb1 and Lcb1ΔTMDl were Cub-tagged at their C-termini by inserting a Cub-LexA-VP16 cassette immediately before their stop codons. Lcb2 was tagged at its C-terminus by insertion of the NubG-HA cassette immediately before the stop codon. HA, FLAG, and GFP epitope-tagged bait and prey proteins were constructed by standard methods and used for coimmunoprecipitation assays.
Immunoprecipitation (IP) was carried out as described (6) with some modifications. In brief, microsomal membrane proteins were resuspended at 1 mg/ml in IP buffer (50 mM HEPES-KOH, pH 6.8, 150 mM KOAc, 2 mM MgOAc, 1 mM CaCl2, and 15% glycerol) with protease inhibitors, and solubilized in 1% digitonin at 4 °C for 2.5 h. After centrifugation at 100,000 x g for 30 min, 1 ml of the supernatant was incubated with 50 μl of anti-FLAG beads (Sigma-Aldrich) or 25 μl of GFP-Trap (ChromoTek) at 4 °C for 4 h. The beads were pelleted and washed four times with IP buffer containing 0.1% digitonin. For anti-FLAG IPs, bound proteins were eluted in 130 μl IP buffer containing 0.25% digitonin and 200 μg/ml of FLAG peptide. For anti-GFP IPs, bound proteins were eluted by boiling in 130 μl SDS sample buffer. 17.5 μl of solubilized membranes (input), 17.5 μl of unbound and 5 μl of bound fractions were loaded on a NuPAGE 4 -12% Bis-Tris gel (Invitrogen), and the resolved proteins were detected by immunoblotting as described below.
For the proximity-dependent biotin identification (BioID) assay (23), yeast orm1Δorm2Δ1cb1Δ mutant cells expressing Lcb1-BirA and the Orm1/2 phosphorylation mutants were grown in SD medium to an OD600 of 1.5 and then split and diluted to an OD of 0.02 for incubation without and with biotin (50 μM) for 18 h. Cells were harvested, microsomal proteins were prepared, separated by SDS-PAGE, and transferred to nitrocellulose for immunoblotting as described above.
Immunofluorescent Microscopy
LCB2GFP@82 was constructed by an in-frame insertion of a multimerizing GFP cassette between codons 82 and 83 of Lcb2 using standard procedures. The other RFP and GFP-tagged proteins were constructed by inserting cassettes expressing the non-polymerizing variants of the fluorescent proteins either immediately after the start codon (for N-terminal tagging) or immediately before the stop codon (for C-terminal tagging). Imaging was performed with an Olympus IX70 imaging system using a 100x objective, an EXi-CCD camera (Q-imaging) and IPLab 3.9 software. Excitation and emission wavelengths: 460-490 nm/510nm for the GFP filter, 510-550 nm/590nm for the RFP filter.
The fluorescence distributions (GFP/RFP) and their intensities were analyzed using Fiji ImageJ software (National Institutes of Health). In brief, the merged image was used to generate an LCB1-RFP and LCB1ΔTMD1-GFP grayscale panel. Straight lines were drawn at 0°, 120° and 240° through the same 20 cells in each panel. The intensity of fluorescence at the periphery was determined by averaging the fluorescence intensity at each of the points where the lines intersected the peripheral ER of each cell. The fluorescence in the cell was determined by averaging the intensity along points (0.06250 μm intervals, 52 to 102 depending on cell size) of each of the three bisecting lines, including the points of intersection with the peripheral ER, using the analytic tool “Plot Profile.” After subtracting the background, the average fluorescence intensity at the periphery (Fperi) and the average fluorescence intensity in the cell (Fcell) were used to calculate Fperi/Fcell.
Assessing Membrane Topology
Membrane topology of the yeast Lcb1-TMD1FVT protein was determined by inserting a 50 amino acid cassette containing 3 consensus N-glycosylation sites derived from yeast invertase between residues 4 and 5 as previously described (15). Whether the N-terminus of the protein was in the cytosol or the lumen was determined from the electrophoretic mobilities of the proteins with and without endoglycosidase H (EndoH) treatment as previously described (15).
Gel electrophoresis and immunoblotting
SDS-PAGE and immunoblotting using antibodies directed against various components of the SPOTS complex, or the relevant epitope tags, were performed as previously described (12). To assess the phosphorylation status of the yeast ORMs, the Phos-tag™ reagent was employed as recommended by the manufacturer. Protein expression was quantified using Fiji ImageJ software.
Data analysis
All results are presented as mean ± standard deviation. p values <0.05 were considered significant as assessed by single factor ANOVA within Microsoft Excel. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
The first transmembrane domain of Lcb1 is essential for ORM regulation of SPT
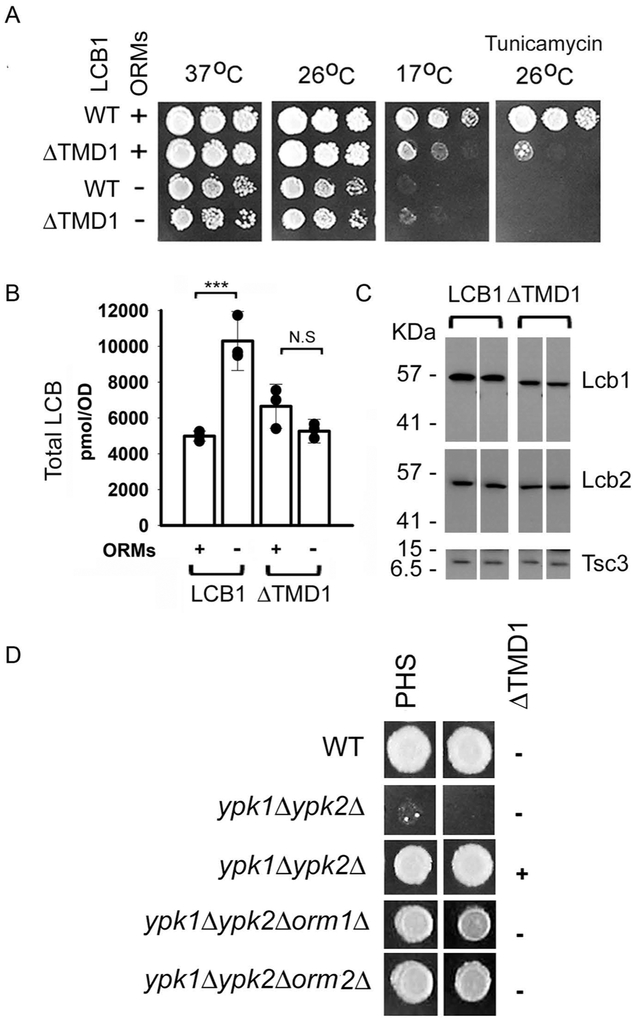
Our previous observation that TMD1 of Lcb1 was not required for ER localization or for catalytic activity of SPT (15) suggested that it might play a regulatory role. Consistent with this hypothesis, yeast cells expressing Lcb1 from which TMD1 has been deleted (lcb1ΔTMD1) are both cold and tunicamycin sensitive (Fig. 1A), phenotypes that also result from elevated LCB levels (6). Similar phenotypes are seen in an orm1Δorm2Δ mutant, raising the possibility that TMD1 is required for ORM regulation of SPT. This is consistent with the observation that the ORMs interact with Lcb1 in the absence of any other components of the SPOTS complex (7) and suggests that LCB levels in an Lcb1ΔTMD1orm1Δorm2Δ triple mutant should be no higher than in the LCB1ΔTMD1 single mutant. Comparison of total LCB levels in cells containing Lcb1 and Lcb1ΔTMD1 with and without the ORMs (Fig. 1B) confirmed this prediction, showing that SPT containing Lcb1ΔTMD1 is not regulated by the ORMs. In these experiments, the LCB levels were lower in the lcb1ΔTMD1orm1Δorm2Δ triple mutant than in the orm1Δorm2Δ double mutant. This is not the result of a lower intrinsic activity of SPT containing an Lcb1ΔTMD1 subunit. Rather, the expression of the Lcb1ΔTMD1. and consequently Lcb2 and Tsc3 (4), are reduced by 10-30% compared to their wild-type counterparts (Fig. 1C).
