Summary
The adult CNS has poor ability to replace degenerated neurons following injury or disease. Recently, direct reprogramming of astrocytes into induced neurons has been proposed as an innovative strategy toward CNS repair. As a cell population that shows high diversity on physiological properties and functions depending on their spatiotemporal distribution, however, whether the astrocyte heterogeneity affect neuronal reprogramming is not clear. Here, we show that astrocytes derived from cortex, cerebellum, and spinal cord exhibit biological heterogeneity and possess distinct susceptibility to transcription factor-induced neuronal reprogramming. The heterogeneous expression level of NOTCH1 signaling in the different CNS regions-derived astrocytes is shown to be responsible for the neuronal reprogramming diversity. Taken together, our findings demonstrate that region-restricted astrocytes reveal different intrinsic limitation of the response to neuronal reprogramming.
Keywords: astrocytes, heterogeneity, neurons, ASCL1, NGN2, reprogramming
Highlights
-
•
Region-restrict astrocytes (ACs) exhibit obvious heterogeneity
-
•
Region-restrict ACs show distinct susceptibility to neuronal reprogramming
-
•
AC heterogeneity does not affect the maturation of induced neurons
-
•
Notch1 is involved in the neuronal reprogramming diversity of region-restrict ACs
Su and colleagues show that astrocytes derived from cortex, cerebellum, and spinal cord exhibit biological heterogeneity and possess distinct susceptibility to transcription factor-induced neuronal reprogramming. The heterogeneous expression level of Notch1 signaling in the different CNS regions-derived astrocytes is shown to be responsible for the neuronal reprogramming diversity.
Introduction
Astrocytes, the most abundant cells in mammalian CNS, play crucial roles in nearly all aspects of CNS function from regulating the blood flow and modulation of synaptic transmission to participation in the maintenance of blood-brain barrier (Matyash and Kettenmann, 2010, Oberheim et al., 2012, Okuda, 2018). Historically, astrocytes were regarded as homogeneous cells defined by their star-shaped morphology, and then they were divided into many types, including protoplasmic and fibrous astrocytes, by their different appearances (Chaboub and Deneen, 2012, Molofsky et al., 2012, Zhang and Barres, 2010). In the last decades, however, it was appreciated that astrocytes were much more heterogeneous in their morphological and functional properties than previously thought, which contributes to the identification of accurate subpopulations of astrocytes in the CNS (Buosi et al., 2018, Denis-Donini et al., 1984, Matyash and Kettenmann, 2010, Okuda, 2018). Based on their spatiotemporal distribution and activation level, astrocytes have been identified as a complex colony with heterogeneity on morphology, gene expression, function, and many other aspects in physiological or pathological conditions (Hu et al., 2016). For example, Emsley and Macklis (2006) depicted nine different classes of astrocytes which displayed significant dissimilarities in morphology, density, and proliferation, depending on their various distribution regions across the CNS. The heterogeneous astrocytes derived from spatiotemporal location enable us to speculate that they may fulfill specific roles for diverse neuron populations, which is indispensable for the maintenance of normal neurological activity (Ben Haim and Rowitch, 2017, Buosi et al., 2018). Furthermore, it was reported that astrocytes at different developmental stage exerted distinct effects on neuronal growth, suggesting that astrocytes also process dynamic functions and properties in term of their temporal distribution (Ben Haim and Rowitch, 2017, Buosi et al., 2018).
The irreversible loss of neurons is an important pathological feature of CNS injury and disease, resulting in persistent neurological disability. Due to the limited intrinsic neurogenic capacity and the presence of the hostile environment at the lesion site, the injured CNS resulted from wound or disease cannot self-repair and self-restore to an original degree (Hashemian et al., 2015). Since the generation of induced pluripotent stem cells (iPSCs) by Takahashi and Yamanaka (2006), cell reprogramming has attracted the most attention as a potential therapeutic strategy for CNS injury and disease. Because of the ubiquitous distribution in the CNS and the close lineage to neuron, astrocytes have been selected as an ideal candidate for neuronal reprogramming. Currently, astrocytes have been successfully reprogrammed into different types of functional mature neurons using defined transcription factors in vitro (Berninger et al., 2007, Heinrich et al., 2010). For example, Gotz and colleagues showed that the early postnatal cortical astrocytes in culture could be induced to adopt a neuronal fate after forced expression of proneural gene NGN2 (neurogenin-2) and ASCL1 (achaete-scute homolog 1 or achaete-scute complex homolog 1) (Berninger et al., 2007). Importantly, some studies have further demonstrated that the functional neurons can also be directly generated in vivo from resident astrocytes (Guo et al., 2014, Niu et al., 2013, Su et al., 2014a). In adult mammal brain and spinal cord, transcription factor SOX2, NEUROD1 or ASCL1 was sufficient to convert astrocytes into mature neurons (Guo et al., 2014, Niu et al., 2013, Su et al., 2014a). In spite of these advancements, little is known about the extracellular and intracellular factors that regulate the direct lineage switching of astrocytes to induced neurons. It was reported that the local microenvironments including injury conditions have significant influence on the efficacy of reprogramming and subsequent survival of newly generated neurons in the adult rodent brain (Grande et al., 2013). However, whether the neuronal reprogramming is affected by the heterogeneity of astrocytes remains an open question.
To tackle this issue, astrocytes derived from different CNS regions, including cortex, cerebellum, and spinal cord, were used for direct lineage reprogramming. Here, we found that region-restrict astrocytes exhibit marked biological heterogeneity and showed distinct susceptibility to transcription factor-induced neuronal reprogramming. Of note, heterogeneous expression level of NOTCH1 signaling was identified in the different CNS regions-derived astrocytes, which is responsible for the neuronal reprogramming diversity. Taken together, our findings suggest that region-restricted astrocytes reveal diverse intrinsic limitation of the response to neuronal reprogramming. This proof-of-principle study will significantly expand our understanding of the regulation of neuronal reprogramming.
Results
Region-Restrict Astrocytes Exhibit Heterogeneous Gene Expression and Proliferation Activity
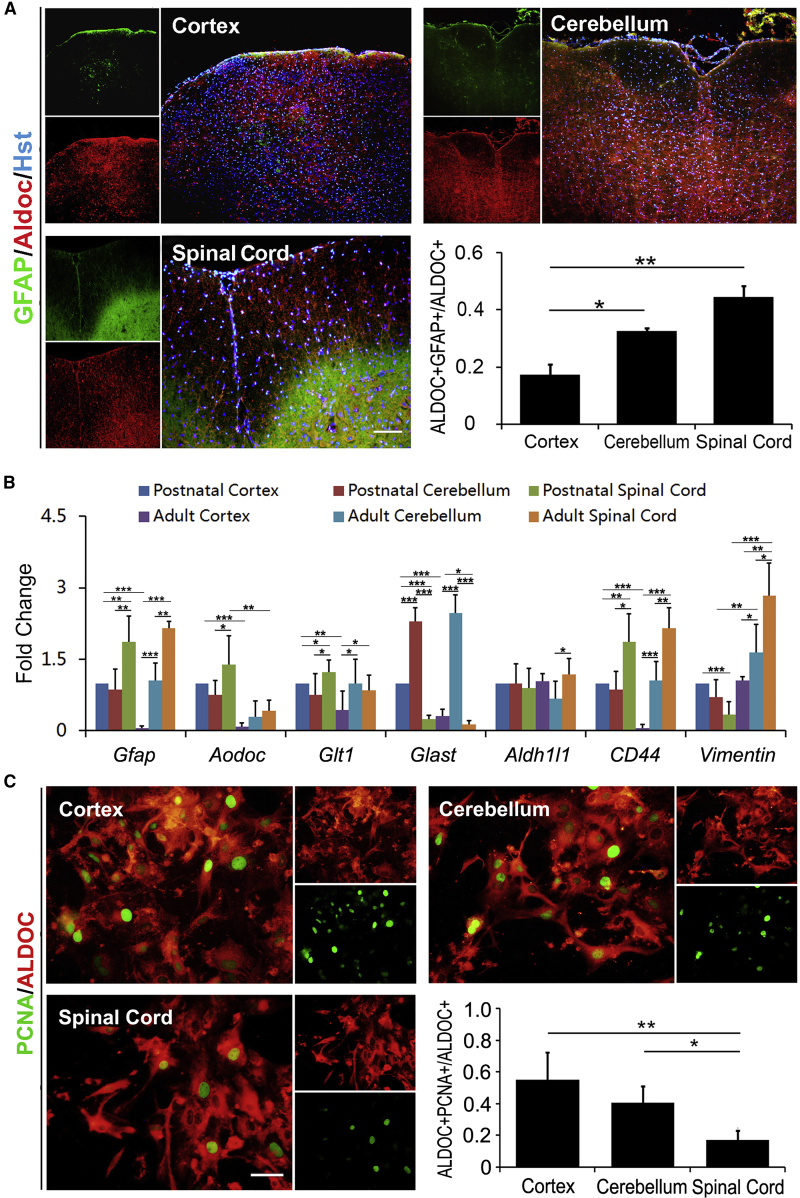
Traditionally, protoplasmic and fibrous astrocytes have been identified in gray matter and white matter of the CNS, respectively. However, in addition to the differences between protoplasmic and fibrous astrocytes, conspicuous molecular heterogeneity has been described within the astrocytes of different CNS regions (Hu et al., 2016). Glial fibrillary acidic protein (GFAP) has been used as a regular marker to identify astrocytes for several decades. However, there is growing evidence that the GFAP-labeled cells cannot represent all the astrocytes in the CNS (Hu et al., 2016). GFAP is mainly expressed in mature fibrous astrocytes and reactive astrocytes. To investigate the molecular heterogeneity of astrocytes from different CNS regions, immunohistochemistry was performed using antibodies against GFAP and Aldoc (aldolase C, a universal astrocyte marker) (Bachoo et al., 2004, Tsai et al., 2012). Figure 1A shows that Aldoc+ cells dispersed consistently across the cerebral cortex, cerebellum, and spinal cord, whereas GFAP+ cells tended to be enriched in spinal cord compared with the other two brain regions. Quantitative analysis indicated that the ratio of GFAP+/Aldoc+ cells to Aldoc+ cells in the cerebral cortex, cerebellum, and spinal cord was 17.3%, 32.5%, and 44.5%, respectively (Figure 1A). To further demonstrate the molecular heterogeneity of region-restrict astrocytes, we isolated and cultured postnatal astrocytes from the cerebral cortex, cerebellum, and spinal cord. Using a protocol we successfully established recently (Sun et al., 2017), correspondingly, adult astrocytes were also prepared from the three different regions (Figure S1A). Then, the expression level of astrocyte characteristic genes, including Gfap, Aldoc, Glt1, Glast, Aldh1l1, CD44, and Vimentin, was detected in these postnatal or adult astrocytes by real-time qPCR. As shown in Figure 1B, the astrocytes of different origins exhibited distinct expression patterns in the selected genes, suggestive of their molecular heterogeneity.
Figure 1.
Astrocyte Heterogeneity in Gene Expression and Proliferation
(A) Immunohistochemical analysis of GFAP and Aldoc in astrocytes from adult mouse cortex, cerebellum, and spinal cord (n = 3; cells = 1,500–2,000 for each condition).
(B) The heterogeneous gene expression in cortex-, cerebellum-, and spinal cord-derived cultured postnatal or adult astrocytes determined by real-time PCR (a representative result from three independent experiments).
(C) The heterogeneous cell proliferation of regional postnatal astrocytes (n = 3; cells = 1,000–1,500 for each condition).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by ANOVA Tukey's post hoc test. Scale bars, 100 μm (A) and 50 μm (C).
In addition, the proliferative activity of postnatal astrocytes was assessed in culture by staining for PCNA, a nuclear protein expressed at high levels in proliferating cells. Immunocytochemical analysis revealed that cortex-derived astrocytes possessed a high proliferation rate, while spinal cord-derived astrocytes showed a low proliferation rate (Figure 1C). There was more than 55% proliferating astrocytes in the cortex but only less than 20% in spinal cord (Figure 1C). Meanwhile, we also analyzed the proliferative activity in cultured astrocytes from adult mice. Similarly, cortex-derived astrocytes showed a higher proliferation rate than cerebellum- or spinal cord-derived astrocytes (Figure S1B). Taken together, the region-restrict astrocytes exhibit significant heterogeneities in characteristic molecule expression and proliferative activity.
Region-Restrict Astrocytes Possess Distinct Susceptibility to Neuronal Reprogramming
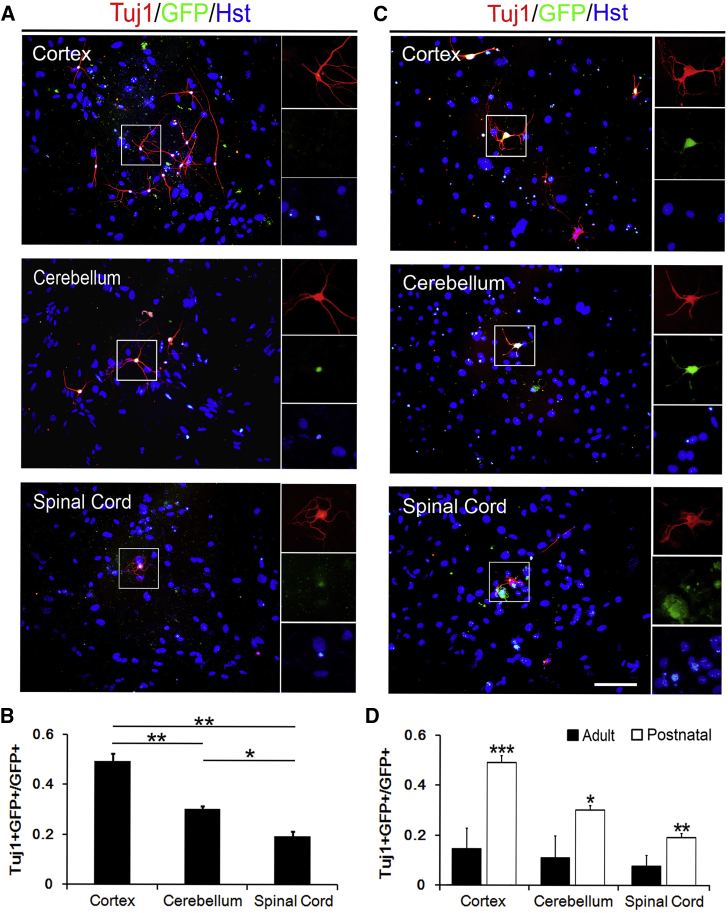
Astrocytes can be converted to neuron-like cells by forced expression of transcription factors in vitro (Berninger et al., 2007, Sun et al., 2017). It was reported that a single transcription factor, such as NGN2 or ASCL1, is sufficient to reprogram astrocytes into neurons (Berninger et al., 2007). To investigate the effects of the heterogeneity of region-restrict astrocytes on the transcription factor-mediated neuronal reprogramming, a lentiviral delivery system in which gene expression was under the control of a human GFAP promoter was used to specifically target astrocytes. Twelve days after infected with lentivirus expressing NGN2, both postnatal and adult astrocytes were induced to obtain a neuron-like morphology (Figures 2A and 2C). Immunostaining showed that these NGN2-induced cells were positive for Tuj1, a pan-neuronal marker (Figures 2A and 2C). For the postnatal astrocytes from different regions, the reprogramming efficiency was estimated at about 50% for cortex, 30% for cerebellum, and 20% for spinal cord (Figure 2B). The similar heterogeneity of the NGN2-mediated reprogramming efficiency was observed in adult astrocytes cultured from cortex, cerebellum, and spinal cord (Figure 2D). As shown in Figure 2D, strikingly, the neuronal induction efficiency of adult astrocytes from all these three regions was much lower than that of postnatal astrocytes. Therefore, astrocytes from spinal cord, especially those from adult animals, tend to be more difficult to be reprogrammed into neurons by NGN2. Of note, terminal deoxynucleotidyl transferase dUTP nick-end labeling staining showed no significant difference in the apoptosis of region-restrict astrocytes-derived immature neurons, suggesting that the heterogeneity of the NGN2-mediated reprogramming efficiency did not result from heterogeneous cell apoptosis (Figure S2).