Figure 1. Deletion of TMD1 of Lcb1 results in phenotypes similar to deletion of the ORMs.
A. Serial dilutions of cells with or without orm1 and ORM2 (ORMs) and expressing either full-length Lcb1 (WT) or Lcb1ΔTMD1 (ΔTMD1) were spotted onto YPD, or YPD + tunicamycin (1 μg/ml) and the plates were incubated at 37 °C (2 days), 26 °C (3 days), or 17 °C (7 days) as indicated. B. Total LCBs were extracted from the indicated strains (as in panel A) and quantified by HPLC as described in Materials and Methods. Results are the average ± SD of LCB levels from three independent colonies (*** p<0.001, n=3; N.S, not significant, n=3). Results are representative of multiple independent experiments. C. Twenty-five μg of microsomal protein from the samples used for LCB analysis (panel B) were separated by SDS-PAGE, transferred to nitrocellulose and Lcb1, Lcb2 and Tsc3 were detected by immunoblotting. Protein expression was quantitatively analyzed with Fiji ImageJ software (n=6). D. Wild-type (WT), ypk1Δypk2Δ, ypk1Δypk2Δ with a plasmid expressing Lcb1ΔTMD1, ypk1Δypk2Δorm1Δ. or ypk1Δypk2Δorm2Δ cells were spotted on plates containing YPD medium with or without PHS. Plates were incubated at 26° C for 3 days.
Deletion of the YPK kinases, which phosphorylate and thereby derepress the ORMs, is lethal, possibly due to constitutive repression of SPT since deletion of the ORMs restores viability (10). If TMD1 of LCB1 is required for ORM binding and regulation, then expression of Lcb1ΔTMD1 in the ypk1Δypk2Δ double mutant should create a subset of SPT not subject to repression and, like deletion of the ORMs, rescue the double mutant. To test this prediction, we crossed a ypk1Δ mutant to a ypk2Δ mutant containing a plasmid expressing Lcb1ΔTMD1. Following sporulation of the diploid, the viability of the meiotic products was tested. The results showed that the lethality of the ypk1Δypk2Δ double mutant was indeed rescued by the Lcb1ΔTMD1 expression plasmid (Fig. 1D), providing additional support for the conclusion that TMD1 mediates ORM regulation.
The first transmembrane domain of Lcb1 is essential for ORM binding to SPT
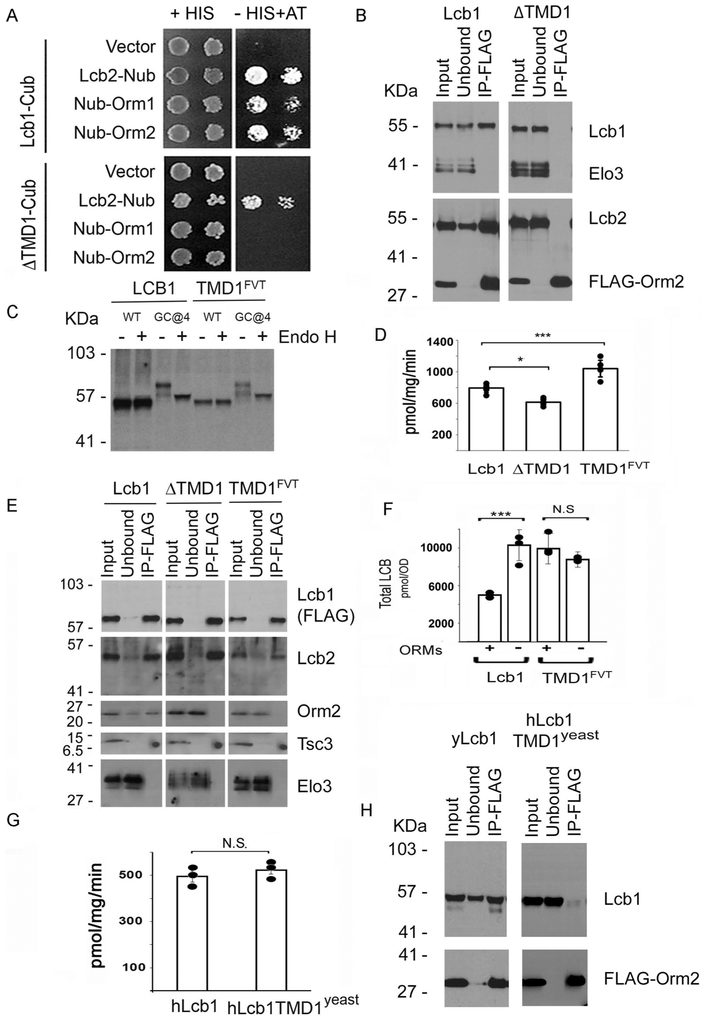
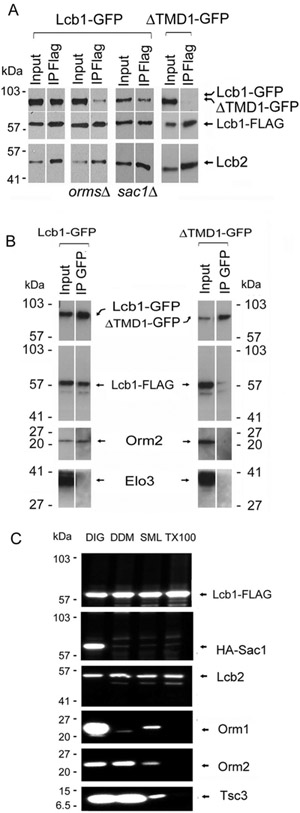
Two lines of evidence suggest that not only does deletion of TMD1 ablate ORM regulation, but that it also eliminates ORM binding to Lcb1. First, there was a positive split ubiquitin 2-hybrid interaction between Lcb1-Cub and either Nub-Orm1 or Nub-Orm2, but not between Lcb1ΔTMD1-Cub and the Nub-tagged Orms (Fig. 2A). The lack of interaction between Lcb1ΔTMD1-Cub and the Nub-tagged ORM proteins cannot be attributed to a general perturbation of Lcb1 since both the full-length and ΔTMD1 mutant Cub-tagged Lcb1s showed a positive interaction with Lcb2-Nub (Fig. 2A), and SPTs from which TMD1 of Lcb1 had been deleted were active (15). The second line of evidence that TMD1 is required for ORM binding comes from experiments where FLAG-tagged Orm2 was used to coimmunoprecipitate associated proteins. Full length Lcb1, and Lcb2, coimmunoprecipitated in anti-FLAG pull downs of FLAG-Orm2 when TMD1 was present, whereas neither copurified when it was deleted (Fig. 2B).
Figure 2. TMD1 of Lcb1 is required for ORM binding to SPT.
A. Split-ubiquitin 2-hybrid interactions between yeast Lcb1-Cub or Lcb1ΔTMD1-Cub and Lcb2-Nub, Nub-Orm1, or Nub-Orm2 were assessed by growth on medium lacking histidine (HIS) and containing 50 mM 3-AT. Plates were incubated at 26 °C for 3 days. B. Cells expressing full-length Lcb1 or Lcb1ΔTMD1 were transformed with a plasmid expressing FLAG-Orm2. Solubilized microsomes were prepared, and following incubation with anti-FLAG beads adsorbed proteins were resolved by SDS-PAGE and visualized by immunoblotting with the indicated antibodies as described in Materials and Methods. Elo3, a component of the ER-associated fatty acid elongase complex, was used as a negative control. C. A glycosylation cassette was inserted after residue 4 (GC@4) into full-length Lcb1 or Lcb1 in which TMD1 had been replaced by the first TMD of FVT1 (TMD1FVT). Solubilized microsomes were resolved by SDS-PAGE before or after treatment with endoglycosidase H (Endo H) and Lcb1 was visualized by immunoblotting with an anti-Lcb1 antibody. D. SPT activity was measured in microsomes prepared from cells expressing full-length Lcb1, Lcb1ΔTMD1. or Lcb1-TMD1FVT. Results are the average ± SD from two independent colonies assayed in triplicate (* p< 0.05, *** p< 0.001, n=6). E. Microsomes were prepared from lcb1Δ mutant yeast cells expressing Lcb1-FLAG, Lcb1ΔTMD1-FLAG, or Lcb1-TMD1FVT-FLAG, solubilized, incubated with anti-FLAG beads and adsorbed proteins resolved by SDS-PAGE. Coimmunoprecipitated proteins were visualized by immunoblotting with the indicated antibodies. Elo3 was used as a negative control. F. Total LCB levels were determined from cells expressing wild-type Lcb1 or Lcb1-TMD1FVT with and without the ORM proteins as in Fig. 1B. The results are the average ± SD of three independent colonies for each strain (*** p<0.001, n=3; N.S, not significant, n=3). G. SPT activity was determined in microsomes prepared from yeast lcb1Δ mutant cells and expressing hLCB2a and ssSPTa with either hLCB1 or hLCB1TMD1yeast, a chimera in which TMD1 of hLCB1 was replaced with TMD1 from yeast Lcb1. SPT activity was measured in triplicate from microsomes prepared from three colonies of each strain and the results are the average ± SD; (N.S, not significant, n=9). H. Solubilized microsomes were prepared from yeast lcb1Δ mutant cells expressing FLAG-Orm2 and either yeast Lcb1 or hLCB1 TMD1yeast, hLCB2a, and ssSPTa. After adsorption on anti-FLAG beads, the interaction of Orm2 with the wild-type and chimeric proteins was assessed as in Fig. 2E.
We have previously shown the N-terminus of Lcb1 is in the lumen of the ER and that deletion of TMD1 (residues 50-85) results in an active protein whose N-terminus is cytosolic (15). To exclude the possibility that the failure of the ORMs to bind to Lcb1ΔTMD1 was the result of the absence of an N-terminal lumenal domain, residues 50-85 of Lcb1 were replaced with the first TMD of FVT1 (residues 2-26), whose N-terminus has been shown to reside in the ER lumen (24). Insertion of a glycosylation cassette (GC) after residue 4 demonstrated that, like full-length Lcb1, the N-terminus of the chimeric protein was lumenal (Fig. 2C). Nevertheless, despite restoring membrane topology and actually increasing in vitro SPT activity (Fig 2D), the ORMs neither bound (Fig. 2E) nor repressed (Fig. 2F) SPT containing the chimeric Lcb1 subunit. Indeed, basal LCB levels were markedly elevated when TMD1 was replaced by the first membrane-spanning domain of FVT1 (Fig. 1F) as would be expected if the chimeric protein could not bind the ORMs. Thus, TMD1 of Lcb1 is necessary for ORM binding and regulation. However, it is not sufficient; a chimera in which TMD1 (residues 1-40) of human LCB1 (hLcb1) was replaced with residues 1-85 of yeast was catalytically active when co-expressed with hLCB2a and ssSPTa (Fig. 2G) but did not coimmunoprecipitate with FLAG-tagged yeast ORMs (Fig. 2H). Taken together, these results show that either the ORMs bind to multiple regions of SPT or that the role of TMD1 is to create a conformation competent for ORM binding to another region of Lcb1.
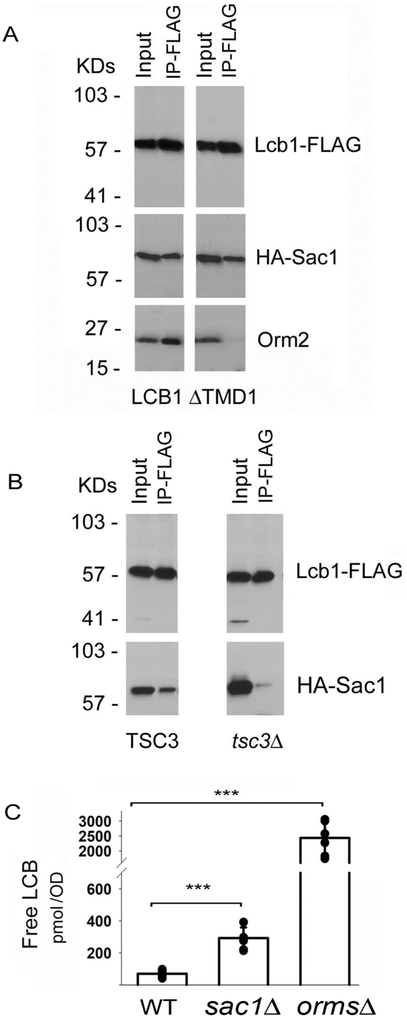
The yeast SPOTS complex also contains Sac1, a PI4P phosphatase that binds to SPT regardless of whether the ORMs are present (6). However, unlike the ORMs, Sac1 binding to SPT does not require TMD1 of Lcb1 (Fig 3A). Rather, Sac1 binding to SPT was significantly reduced when Tsc3 was deleted (Fig. 3B). Thus, association of different components of the SPOTS complex is mediated by different subunits of SPT. In addition, while an orm1Δorm2Δsac1Δ mutant is synthetically lethal, and deletion of the ORMs increased free LCB levels nearly 50-fold (6), deletion of Sac1 had only a modest (2.5-fold) effect on free LCB levels (Fig. 3C) suggesting that it has little if any effect on in vivo SPT activity. In this figure, free, rather than total, LCBs are shown because whereas in the orm1Δorm2Δ mutant total LCBs are nearly doubled, the increase in total LCBs in the sac1Δ mutant is not statistically significant; the effect of deleting Sac1 can only be seen by analyzing free LCBs.
Figure 3. Sac1 binding to SPT requires Tsc3.
A. Solubilized microsomes prepared from yeast lcb1Δ mutant cells expressing HA-Sac1 and either Lcb1-FLAG or Lcb1ΔTMD1 - FLAG were incubated with anti-FLAG antibodies and coimmunoprecipitation of HA-Sac1 and Orm2 was assessed. B. Lcb1- FLAG and HA-Sac1 were co-expressed in wild-type or tsc3Δ mutant cells and the coimmunoprecipitation of HA-Sac1 with Lcb1- FLAG was evaluated as in Panel A. C. Levels of free LCBs in wild-type, Sac1Δ, andorm1Δorm2Δ cells (seven independent colonies of each) were determined as described in Materials and Methods. Results are the average ± SD, *** p<0.001, n=7).