Figure 2.
NGN2-Induced Neuronal Reprogramming Efficiency of Postnatal and Adult Astrocytes
(A) Representative pictures of NGN2-induced neurons from postnatal astrocytes by staining with Tuj1 at 12 dpi.
(B) NGN2-induced neuronal reprogramming efficiency of postnatal astrocytes (n = 3; cells = 900–1,500 for each condition).
(C) Representative pictures of NGN2-induced neurons from adult astrocytes by staining with Tuj1 at 12 dpi.
(D) Comparing NGN2-induced neuronal reprogramming efficiency of adult astrocytes compared with that of postnatal astrocytes (n = 3; cells = 700–1,500 for each condition).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by ANOVA Tukey's post hoc test. Scale bar, 100 μm.
To determine whether the phenotype was also applicable to other transcription factor-mediated neuronal conversion, we also tested the effects of region-restrict astrocyte heterogeneity on the neuronal reprogramming induced by ASCL1. Figure S3A showed that ASCL1 alone can result in the generation of induced neurons expressing Tuj1. Consistent with NGN2, the efficiency of ASCL1-mediated astrocyte-to-neuron conversion was high in the cortex group but rather low in the spinal cord group (Figures S3A and S3B). Taken together, these results demonstrate that no matter what transcription factor is used, region-restrict astrocytes show distinct susceptibility to neuronal reprogramming.
Maturation of Region-Restrict Astrocyte-Derived Induced Neurons
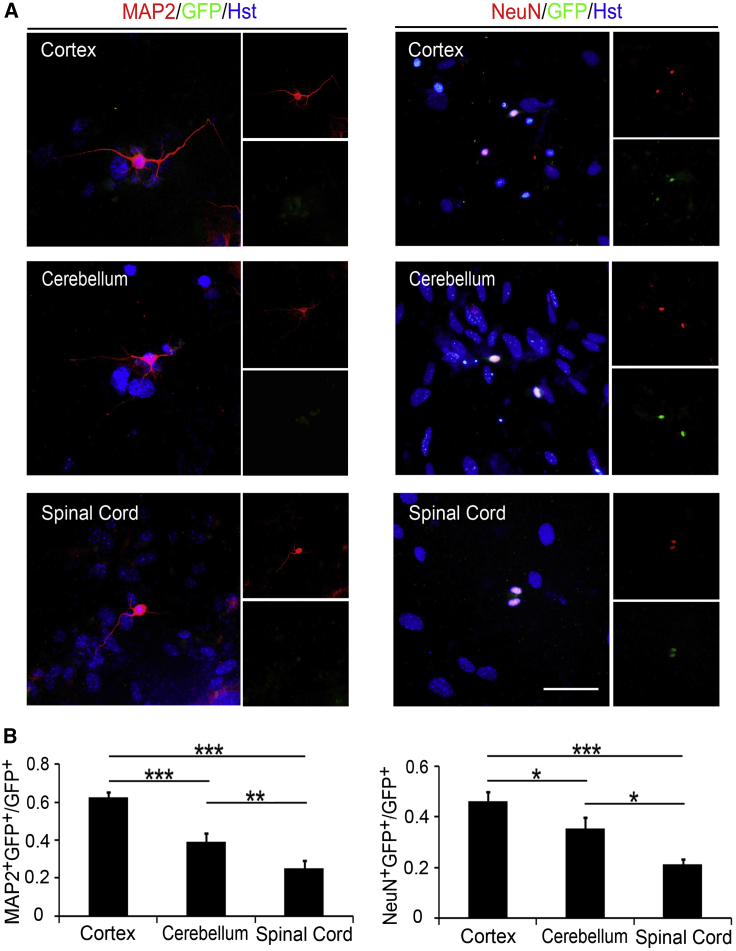
As a neuronal marker, Tuj1 is broadly expressed in both immature and mature neurons. Therefore, the maturation of induced neurons was immunocytochemically analyzed by staining with mature neuronal markers, MAP2 and NeuN. Eighteen days after being induced by NGN2, MAP2 and NeuN were detected in GFP-labeled cells (Figure 3A), indicating that all cortex-, cerebellum-, and spinal cord-derived astrocytes could be reprogrammed into mature neurons. Just as the distinct reprogramming efficiency observed in astrocytes cultured from the three different CNS regions, quantitative analysis also showed that the ratio of either MAP2+ or NeuN+ cells was highest in cortex group and lowest in spinal cord group (Figure 3B). This result suggested that the maturation of induced neurons was not affected by the heterogeneity of region-restrict astrocytes.
Figure 3.
Maturation of NGN2-Induced Neurons
(A) Representative micrographs of mature NGN2-induced neurons from postnatal astrocytes by staining with mature neuronal marker MAP2 and NeuN at 18 dpi.
(B) Quantification of mature neurons induced from different region-derived postnatal astrocytes (n = 3; cells = 500–1,000 for each condition).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by ANOVA Tukey's post hoc test. Scale bar, 50 μm.
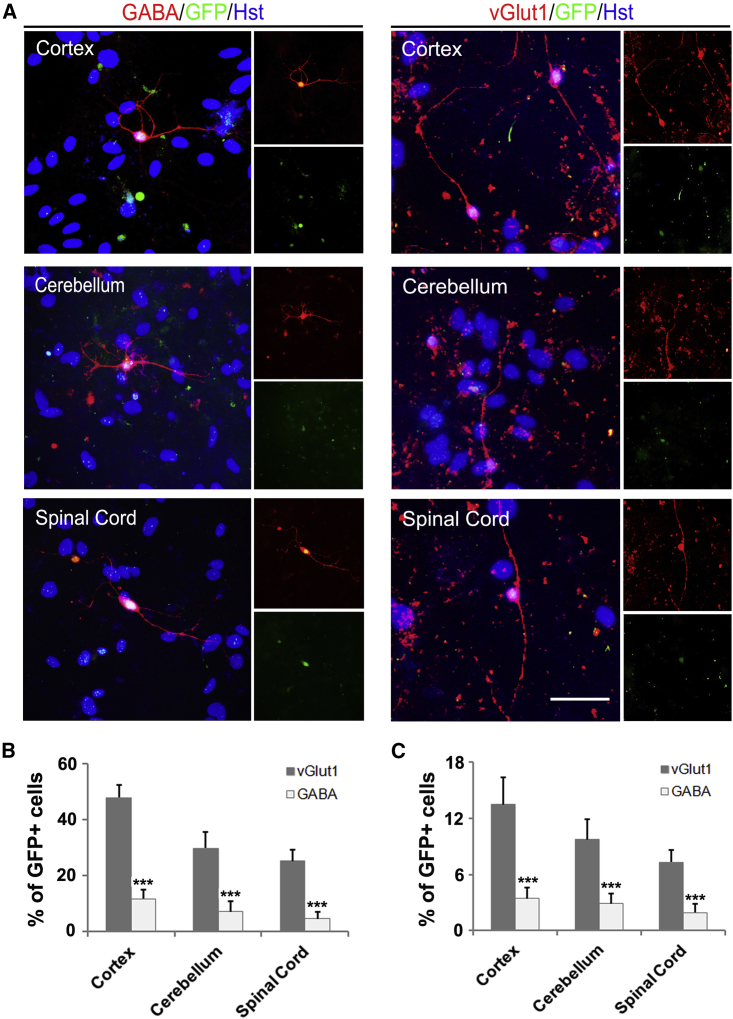
It was reported that NGN2 and ASCL1 directed cultured astrocytes to mainly generate glutamatergic and GABAergic neurons, respectively (Berninger et al., 2007, Heinrich et al., 2010). In our study, the subtype of NGN2-induced neurons was identified by immunocytochemical analysis using antibody against vGlut1 or GABA, specific markers for excitatory or inhibitory interneurons, respectively. Figure 4A showed that both vGlut1+ and GABA+ neurons could be detected in the NGN2-mediated astrocyte-to-neuron system, suggestive of mixed neurons. In parallel with previous reports, both postnatal and adult cortex-derived astrocytes were primarily converted into vGlut1-labeled excitatory neurons (Figures 4B and 4C). Similarly, the majority of NGN2-induced neurons from either cerebella or spinal cord-derived astrocytes were also glutamatergic (Figures 4B and 4C). There was only a small proportion of the NGN2-induced neurons that obtained a GABAergic identity. For ASCL1-mediated reprogramming, in contrast, astrocytes cultured from cortex, cerebella, and spinal cord were all mainly converted into GABAergic neurons (Figures S3C and S3D). Therefore, it seemed that the cell identity preference of NGN2- or ASCL1-mediated neuronal reprogramming from astrocytes was not affected by their local origin heterogeneity. Together, all these findings suggest that although region-restrict astrocytes show distinct susceptibility to neuronal reprogramming, the induced neurons can become mature and tend to acquire a glutamatergic or GABAergic identity.
Figure 4.
NGN2-Induced Neurons Mainly Matured into Glutamatergic Neurons
(A) Representative pictures of mature GABAergic and glutamatergic neurons induced from different region-derived postnatal astrocytes at 18 dpi.
(B and C) Quantification of GABAergic and glutamatergic neurons reprogrammed from different region-derived postnatal (B) or adult (C) astrocytes (n = 3; cells = 300–600 for each condition).
∗∗∗p < 0.001 by ANOVA Tukey's post hoc test. Scale bar, 50 μm.
Identification of Signaling Pathways and Molecules Involved in Mediating the Distinct Susceptibility of Region-Restrict Astrocytes to Neuronal Reprogramming
Because the region-restrict astrocytes exhibit significant heterogeneities in proliferative activity (Figures 1C and S1B), we wanted to know whether the heterogeneous proliferative activity has an effect on the efficiency of NGN2- or ASCL1-mediated neuronal reprogramming. Astrocyte proliferation was blocked with aphidicolin (Abcam ab142400; 5 μg/mL, dissolved in DMSO) and infected with NGN2- or ASCL1-expressing lentivirus to induce neuronal reprogramming. As shown in Figure S4, no significant difference was observed in the reprogramming efficiency between aphidicolin- and DMSO-treated astrocytes, suggesting that the proliferation of both postnatal and adult astrocytes did not affect the susceptibility of region-restrict astrocytes to NGN2- or ASCL1-mediated neuronal reprogramming. In addition, it was reported that the metabolic status of the astrocytes is a key parameter for direct neuronal reprogramming (Gascon et al., 2016). To determine whether the heterogeneous neuronal reprogramming efficiency of region-restrict astrocytes results from different metabolic status in these astrocytes, metabolic factors such as reactive oxygen species (ROS), ATP, and lipid peroxidation were examined in these cultured region-restrict astrocytes. Figure S5A–S5E show that no significant difference was observed in the ROS, ATP, or lipid peroxidation level between cortex, cerebellum, and spinal cord astrocytes.