ORMDL-mediated regulation of mammalian SPT also requires TMD1 of mammalian LCB1
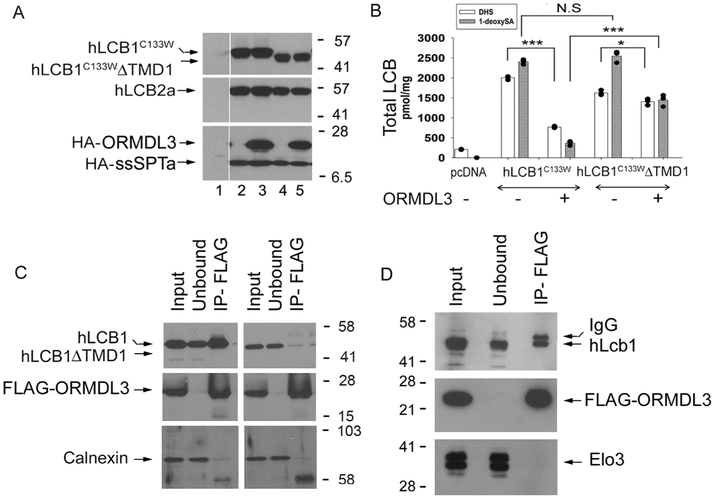
Given the results described above, it was of interest to determine if ORMDL binding to, and regulation of, human SPT depends on the presence of TMD1 of hLCB1. The effect of deleting TMD1 (residues 9-40) of hLCB1 on the ability of the ORMDLs to repress SPT was therefore examined. For these experiments, we took advantage of the hLCB1CC133W HSAN1 mutant that confers increased promiscuity of heterotrimeric SPT for utilization of alanine as substrate, thereby increasing synthesis of deoxy-LCBs (25,26). Deoxy-LCBs provide an accurate measure of in vivo SPT activity because these compounds are not present in the tissue culture medium, arise only from de novo synthesis, and lack the C1-OH that is phosphorylated to form LCB-Ps that are subsequently degraded by the LCB-1-P lyase. Thus, unlike the serine-containing LCBs, their levels are not affected by turnover (12,16). Accordingly, CHO-LyB cells (which lack endogenous SPT activity (27)) were transfected with hLCBC133W or hLCB1C133W ΔTMD1. hLCB2a and ssSPTa, with or without ORMDL3 (Fig. 4A). Thirty hours post-transfection, total LCBs were extracted and the levels of dihydrosphinganine (DHS) and 1-deoxysphinganine (1-deoxySA) were quantified (Fig. 4B). Similar to results we have previously reported from HEK cells (12), co-expression of ORMDL3 reduced the in vivo activity of SPT by nearly 90%. In contrast, when cells were transfected with hLCB1C133W lacking TMD1, hLCB2a and ssSPTa, cotransfection with ORMDL3 reduced in vivo SPT activity by only about 50% (Fig. 4B). Not only did deletion of TMD1 of hLcb1 reduce the ability of ORMDL3 to repress SPT activity, it also reduced binding of ORMDL3 to hLCB1; full length hLCB1, but not hLCB1ΔTMD1, coimmunoprecipitated with FLAG-ORMDL3 (Fig. 4C). Notably, the interaction between hLCB1 and the ORMDLs was seen whether or not hLCB2a and the ssSPT subunit were present (Fig 4D), indicating that ORMDL binding to hLCB1 is independent of other components of mammalian SPT. ORMDL3 was chosen for use in these experiments because of its association with asthma (28) and because it is the most widely studied of the three highly homologous ORMDLs. However, we have previously shown that all three ORMDLs bind and repress mammalian SPT (12).
Figure 4. TMD1 of human LCB1 is required for ORMDL binding.
A. CHO-LyB cells were transfected with empty vector (lane 1), hLCB1C133W (lanes 2 and 3) or hLCB1C133W ΔTMD1 (lanes 4 and 5), and hLCB2a (lanes 2-5), ssSPTa (lanes 2-5) and HA-ORMDL3 (lanes 3 and 5). After thirty hours, microsomal proteins were prepared, resolved by SDS-PAGE and visualized by immunoblotting with the indicated antibodies. B. Total levels of DHS and 1-deoxySa were measured from the samples used in Panel A. Results are the average ± SD of LCB levels from three independent transfections (* p< 0.05, *** p< 0.001, N.S, not significant, n=3). C. hLCB1 (left panel) or hLCB1ΔTMD1 (right panel) was co-expressed with hLCB2a, ssSPTa and FLAG-ORMDL3 in CHO-LyB cells. Solubilized microsomes were prepared and the coimmunoprecipitation of the LCB1 proteins with FLAG-ORMDL3 was determined. Calnexin was used as a negative control. D. hLCB1 and FLAG-ORMDL3 were co-expressed in yeast. Solubilized microsomes were prepared and the coimmunoprecipitation of hLCB1 with FLAG-ORMDL3 was determined. Elo3 was used as a negative control.
The ORMs mediate oligomerization and localization of SPT
Breslow et al. reported that Orm1 associates with Orm2 and suggested that SPT may exist in higher order complexes (6). In addition, Hornemann et al. proposed that mammalian SPT is oligomeric (18). To investigate whether the ORMs mediate the formation of higher order structures, Lcb1-FLAG and Lcb1-GFP were co-expressed in cells with and without the ORMs and the ability of Lcb1-FLAG to coimmunoprecipitate Lcb1-GFP after digitonin (DIG) solubilization was determined. The results showed that in the presence of the ORMs, Lcb1-FLAG and Lcb1-GFP coimmunoprecipitated, while in their absence they did not (Fig. 5A). Similarly, even when the ORMs are present, Lcb1ΔTMD1-GFP, which cannot bind the ORMs (see above), does not pull down with Lcb1-FLAG (Fig. 5A). Reciprocal coimmunoprecipitation experiments confirmed that Lcb1-GFP pulled down both Lcb1-FLAG and Orm2, whereas Lcb1ΔTMD1-GFP pulled down neither (Fig. 5B). While these results clearly show ORM-mediated oligomerization of SPT, formation of higher order structures is independent of Sac1; Lcb1-GFP coimmunoprecipitated with Lcb1-FLAG whether or not Sac1 was present (Fig. 5A). In previous experiments, using sucrose monolaurate (SML) solubilized membranes, no such oligomerization was seen (3). However, in contrast to the situation after digitonin solubilization, there is only weak association between the ORMs and SPT after SML solubilization (Fig. 5C). This provides additional support for the role of the ORMs in the formation of higher order SPT complexes and demonstrates that the stability of the SPOTS complex is highly dependent upon solubilization conditions. Indeed, only after digitonin solubilization was Sac1 associated with the SPOTS complex (Fig. 5C).
Figure 5. The ORM proteins mediate oligomerization of SPT.
A. Yeast Lcb1-GFP was co-expressed with either Lcb1-FLAG (Panels 1-3) or Lcb1ΔTMD1-FLAG (Panel 4) in wild-type (Panels 1 and 4), orm1Δorm2Δ (Panel 2), or Sac1Δ (Panel 3) mutant cells. The ability of Lcb1-FLAG or Lcb1ΔTMD1-FLAG to coimmunoprecipitate Lcb1-GFP was assessed by immunoblotting after adsorption of solubilized microsomal proteins to anti-FLAG beads. B. The ability of Lcb1-GFP or Lcb1ΔTMD1-GFP to coimmunoprecipitate Lcb1-FLAG was determined by adsorption of solubilized microsomal proteins from Panel 1 or Panel 4 in A. to anti-GFP beads followed by SDS-PAGE and immunoblotting with anti-FLAG antibodies. C. Yeast microsomes were prepared from lcb1Δsac1Δ mutant cells expressing Lcb1- FLAG and HA-Sac1, solubilized with 2% DIG, 1% DDM, 0.1% SML or 1% Triton X-100 (TX100) at 4 °C and incubated with anti-FLAG beads. Adsorbed proteins were resolved by SDS-PAGE and visualized by immunoblotting with antibodies to the proteins indicated on the right side of the panel.