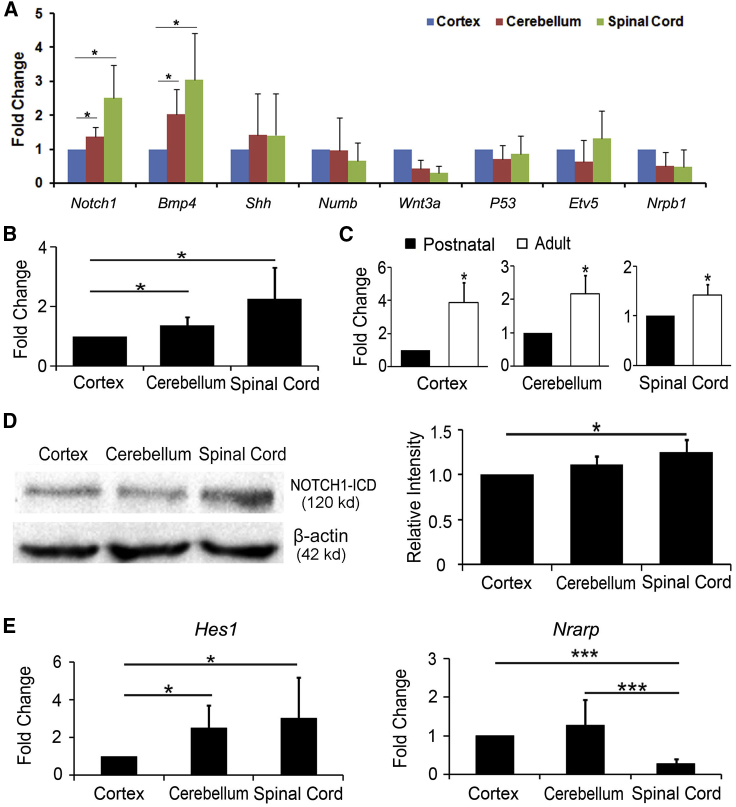
Many factors and signaling pathways closely related to neurogenesis and neural conversion have been identified in previous studies (Hawkins et al., 2014, Lujan et al., 2015, Petersen et al., 2002, Rivetti di Val Cervo et al., 2017, Shan et al., 2011, Wang et al., 2016). For example, INK4a (also known as CDKN2A, cyclin-dependent kinase inhibitor 2A), a tumor suppressor protein that plays an important role in cell-cycle regulation, was identified as a cell-intrinsic barrier for in vitro reprogramming (Li et al., 2009). As a transcription factor, ectopic SOX2 could induce neuronal reprogramming of resident astrocytes in the adult mouse brain and spinal cord (Niu et al., 2013, Su et al., 2014a). For signaling pathways, activating the Shh and Wnt pathways was reported to facilitate neural reprogramming from human astrocytes (Rivetti di Val Cervo et al., 2017). BMP4 played key roles in the neural specification and fate decisions at various stages of development (Shan et al., 2011). Moreover, our previous study revealed that the p53-dependent pathway inhibited the overall production of SOX2-induced neuroblasts after spinal cord injury (Wang et al., 2016). To elucidate the mechanism underlying the heterogeneous reprogramming efficiency from region-restrict astrocytes, we firstly examined the expression of SOX2 and INK4a in astrocytes. However, immunocytochemical analysis showed no significant difference in the expression levels of SOX2 and INK4a in astrocytes cultured from cortex, cerebellum, and spinal cord (Figures S5F–S5I). Furthermore, we also estimated and screened a set of signaling pathways and transcriptional molecules, including Notch1, Shh, Bmp4, Numb, Wnt3a, P53, Etv5, and Nr0b1 (Hawkins et al., 2014, Lujan et al., 2015, Petersen et al., 2002). Real-time PCR analysis showed that the transcriptional level of these molecules varied in astrocytes from postnatal cortex, cerebellum, and spinal cord, reminiscent of the intrinsic diversity among the region-restrict astrocytes (Figure 5A). In the three groups, of note, Notch1 expression was significantly highest in the spinal astrocytes and lowest in the cortical astrocytes (Figure 5B). Moreover, adult astrocytes showed higher Notch1 expression level compared with postnatal astrocytes originated from the same region (Figure 5C). The differential expression of NOTCH1 in region-restrict astrocytes was also confirmed at protein level. Consistent with the result of real-time PCR, western blotting revealed that the NOTCH1 protein expressed preferentially in astrocytes from spinal cord compared with that from cortex and cerebellum (Figure 5D). HES1 is a classic target of Notch1 signaling, while NRARP is an upstream negative regulator of NOTCH1 signaling (Yun and Bevan, 2003, Zine et al., 2001). As shown in Figure 5E, Hes1 gene highly expressed in spinal cord-derived astrocytes compared with that from cortex and cerebellum, which was the opposite to the expression of Nrarp gene. Based on these expression patterns, we speculated that the NOTCH1 signaling pathway may be involved in mediating the distinct susceptibility of region-restrict astrocytes to neuronal reprogramming.
Figure 5.
Expression of Notch1 Signaling Molecules in Heterogeneous Astrocytes
(A) Heterogeneous expression of molecules associated with neuronal differentiation and specialization in postnatal astrocytes (n = 3 for each group, ∗p < 0.05 by ANOVA Tukey's post hoc test).
(B) Notch1 expression in regional postnatal astrocytes determined by real-time PCR (n = 3 for each group, ∗p < 0.05 by ANOVA Tukey's post hoc test).
(C) Comparison of Notch1 expression between postnatal and adult astrocytes by real-time PCR (n = 3 for each group, ∗p < 0.05 by Student's t test).
(D) NOTCH1 expression in regional postnatal astrocytes determined by western blotting.
(E) Notch1 signaling molecules Hes1 and Nrarp expression in postnatal astrocytes determined by real-time PCR (n = 3 for each group; ∗p < 0.05, ∗∗∗p < 0.001 by ANOVA Tukey's post hoc test).
Blocking of NOTCH1 Signaling Can Increase the Efficiency of Neuronal Reprogramming from Astrocytes
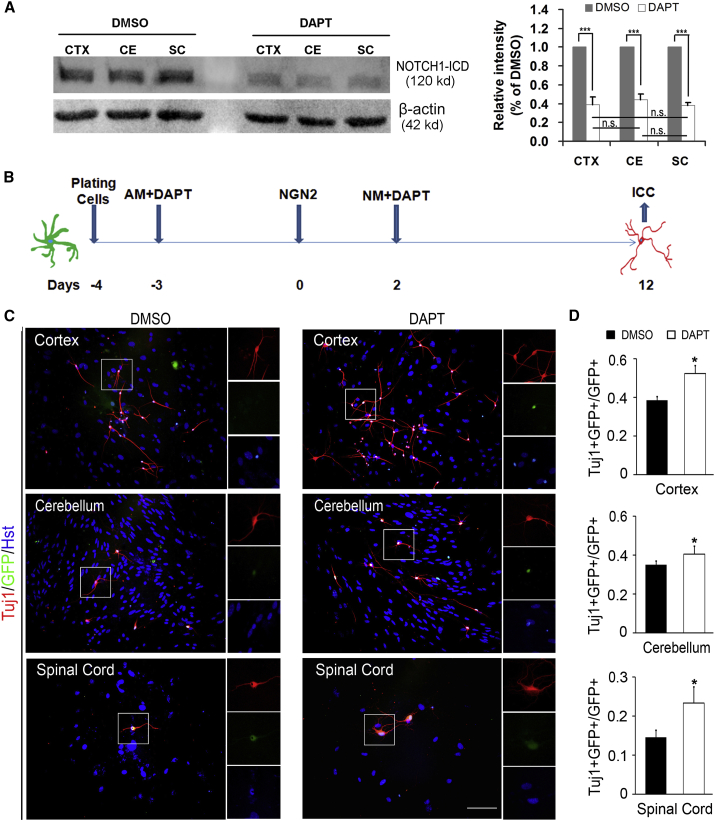
To investigate the roles of NOTCH1 signaling in neuronal reprogramming, the γ-secretase inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester) was used to block the pathway (Liu et al., 2014). DAPT can decrease the activity of γ-secretase and inhibit the generation of NICD (NOTCH1 intracellular domain), which is capable of getting through the nuclear membrane to target corresponding downstream genes (Farnie and Clarke., 2007). As shown in Figure 6A, DAPT (Sigma D5942, 10 μM, dissolved in DMSO) treatment significantly reduced NOTCH1 level in the astrocytes derived from cortex, cerebellum, and spinal cord, while there was no significant difference in the inhibitory efficiency between the three groups. After treated with DAPT, astrocytes were infected with NGN2- or ASCL1-expressing lentivirus to induce neuronal reprogramming (Figures 6B and S6A). Figures 6C and 6D showed that inhibition of NOTCH1 with DAPT resulted in a significant increase in the NGN2-mediated neuronal reprogramming efficiency of postnatal astrocytes from cortex, cerebellum, and spinal cord. Similarly, the efficiency of ASCL1-induced neuronal reprogramming was also significantly increased by treatment of region-restrict astrocytes with DAPT (Figures S6B and S6C). Of note, DAPT treatment did not affect the maturation and cell subtype of the induced neurons from region-restrict astrocytes (Figure S7).
Figure 6.
DAPT Treatment Increased the Efficiency of NGN2-Induced Neuronal Reprogramming from Postnatal Astrocytes
(A) The knockdown efficiency of DATP in postnatal astrocytes was determined by western blotting. Quantitative analysis showed no significant difference in the knockdown efficiency of DAPT between cortex (CTX), cerebellum (CE), and spinal cord (SC) astrocytes. n = 3 for each group. ∗∗∗p < 0.001 (Student's t test); p ≥ 0.05, no statistically significant difference (n.s.) (ANOVA Tukey's post hoc test).
(B) Experimental scheme of blocking NOTCH1 signaling in astrocytes by DAPT during neuronal reprogramming. AM, astrocyte culture medium; NM, neuronal induction medium.
(C) Representative pictures of NGN2-induced neurons with or without DAPT treatment by staining with Tuj1 at 12 dpi.
(D) The efficiency of NGN2-induced neuronal reprogramming in the DAPT group compared with control group (n = 3; cells = 1,000–1,700 for each condition). ∗p < 0.05 by ANOVA Tukey's post hoc test. Scale bar, 100 μm.
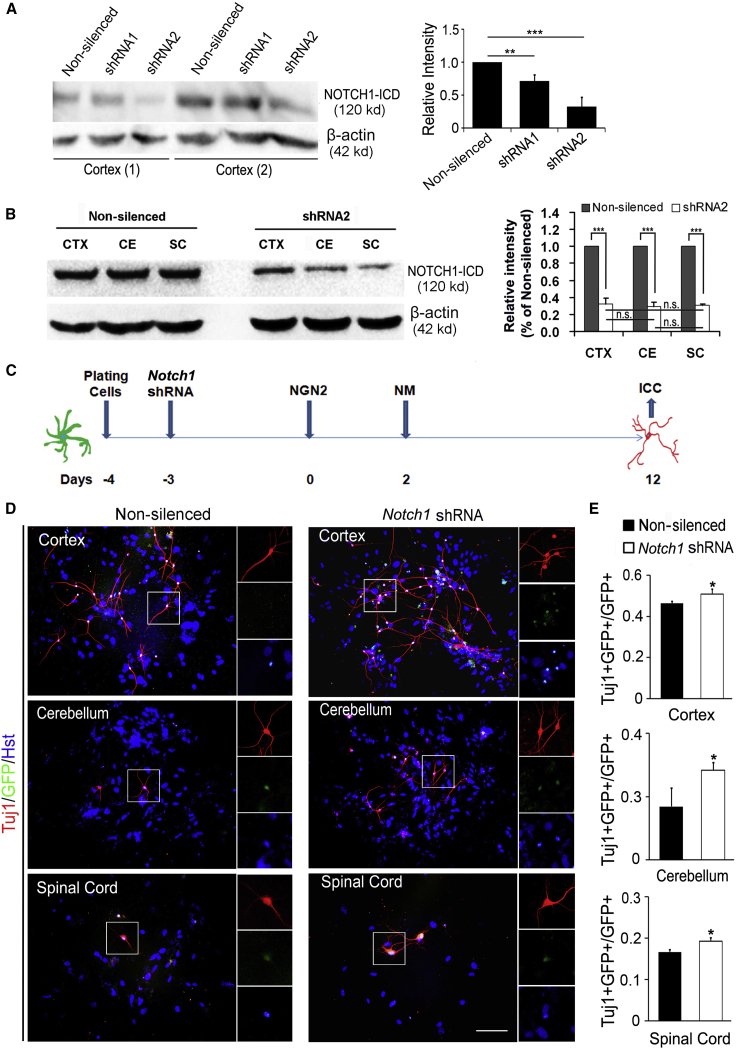
In addition, RNA interference was also used to block Notch1 signaling. Two short hairpin RNA (shRNA) against Notch1 were constructed in the lentiviral vectors. The gene expression was under the control of a human GFAP promoter to specifically target astrocytes. Western blotting indicated that about 30% and 70% of the expression of NOTCH1 in cortical astrocytes were knocked down by shRNA1 and shRNA2, respectively (Figure 7A). Thus, we used shRNA2 to silence Notch1 signaling in astrocytes. Figure 7B showed that shRNA2 obviously downregulated NOTCH1 level in the astrocytes derived from cortex, cerebellum, and spinal cord, whereas no significant difference was observed in the inhibitory efficiency between the three groups. Astrocytes from cortex, cerebellum, and spinal cord were infected with shRNA2-expressing lentivirus and then subjected to NGN2- or ASCL1-induced neuronal reprogramming (Figures 7C and S6D). Similar to DAPT, blocking the Notch1 signaling with shRNA2 markedly increased the yield of neurons induced by NGN2 or ASCL1 (Figures 7D, 7E, S6E, and S6F). Together, these results suggest that the NOTCH1 signaling pathway is involved in mediating the distinct susceptibility of region-restrict astrocytes to transcription factor-mediated neuronal reprogramming.
Figure 7.
Interference of NOTCH1 Signaling Increased the Efficiency of NGN2-Induced Neuronal Reprogramming from Postnatal Astrocytes
(A) Western blotting analysis of the knockdown efficiency of Notch1 shRNA in postnatal cortex astrocytes. n = 3 for each group. ∗∗p < 0.01, ∗∗∗p < 0.001 by ANOVA Tukey's post hoc test.
(B) The knockdown efficiency of Notch1 shRNA in postnatal astrocytes was determined by western blotting. Quantitative analysis showed no significant difference in the knockdown efficiency of Notch1 shRNA between cortex (CTX), cerebellum (CE), and spinal cord (SC) astrocytes. n = 3 for each group. ∗∗∗p < 0.001 (Student's t test); p ≥ 0.05, no statistically significant difference (n.s.) (ANOVA Tukey's post hoc test).
(C) Experimental scheme of Notch1 knockdown in astrocytes by lentivirus-mediated shRNA during neuronal reprogramming.
(D) Representative pictures of NGN2-induced neurons with or without shRNA interference by staining with Tuj1 at 12 dpi.
(E) The efficiency of NGN2-induced neuronal reprogramming in Notch1 shRNA group compared with control group (n = 3; cells = 1,000–1,700 for each condition). ∗p < 0.05 by ANOVA Tukey's post hoc test. Scale bar, 100 μm.
Discussion
Although astrocytes have been considered to be a homogeneous and non-excitable cell type in the CNS, accumulating evidence suggests that they differ in their morphology, developmental origin, gene expression profile, physiological properties, function, and response to injury and disease. For example, GFAP is mainly expressed in astrocytes located in white matter and S100β tends to be expressed in gray matter astrocytes (Ben Haim and Rowitch, 2017, Rusnakova et al., 2013). In contrast to protoplasmic astrocytes in cerebral cortex and striatum, high levels of GFAP are expressed in hippocampus-derived astrocytes and Bergmann glia in the cerebellum (Lein et al., 2007, Wilhelmsson et al., 2006). The regional differences are also observed in the expression patterns of EAAT isoforms within the gray matter astrocytes (Lehre et al., 1995). Therefore, a better understanding of the heterogeneity of astrocytes will undoubtedly contribute to identification of astroglial function and potential application. By immunohistochemistry and real-time PCR, in present study, distinct expression patterns of the characteristic molecules were observed in astrocytes from postnatal and adult cortex, cerebellum, and spinal cord, suggestive of their molecular heterogeneity. In addition, either postnatal or adult astrocytes from different CNS regions also exhibited significant heterogeneities in the proliferative activity.