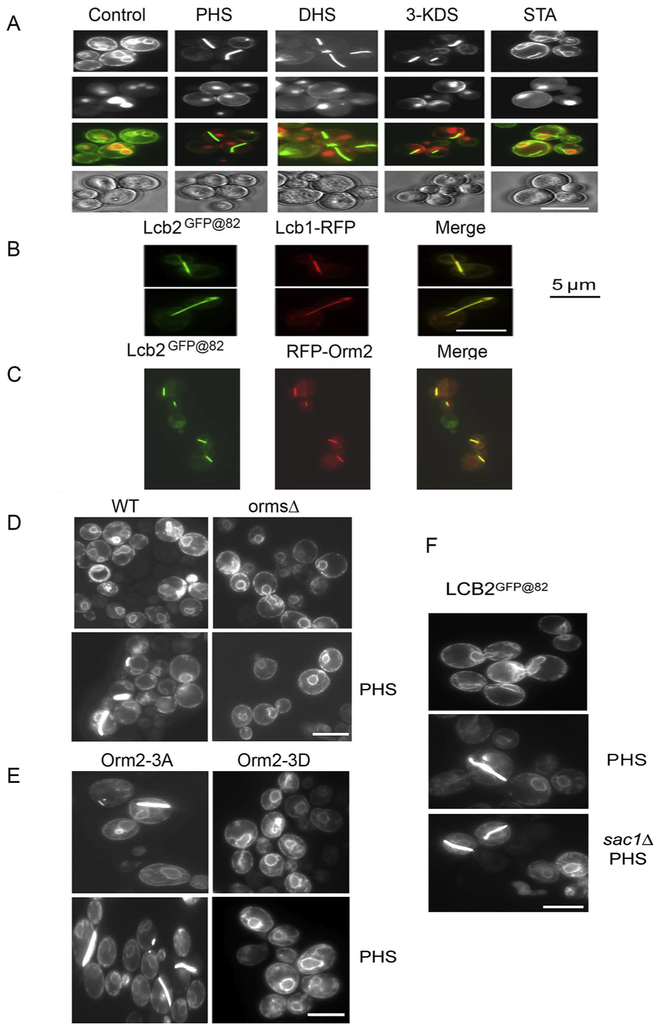
While characterizing regions of Lcb2 that would tolerate insertion of epitope tags, we observed that when a multimerizing form of GFP was inserted into the lumenal loop of Lcb2, treatment of cells with phytosphingosine (PHS) and other physiological LCBs resulted in relocalization of Lcb2GFP@82 into unusual bar-like structures (Fig. 6A). These structures also contain Lcb1 (Fig. 6B). Given that they are only seen in the presence of high levels of LCBs and that cells expressing Lcb2GFP@82 have significant SPT activity, the bars do not appear to be simple aggregates of an altered Lcb2. Notably, the ORMs are also present in these structures (Fig. 6C). Indeed, their formation is dependent upon the presence of the ORMs (Fig. 6D); 51 ± 4% of cells containing the ORMs had bars in the presence of PHS while cells lacking the ORMs failed to form bars in the presence of PHS. These results are entirely consistent with the fact that oligomerization is ORM-dependent (Fig. 5A). More importantly, the phosphorylation state of the ORMs appears to control bar formation; in cells expressing a constitutively-negative Orm2 mutant (Orm2-3A) that contains alanine substitutions at the three serine residues (serines 46, 47 and 48) subject to derepressive phosphorylation by the YPK kinases (6,10), the structures form spontaneously, with 49 ± 9% of cells showing bars without PHS and 52 ± 16% with PHS (Fig. 6E). Conversely, the structures cannot be induced by PHS in cells expressing an Orm2 mutant (Orm2-3D) containing phosphomimetic mutations at the same sites (Fig. 6E). Taken together, these results indicate that when sphingolipid levels are high, hypophosphorylated ORMs favor oligomerization of SPT that may directly or indirectly inhibit its activity. In contrast to the ORMS, Sac1 is not required for bar formation (Fig. 6F). Thus, the ORMs are required for both oligomerization and bar formation while Sac1 is required for neither. This strongly suggests that the bars, though only seen when SPT contains Lcb2 with a multimerizing GFP inserted after residue 82, are nevertheless reporting a physiological change in the intracellular localization/organization of SPT.
Figure 6. ORM-dependent relocalization of SPT is induced by LCBs.
A. Yeast cells expressing Lcb2 containing a multimerizing version of GFP inserted between residues 82 and 83 (Lcb2GFP@82) were grown in YPD medium with and without LCBs (PHS, DHS, or 3-ketosphinganine (3-KDS) and the localization of Lcb2 was observed using fluorescence microscopy. The non-physiological LCB stearylamine (STA) served as a negative control. B. Lcb1-RFP was co-expressed with Lcb2GFP@82, and the localization of the proteins was compared by fluorescence microscopy. C. RFP-Orm2 and Lcb2GFP@82 were co-expressed and their localization visualized by fluorescence microscopy. D. PHS-induced relocalization of SPT in wild-type (-PHS, no bars, n>500 cells; +PHS, 51 ± 4% with bars, n=247 cells, 5 experiments) and orm1Δorm2Δ mutant cells (-PHS, no bars, n=93 cells; +PHS, no bars, n=187 cells, 3 experiments) expressing Lcb2GFP@82 was compared as in Panel A. E. PHS-induced relocalization of SPT in cells expressing Lcb2GFP@82 and the non-phosphorylatable Orm2-3A (-PHS, 49 ± 9% with bars, n=116 cells; +PHS, 52 ± 16% with bars, n=67 cells, 3 experiments) or the phosphomimetic Orm2-3D mutant (-PHS, no bars, n=80 cells; +PHS, no bars, n=118 cells, 3 experiments) was determined as in Panel A. F. PHS-induced relocalization of SPT in wild-type and sac1Δ cells (-PHS, no bars, n>100 cells; +PHS, 23 ± 10% with bars, n=99 cells, 4 experiments) expressing Lcb2GFP@82 was determined as in Panel A. Scale bar, 5 microns.
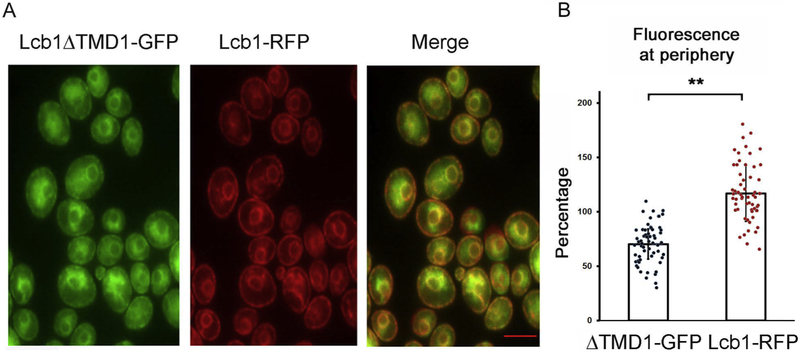
Breslow et al. presented evidence that the phosphorylation state of the ORMs shifted the distribution of the SPOTs complex within the ER (6). Since deletion of TMD1 of Lcb1 results in loss of ORM binding, a situation comparable to full derepression of the ORMs, we co-expressed full-length Lcb1-RFP and Lcb1ΔTMD1-GFP and analyzed their distribution by fluorescent microscopy. The results clearly showed that a greater fraction of Lcb1-RFP was localized to the peripheral ER (Fig. 7). Thus, not only does ORM binding influence the oligomeric state of the enzyme, but also its localization, implying that oligomerization is a determinant of subcellular localization.
Figure 7. Deletion of TMD1 of Lcb1 results in altered localization of SPT.
A. The localization of Lcb1-RFP and Lcb1ΔTMD1-GFP were compared by fluorescence microscopy. The merged image (A, right hand panel) clearly shows lack of complete colocalization. B. The average intensity of Lcb1ΔTMD1GFP or Lcb1RFP at the periphery, compared to the average fluorescence intensity in the cell, were determined using Fiji ImageJ software as described in Materials and Methods. Results are expressed as the quotient of the average fluorescence intensity at two points in the peripheral ER at opposite ends of the cell divided by the average fluorescence intensity at any point on a line connecting those two points times 100%. For each of 20 cells, three lines were used; ** p<0.01, n=60. Scale bar, 5 microns.
The ORMs directly repress the Lcb1/Lcb2 heterodimer
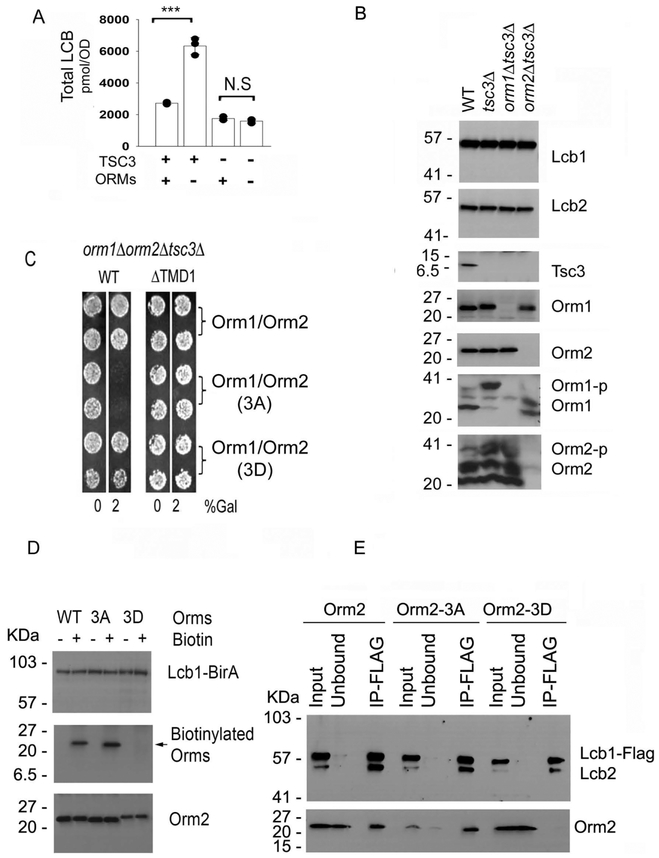
Since Tsc3 enhances (4), and the ORMs, in their unphosphorylated state, repress SPT activity, it was important to determine whether the ORMs antagonize the activity of Tsc3 or act directly on the Lcb1-Lcb2 heterodimer. If the role of the ORMs were to antagonize Tsc3, then they should have no effect on in vivo SPT activity when Tsc3 is deleted. Consistent with this, the levels of LCBs were not significantly different in a tsc3Δorm1Δorm2Δ triple mutant than in a tsc3Δ single mutant (Fig. 8A). However, in the tsc3Δ mutant the ORMs are, as expected, hyperphosphorylated and likely derepressed (Fig. 8B) making it difficult to interpret the lack of change in LCB levels. We therefore used the constitutively negative Orm1-3A and Orm2-3A mutants to insure maximum repression (10,11). The results showed that overexpression of the non-phosphorylatable Orm1-3A and Orm2-3A proteins results in death of the tsc3Δ mutant, and that this effect is eliminated by deleting TMD1 of Lcb1 (Fig. 8C). Thus, the ORMs directly inhibit the Lcb1/Lcb2 catalytic heterodimer.
Figure 8. ORMs directly repress the Lcb1-Lcb2 heterodimer.
A. Total LCB levels in wild-type, tsc3Δ, orm1Δorm2Δ, and orm1Δorm2Δtsc3Δ mutant cells were determined as described in Materials and Methods. Results are the average ± SD from three independent colonies of each strain, *** p<0.001, n=3; N.S, not significant, n=3. B. Microsomal protein extracts were prepared from wild-type (WT), tsc3Δ,orm1Δtsc3Δ and orm2Δtsc3Δ mutant cells, resolved by SDS-PAGE with or without the Phos-tag™ reagent, and the indicated proteins were detected by immunoblotting using antibodies to the proteins indicated on the right side of the panel. C. Plasmids co-expressing wild-type Orm1 and Orm2, the non-phosphorylatable Orm1-3 A and Orm2-3A, or the phosphomimetic Orm1-3D and Orm2-3D mutant proteins from divergent galactose inducible GAL1 and GAL10 promoters were introduced into an orm1Δorm2Δtsc3Δ mutant or an orm1Δorm2Δtsc3Δ mutant with LCB1 replaced by LCB1ΔTMD1. Growth of two independent colonies of each was assessed in the absence or presence of 2% (w/v) galactose. D. orm1Δorm2ΔLcb1Δ mutants co-expressing Lcb1-BirA and either wild-type Orm2, the non-phosphorylatable Orm2-3A, or the phosphomimetic Orm2-3D mutant proteins were grown with or without biotin as described in Materials and Methods. 5 μg of microsomal protein were separated by SDS-PAGE, transferred to nitrocellulose and immunoblotted for detection with anti-Lcb1, anti-Orm2 and streptavidin-HRP (Thermo Scientific). E. Microsomal proteins were prepared from orm1Δorm2ΔLcb1Δ mutants co-expressing Lcb1-FLAG and either wild-type Orm2, the non-phosphorylatable Orm2-3A, or the phosphomimetic Orm2-3D mutant proteins. Anti-FLAG pulldowns followed by anti-Lcb1, Lcb2 and Orm2 immunoblotting were performed as described in the Materials and Methods.
Using wild-type Orm2, the non-phosphorylatable Orm2-3A, and the phosphomimetic Orm2-3D mutants and the biotin ligase (BirA) proximity assay (23), we also examined the effects of ORM phosphorylation on binding to Lcb1. The results showed that Lcb1-BirA strongly biotinylated both wild-type Orm2 and Orm2-3A. However, there was no biotinylation of Orm2-3D (Fig. 8D). These data strongly suggest that phosphorylated ORMs do not bind to SPT. This conclusion was confirmed by direct co-immunoprecipitation experiments in which Lcb1-FLAG was co-expressed with the either Orm2, Orm2-3A or Orm2-3D and captured with anti-FLAG antibodies; both Orm2 and Orm2-3A co-immunoprecipitated with Lcb1-FLAG whereas Orm2-3D did not (Fig. 8E). These results are consistent with the decreased binding of ORMs to Lcb1 observed after treatment of cells with myriocin, which has been shown to increase ORM phosphorylation (6,29).
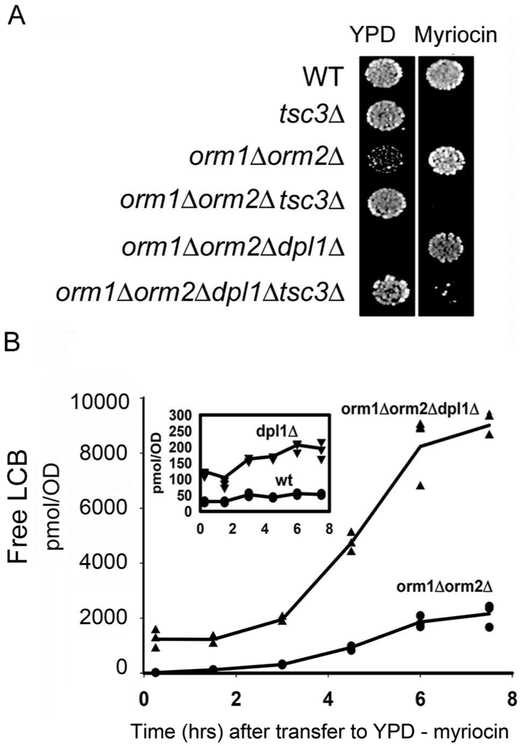
The LCB-P lyase, Dpl1, is essential for maintaining LCBs at sublethal levels in yeast orm1Δorm2Δ mutants
Previous studies have shown that cells lacking Dpl1, the lyase that degrades LCB-Ps to palmitaldehyde and phosphoethanolamine, accumulate high LCB levels indicating that this degradative pathway is important for maintaining LCB homeostasis (30). It was therefore of interest to investigate the effect of combining the loss of this degradative pathway with the loss of ORM regulation. Accordingly, an orm1Δorm2Δ mutant was crossed to a dpl1Δtsc3Δ mutant, the resulting diploid was sporulated, and tetrads were dissected. From 42 tetrads, not a single orm1Δorm2Δdpl1Δ triple mutant was recovered. In contrast, 8 orm1Δorm2Δdpl1Δtsc3Δ quadruple mutant products of meiosis were recovered. However, when the tetrads were dissected on plates containing myriocin, which reduces SPT activity, the vast majority of the orm1Δorm2Δdpl1Δ triple mutants survived but failed to grow when transferred to medium lacking myriocin (Fig. 9A). In addition, when cells were grown in the presence of myriocin and then diluted into YPD, the levels of free LCBs in the orm1Δorm2Δdpl1Δ triple mutant rose faster and reached a higher level than those in the orm1Δorm2 mutant (Fig. 9B). Since the dpl1Δ single mutant showed only a modest increase in LCB levels following transfer to medium without myriocin (Fig. 9B, inset), these results indicate that the combined loss of the ORMs and Dpl1 results in LCB levels incompatible with cell viability and that the reduction in SPT activity due to loss of Tsc3 or inhibition by myriocin reduces LCBs to a permissive level.
Figure 9. The LCB-P lyase, Dpl1, is essential for viability of an orm1Δorm2Δ mutant.