Currently, direct lineage reprogramming of astrocytes into induced neurons provides a valuable cell source for regenerative medicine, drug discovery, and disease modeling. It has been documented that cerebral cortex-derived postnatal astrocytes are susceptible to lineage reprogramming into neurons. However, it remains unknown whether astrocytes from other age or other CNS regions are susceptible to neuronal reprogramming. Moreover, whether the astrocyte heterogeneity affect neuronal reprogramming is also not clear. Here, we showed that astrocytes derived from cortex, cerebellum, and spinal cord of postnatal or adult mice could all be converted into neurons by defined transcription factors. Of note, postnatal and adult astrocytes from the same spatial domain showed distinct susceptibility to transcription factor-induced neuronal reprogramming. Likewise, the heterogeneous susceptibility was also observed in the neuronal reprogramming of different region-derived astrocytes. However, it seemed that the maturation and cell subtype of the induced neurons were not affected by the heterogeneity of region-restrict astrocytes.
Notch signaling is an evolutionarily conserved cell-cell contact pathway that was reported to be responsible for neural stem cell maintenance and neurogenesis in the embryonic brain as well as in the adult brain (Imayoshi et al., 2013, Zhang et al., 2018). A key feature of Notch signaling is its remarkable cell-context dependency (Schwanbeck et al., 2011). In the adult brain, Notch signaling components are highly expressed in astrocytes, which is necessary for maintaining a mature and differentiated astrocyte fate (Cahoy et al., 2008). Intriguingly, NOTCH1 signaling is reduced in astrocytes after stroke, and blocking NOTCH1 signaling triggers astrocytes in the striatum and the medial cortex to enter a neurogenic program, resulting in the generation of new neurons (Magnusson et al., 2014). Thus, we here speculated that region-specific astrocytes might maintain different levels of NOTCH1 signaling that are responsible for the distinct efficiency of neuronal reprogramming. Indeed, our results showed that differential expression of NOTCH1 signaling was observed in astrocytes from cortex, cerebellum, and spinal cord. Spinal cord-derived astrocytes (especially adult astrocytes) expressed high level of NOTCH1 signaling and are not susceptible to neuronal reprogramming. In contrast, low level of NOTCH1 signaling was expressed in cortex-derived astrocytes (especially postnatal astrocytes) that are inclined to neuronal reprogramming. Moreover, blocking NOTCH1 signaling with γ-secretase inhibitor DAPT or lentiviral vector-mediated shRNA resulted in an increase in the efficiency of lineage reprogramming into neurons. All these data suggested that the NOTCH1 signaling pathway might be involved in mediating the distinct susceptibility of region-restrict astrocytes to neuronal reprogramming. Of note, although no significant difference was observed in the knockdown efficiency of DAPT and Notch1 shRNA between cortical, cerebellum, and spinal cord astrocytes, the increase of neuronal reprogramming efficiency was relatively small in DAPT-treated cerebellum astrocytes and Notch1 shRNA-treated cortex astrocytes, suggesting that, in addition to the NOTCH1 signaling pathway, other factors might also contribute to the heterogeneous susceptibility of region-restrict astrocytes to neuronal reprogramming.
Although our study provided evidence that the distinct susceptibility of region-restrict astrocytes to neuronal reprogramming might resulted from the different level of NOTCH1 signaling pathway in these cells, the underlying mechanisms remained to be elucidated. As a single-pass transmembrane receptor, Notch is activated by interaction with its ligands (Jagged or Delta). After undergoing two sequential cleavages mediated by a metalloprotease (ADAM) and a γ-secretase, the NICD is released and then translocates into the nucleus, ultimately activating the transcriptional complex (Andersson et al., 2011). Recently, Notch has been identified as a strong inducer of chromatin remodeling, suggestive of a possible role in regulating higher-order chromatin structures (Wang et al., 2015). It was reported that NICD could block the binding of MEF2C (myocyte enhancer factor 2C) to chromatin and suppress its transcriptional activity in myotubes (Wilson-Rawls et al., 1999). Notch inactivation was shown to increase binding of the transcription factor MEF2C to the promoter regions of cardiac structural genes and promote the reprogramming of mouse fibroblasts into induced cardiac-like myocytes (Abad et al., 2017). These results raise the question of whether the NOTCH1 in region-restrict astrocytes reduce the chromatin accessibility of proneural factors (such as NGN2 and ASCL1) to target genes and make them refractory to neuronal reprogramming. Therefore, further study is needed to fully understand the underlying mechanism.
In summary, our study provided evidence that cortex-, cerebellum-, and spinal cord-derived postnatal or adult astrocytes are heterogeneous populations and they showed distinct susceptibility to transcription factor-mediated neuronal reprogramming. The heterogeneous expression level of NOTCH1 signaling in these region-restrict astrocytes was shown to be responsible for the neuronal reprogramming diversity.
Experimental Procedures
Animals
Wild-type newborn (P2–P4) and adult (8–12 weeks old) C57/BL6J mice were obtained from Shanghai Ling Chang Biotech, and used for primary astrocyte culture and histological analysis. The mice were maintained in a conventional animal facility under a controlled temperature and a 12-h light/dark cycle with access to sufficient food and water. All animal care and experimental procedures were performed according to the guidelines recommended by the NIH and were approved by the Animal Experimentation Ethics Committee of the Second Military Medical University.
Primary Astrocyte Culture
Highly enriched primary postnatal astrocytes were isolated from the cerebral cortex, cerebellum, and spinal cord of newborn mice as described previously (Su et al., 2014a). After removal of the meninges, the tissues were dissociated into a single-cell suspension by trypsinization and mechanical disruption. Cells were pelleted for 4 min at 800 rpm, re-suspended and plated in DMEM medium containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin at 37°C with 5% CO2. To prepare primary adult astrocyte, a culture system we previously established was used. The single-cell suspension derived from cerebral cortex, cerebellum, and spinal cord of adult mice was seeded on the culture plate pre-coated with gelatin and Matrigel in DMEM medium containing 20% FBS, forskolin (Fsk) (10 μM), glial-derived neurotrophic factor (10 ng/mL), and 1% penicillin-streptomycin at 37°C with 5% CO2. For both postnatal and adult astrocytes, the medium was changed every 3 days. When cultured cells grew to confluence, loosely attached microglia and oligodendrocyte precursor cells were removed from the cell monolayer by shaking vigorously. The attached enriched astrocytes were subsequently detached using trypsin-EDTA and then subjected to subsequent experiments. Immunocytochemical analysis by Iba-1 staining showed that there was less than 3% contaminating microglia in the cultured primary adult astrocytes (data not shown).