A. The indicated yeast strains were grown in YPD with (orm1Δorm2Δ,orm1Δorm2Δdpl1Δ) or without (wild-type, tsc3Δorm1Δ,orm2Δtsc3Δ, orm1Δorm2Δdpl1Δtsc3Δ) myriocin (270 ng/ml), spotted onto YPD agar plates with or without myriocin and incubated at 26 °C for three days. B. The indicated yeast strains were grown in YPD containing myriocin (90 ng/ml) to ~1.0 OD600n/ml. The cells were collected by centrifugation, washed twice with water and then shifted into fresh YPD medium at 0.1 OD600/ml. At the indicated times, 1 OD600 cells was removed and the free LCB levels were determined by mass spectrometry as described in Materials and Methods. Each time point represents the average of LCBs ± SD from three independent colonies. The rate of increase in LCBs after transfer out of myriocin was much higher in the orm1Δorm2Δdpl1Δ triple mutant (1502 ± 170 mol/OD600/hr) than in the orm1Δorm2Δ double mutant (324 ± 51 pmol/OD600/hr, p < 0.001) or the dpl1Δ single mutant (14 ± 5 pmol/OD600/hr, p < 0.001).
DISCUSSION
Topology mapping of yeast Lcb1 revealed the presence of three TMDs, an N-terminal TMD located between amino acids 50-85 oriented to place the N-terminal 50 amino acids in the ER lumen, and two centrally located TMDs separated by a short lumenal loop (15). Although it has been suggested that the first TMD of yeast Lcb1 targets the protein to the ER (31), its deletion does not abolish ER targeting (15). Nor does it reduce SPT catalytic activity. However, deletion of TMD1 from yeast Lcb1 abolished ORM binding, resulting in phenotypes strikingly similar to an orm1Δorm2Δ double mutant or a constitutively derepressed orm2 phosphomimetic mutant in which the three YPK phosphorylation sites were mutated to aspartates. Thus, in contrast to models in which TMD1 is hypothesized to act simply as a membrane tethering domain, we conclude that TMD1 is a regulatory domain, and that TMD2 and TMD3 of Lcb1 determine membrane topology. Deletion of TMD1 of hLCB1 had the same effect on binding of the mammalian ORMDLs; in the absence of TMD1 there was markedly reduced ORMDL binding to SPT and in vivo enzymatic activity increased. In addition, as reported for yeast ORM binding to Lcb1 (7), only hLCB1 was necessary for ORMDL binding. Unfortunately, neither the N-terminal region of yeast or mammalian LCB1 could be stably expressed. Thus, it was not possible to directly determine whether TMD1 was sufficient for ORM binding. However, a chimeric hLCB1 with its TMD1 replaced by the cognate domain from the yeast protein did not bind the yeast ORMs (Fig. 2H). Consequently, TMD1 is necessary, but not sufficient, for ORM binding. Indeed, none of the data prove that TMD1 is directly involved in ORM binding. Nor do they exclude the possibility that even if TMD1 is involved in binding, other regions of LCB1 are not also involved. In this regard, it is noteworthy that a mutation (R228C) in the cytoplasmic loop between TMD1 and TMD2, identified as a suppressor of the ypk1Δypk2Δ mutant, also reduces ORM binding (Han et al., unpublished results).
The ORMs are believed to have functions beyond regulation of SPT (29). However, the similarity of the phenotype of the orm1Δorm2Δ double mutant to that of the lcb1ΔTMD1 mutant suggests that, at least in yeast, the major role of the ORMs is to regulate SPT activity. In addition, deletion of orm1 or ORM2 suppresses the lethality of the yeast ypk1Δypk2Δ double mutant (10,32). Thus, our finding that the lethality of the ypk1Δypk2Δ double mutant is also suppressed by mutations in Lcb1 that abolish ORM binding indicates that the only essential function of the YPK kinases, at least for growth at 26 °C, is to derepress SPT activity when the ORMs are unphosphorylated and thus constitutively repressive. However, since PHS restores normal growth to an lcb1Δ mutant that has no SPT activity, but only partially rescues a ypk1Δypk2Δ mutant in which SPT is markedly reduced but not abolished (Fig. 1D), there are clearly additional YPK targets that regulate sphingolipid metabolism. Indeed, the fact that ceramide synthase is activated by these kinases (33) raises the possibility that ceramide synthases must be phosphorylated for efficient chemical complementation by PHS of SPT-deficient cells. Alternatively, the YPK kinases may be involved in PHS uptake and/or transport.
Siow and Wattenberg showed that in permeabilized cells addition of C6-ceramide reduced SPT activity in an ORMDL-dependent manner (14), which could reflect a change in conformation of the complex. This is consistent with the proposal that it is the oligomeric state of the ORMDLs, presumably bound to SPT, which regulates in vivo activity (19). Oligomerization and/or subcellular relocalization have also been implicated in ORM regulation of yeast SPT (6). The fortuitous observation that in the presence of high levels of LCBs an SPT with GFP inserted into Lcb2 forms bar-like structures provides compelling evidence that the structural organization of SPT is responsive to changes in sphingolipid availability. Because bars do not form when a non-multimerizing GFP is inserted at the same position in Lcb2 (Han et al., unpublished results), it seems most probable that the bars represent the serendipitous capture, facilitated by the polymerization of GFP, of an intermediate in the physiological oligomerization/relocalization pathway. Regardless of the precise mechanism responsible for bar formation, our hypothesis that sphingolipid availability regulates SPT oligomerization/localization is supported by the findings that the ability of Lcb1-FLAG to coimmunoprecipitate Lcb1-GFP is dependent on the ORMs and that SPT containing full-length Lcb1 localizes differently from SPT containing Lcb1 lacking TMD1.
The ORMs also colocalize with SPT in the bars. More importantly, bar formation is dependent upon the presence of the ORMs, suggesting that the ORMs control the state of oligomerization; the repressive non-phosphorylatable Orm2-3A mutant results in constitutive bar formation whereas the phosphomimetic Orm2-3D mutant cannot form bars, even in the presence of high LCBs. Thus, in contrast to mammalian ORMDLs, which appear to control SPT activity in direct response to sphingolipid availability, the phosphorylation state of the ORMs is a key determinant of SPT regulation in yeast.
Interestingly, although Sac1 is not required for bar formation, its binding to the SPOTS complex is dependent upon Tsc3. Thus, the localization of Sac1, a PI4P phosphatase that participates in maintenance of PI levels (34), may be regulated indirectly by ORM-mediated intracellular relocalization of the SPOTS complex. In addition, while Tsc3 is required for Sac1 binding to the SPOTs complex, it is not required for ORM inhibition of the Lcb1/Lcb2 heterodimer.
We have previously shown that accumulation of 1-deoxySA is a more accurate measure of in vivo SPT activity than is accumulation of the serine-containing LCBs (12,25). This was attributed to the fact that the 1-deoxy-LCBs cannot be phosphorylated to generate the substrate for cleavage by the LCB-P lyase, Dpl1. Indeed, the effect of deletion of the ORMs and deletion of Dpl1 on LCB levels is additive and Dpl1 is essential for survival of the orm1Δorm2Δ double mutant. The observation that when SPT is induced, or there is no binding of ORMDL to SPT, deoxy-LCBs accumulate to a greater extent than serine-containing LCBs indicates that degradation of LCB-Ps by the lyase is also important for maintaining sphingolipid homeostasis in the absence of ORMDL regulation in mammalian cells. Although in mammalian cells there is an alternative pathway for metabolism of deoxy-LCBs (35), it appears to play a relatively minor role, at least in cultured cells, in maintenance of LCB homeostasis.
We have shown that the first transmembrane domain of LCB1 is required for the binding of both mammalian and yeast ORMs. In addition, we have shown that yeast ORMs participate in regulating the oligomeric state and intracellular localization of SPT. Our working hypothesis is that unlike in yeast where the ORMs are regulated by N-terminal phosphorylation, mammalian ORMDLs regulate SPT activity by mass action, and that the levels of ORMDL expression depend upon both the synthesis of sphingolipids and their ability to bind to SPT (12). The differences in ORM regulation of SPT between higher and lower eukaryotes may reflect the need for more complex regulation in multicellular organisms. Nevertheless, while the mechanism(s) by which the yeast and mammalian ORMs regulate SPT are different, it seems likely that in both cases ORM regulation of SPT activity is only a component of the overall regulation of sphingolipid homeostasis. Indeed, there are no ORMs in C. elegans, suggesting that there may even be other mechanisms for the regulation of SPT. Further studies will be required to elucidate such mechanisms.
HIGHLIGHTS.
The first membrane-spanning domain of Lcb1 is required for ORM binding to yeast and mammalian serine palmitoyltransferase (SPT).
Tsc3, the small activating subunit of yeast SPT, mediates Sac1 binding to the yeast SPOTs complex.
The ORMs mediate oligomerization and localization of SPT.
Yeast cells lacking both ORM regulation of SPT and the LCB-P lyase, Dpl1, accumulate lethal levels of sphingolipids.
Acknowledgments
This work was supported by National Institutes of Health Grant R21HD080181, NSF grant MCB-158500, and USUHS grants BIO-71-3052 and CHIRP 70-3155. The authors declare that they have no conflicts of interest with the contents of this article. The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the view of the Department of Defense or the Uniformed Services University of the Health Sciences.
The abbreviations used are:
- 1-deoxySA
1-deoxysphinganine
- 3-AT
3-amino-1,2,4-triazole
- 3-KDS
3-ketosphinganine
- DDM
n-dodecyl-β-d-maltoside
- DHS
dihydrosphinganine
- DIG
digitonin
- EndoH
endoglycosidase H
- ER
endoplasmic reticulum
- hLCB1
human LCB1 subunit (SPTLC1)
- hLCB2a
human LCB2 subunit (SPTLC2)
- hLCB2b
human LCB2 subunit (SPTLC3)
- LCB
long-chain bases
- PHS
phytosphingosine
- SML
sucrose monolaurate
- SPT
serine palmitoyltransferase
- TX100
triton X-100
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Hannun YA, and Obeid LM (2018) Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol 19, 175–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yard BA, Carter LG, Johnson KA, Overton IM, Dorward M, Liu H, McMahon SA, Oke M, Puech D, Barton GJ, Naismith JH, and Campopiano DJ (2007) The structure of serine palmitoyltransferase; gateway to sphingolipid biosynthesis. J Mol Biol 370, 870–886 [DOI] [PubMed] [Google Scholar]
- 3.Gable K, Han G, Monaghan E, Bacikova D, Natarajan M, Williams R, and Dunn TM (2002) Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J Biol Chem 277, 10194–10200 [DOI] [PubMed] [Google Scholar]
- 4.Gable K, Slife H, Bacikova D, Monaghan E, and Dunn TM (2000) Tsc3p Is an 80-Amino Acid Protein Associated with Serine Palmitoyltransferase and Required for Optimal Enzyme Activity. J. Biol. Chem. 275, 7597–7603 [DOI] [PubMed] [Google Scholar]
- 5.Han G, Gupta SD, Gable K, Niranjanakumari S, Moitra P, Eichler F, Brown RH Jr., Harmon JM, and Dunn TM (2009) Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc Natl Acad Sci U S A 106, 8186–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, and Weissman JS (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S, Lone MA, Schneiter R, and Chang A (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci U S A 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, and Loewith R (2012) Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol 14, 542–547 [DOI] [PubMed] [Google Scholar]
- 9.Gururaj C, Federman RS, and Chang A (2013) Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J Biol Chem 288, 20453–20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roelants FM, Breslow DK, Muir A, Weissman JS, and Thorner J (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 108, 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimobayashi M, Oppliger W, Moes S, Jeno P, and Hall MN (2013) TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol Biol Cell 24, 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta SD, Gable K, Alexaki A, Chandris P, Proia RL, Dunn TM, and Harmon JM (2015) Expression of the ORMDLS, modulators of serine palmitoyltransferase, is regulated by sphingolipids in mammalian cells. J Biol Chem 290, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siow D, Sunkara M, Dunn TM, Morris AJ, and Wattenberg B (2015) ORMDL/serine palmitoyltransferase stoichiometry determines effects of ORMDL3 expression on sphingolipid biosynthesis. J Lipid Res 56, 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siow DL, and Wattenberg BW (2012) Mammalian ORMDL Proteins Mediate the Feedback Response in Ceramide Biosynthesis. J Biol Chem 287, 40198–40204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han G, Gable K, Yan L, Natarajan M, Krishnamurthy J, Gupta SD, Borovitskaya A, Harmon JM, and Dunn TM (2004) The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J Biol Chem 279, 53707–53716 [DOI] [PubMed] [Google Scholar]
- 16.Harmon JM, Bacikova D, Gable K, Gupta SD, Han G, Sengupta N, Somashekarappa N, and Dunn TM (2013) Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J Biol Chem 288, 10144–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimberlin AN, Han G, Luttgeharm KD, Chen M, Cahoon RE, Stone JM, Markham JE, Dunn TM, and Cahoon EB (2016) ORM expression alters sphingolipid homeostasis and differentially affects ceramide synthase activity. Plant Physiol 172, 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornemann T, Wei Y, and von Eckardstein A (2007) Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem J 405, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiefer K, Carreras-Sureda A, Garcia-Lopez R, Rubio-Moscardo F, Casas J, Fabrias G, and Vicente R (2015) Coordinated regulation of the orosomucoid-like gene family expression controls de novo ceramide synthesis in mammalian cells. J Biol Chem 290, 2822–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose MW, F Hieter P (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21.Roux KJ, Kim DI, Raida M, and Burke B (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merrill AH Jr., Sullards MC, Allegood JC, Kelly S, and Wang E (2005) Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 36, 207–224 [DOI] [PubMed] [Google Scholar]
- 23.Roux KJ, Kim DI, and Burke B (2013) BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci 74, Unit 19 23. [DOI] [PubMed] [Google Scholar]
- 24.Gupta SD, Gable K, Han G, Borovitskaya A, Selby L, Dunn TM, and Harmon JM (2009) Tsc10p and FVT1: topologically distinct short-chain reductases required for long-chain base synthesis in yeast and mammals. J Lipid Res 50, 1630–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gable K, Gupta SD, Han G, Niranjanakumari S, Harmon JM, and Dunn TM (2010) A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J Biol Chem 285, 22846–22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penno A, Reilly MM, Houlden H, Laura M, Rentsch K, Niederkofler V, Stoeckli ET, Nicholson G, Eichler F, Brown RH Jr., von Eckardstein A, and Hornemann T (2010) Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J Biol Chem 285, 11178–11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanada K, Hara T, Fukasawa M, Yamaji A, Umeda M, and Nishijima M (1998) Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J Biol Chem 273, 33787–33794 [DOI] [PubMed] [Google Scholar]
- 28.Worgall TS (2016) Sphingolipids, ORMDL3 and asthma: what is the evidence? Curr Opin Clin Nutr Metab Care [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Huang C, Polu SR, Schneiter R, and Chang A (2012) Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J Cell Sci 125, 2428–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saba JD, Nara F, Bielawska A, Garrett S, and Hannun YA (1997) The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem 272, 26087–26090 [DOI] [PubMed] [Google Scholar]
- 31.Yasuda S, Nishijima M, and Hanada K (2003) Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J Biol Chem 278, 4176–4183 [DOI] [PubMed] [Google Scholar]
- 32.Roelants FM, Torrance PD, Bezman N, and Thorner J (2002) Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol Biol Cell 13, 3005–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muir A, Ramachandran S, Roelants FM, Timmons G, and Thorner J (2014) TORC2- dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. Elife 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brice SE, Alford CW, and Cowart LA (2009) Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem 284, 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alecu I, Othman A, Penno A, Saied EM, Arenz C, von Eckardstein A, and Hornemann T (2017) Cytotoxic 1-deoxysphingolipids are metabolized by a cytochrome P450-dependent pathway. J Lipid Res 58, 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]