Lentivirus Preparation
Plasmids construction and lentivirus production were performed as described previously (Su et al., 2014a, Wang et al., 2016). In brief, cDNAs encoding human NGN2 or ASCL1 were subcloned into a third-generation lentiviral vector (pCSC-SP-IRES-GFP; IRES: internal ribosomal entry site) to generate pCSC-SP-PW-NGN2-IRES-GFP (abbreviated NGN2) or pCSC-SP-PW-ASCL1-IRES-GFP (abbreviated ASCL1). The gene expression was under the control of a human GFAP promoter and the virus-infected cells were visualized by the co-expressed GFP. HEK293T cells were used to generate replication-deficient lentivirus by transient transfection with the lentiviral vector and packaging plasmids (pMDL, VSV-G, and pREV). Lentivirus was collected, precipitated with polyethylene glycol 8000, and concentrated by centrifugation.
Lineage Reprogramming
For neuronal reprogramming, cultured astrocytes (4 × 104/mL) were seeded on culture vessels or glass coverslips pre-coated with gelatin and Matrigel. Twenty-four hours after plating, astrocytes were infected with the NGN2- or ASCL1-expressing lentivirus (multiplicity of infection of ∼1) in the presence of 6 μg/mL polybrene. The following day, culture medium was switched to neuronal induction medium, DMEM:F12:neurobasal (2: 2: 1) containing 0.8% N-2 (Invitrogen, Carlsbad, CA, USA), and 0.4% B-27 (Invitrogen). Fsk (10 μM) and dorsomorphin (1 μM) were also added to the above induction medium.
RNA Interference
Lentiviral vectors encoding Notch1 shRNA or a scramble shRNA were constructed as described previously (Zhu et al., 2016). The sequences for shRNA are listed below: nonsilencer shRNA (5′-TTCTCCGAACGTGTCACGT-3′ and 5′-ACGTGACACGTTCGGAGAA-3′), Notch1 shRNA1 (5′-CCGGTGCATCAGCCACTTGAATGTTTCAAGAGAACATTCAAGTGGCTGATGCTTTTTTG-3′ and 5′-AATTCAAAAAAGCATCAGCCACTTGAATGTTCTCTTGAAACATTCAAGTGGCTGATGCA-3′), and Notch1 shRNA2 (5′-CCGGTGGGCTATGAATTTCACCGTTTCAAGAGAACGGTGAAATTCATAGCCCTTTTTTG-3′ and 5′-AATTCAAAAAAGGGCTATGAATTTCACCGTTCTCTTGAAACGGTGAAATTCATAGCCCA-3′). The knockdown efficiency of NOTCH1 signaling was measured by western blot. The nonsilencer group was used as a control.
Immunofluorescence
For immunocytochemistry, cells cultured on coverslips were fixed with 4% (w/v) paraformaldehyde (PFA) for 20 min at room temperature and then washed three times with 1 × PBS. For immunohistochemistry, animals were deeply anesthetized with intraperitoneal injection of 10% chloral hydrate and transcardially perfused with 20 mL PBS and 20 mL 4% (w/v) paraformaldehyde (PFA/PBS). The cerebral cortex, cerebellum, and spinal cord were isolated and post-fixed in the perfusion solution overnight at 4°C. After cryoprotection with 30% sucrose in PBS for 48–72 h at 4°C, tissues were frozen in OCT compound and coronal sections were collected on a cryostat (Leica) set at 15–20 μm thickness. Fixed cells or tissue sections were permeabilized and blocked with 0.2% Triton X-100 and 3% BSA in 1× PBS for 1 h, followed by overnight incubation at 4°C with the primary antibodies listed below: GFAP (Sigma G3893, mouse, 1:100), GFAP (Boster M00213, rabbit, 1:100), Aldoc (Santa Cruz sc-12065, goat, 1:50), Tuj-1 (Covance MMS-435P, mouse, 1:500), GFP (Aves GFP-1020, chicken, 1:600), NeuN (Sigma MAB377, mouse, 1:100), Map2 (Abcam ab32454, rabbit, 1:200), GABA (Sigma A2052, rabbit, 1:200), and vGlut1 (SYSY 135302, rabbit, 1:100). The corresponding secondary antibodies conjugated with Alexa Fluor 543, 488, or 647 (Jackson ImmunoResearch Laboratories) were applied at 1:200 dilution for indirect fluorescence. Nuclei were counterstained with Hoechst (Sigma 33,342, 1:1,000). Images were captured with an Olympus fluorescence microscope or a Zeiss LSM 510 confocal microscope.
Cell Proliferation Assays
In vitro, proliferating cells were labeled by incubation with 10 mM bromodeoxyuridine (BrdU) (Sigma B5002) diluted in culture medium for 12–16 h as described previously (Su et al., 2014b). In brief, 4% PFA-fixed cells were treated with 2 M HCl for 30 min at 37°C to denature DNA, rinsed in 0.1 M boric acid for 10 min to neutralize excess HCl, and incubated with blocking solution (0.2% Triton X-100 and 3% BSA in 1× PBS) for 1 h. The BrdU incorporation was detected by fluorescent staining using an anti-BrdU antibody (Sigma B8434, mouse, 1:200). In addition, proliferating cells were also detected directly with antibodies such as Ki67 (Abcam ab6526, mouse, 1:100) and PCNA (Boster BM0104, mouse, 1:100).
Real-Time qPCR
Real-time qPCR was performed as described previously (Sun et al., 2017). In brief, total RNA was extracted from cultured cells using TRIzol Reagent, and then reverse transcribed to cDNA using a RevertAid First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer's instructions. Based on SYBR green I dye detection, real-time PCR assay was carried out using the corresponding primers (detailed in Table S1), and mRNA expression in each sample was normalized to GAPDH. The data were analyzed using the 2-ΔΔCt method.
Western Blot Analysis
Total proteins were extracted from cultured astrocytes at a specified time point. Then the proteins were electrophoresed on a 10% SDS-PAGE gel, followed by being transferred onto nitrocellulose membranes. The membranes were then blocked in 5% nonfat milk and incubated with specific primary antibodies against NOTCH1 (Santa Cruz sc-9170, rabbit, 1:200) and β-actin (Proteintech 60008-1-Ig, mouse, 1:5,000). Immunoreactive bands were visualized using IRDye 700- or 800-conjugated secondary antibodies and chemiluminescence reagents (ECL, Amersham) according to the manufacturer's instructions. The results were scanned using the Odyssey infrared imaging system and analyzed with Image-Pro Plus 6.0 software.
Statistical Analysis
All results are validated by at least three independent experiments. The quantitative data were expressed as mean ± SD. Statistical analysis was performed using Student's t test or one-way ANOVA with Tukey's post hoc test. Differences were considered statistically significant at p < 0.05.
Author Contributions
X. Hu, C.H., and Z.S. conceived and designed the experiments. X. Hu, S.Q., X. Huang, Y.Y., Z.T., Y.G., X.C., and D.W. performed the experiments. X.-F.L. provided critical technical inputs. X. Hu, S.Q., X. Huang, C.H., and Z.S. analyzed the data and prepared the manuscript. All authors reviewed and approved the manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation (31671110), the National Key Research and Development Program of China (2016YFA0100802) and the Science and Technology Commission of Shanghai Municipality (15JC1400202).
Published: January 31, 2019
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.12.017.
Contributor Information
Cheng He, Email: chenghe@smmu.edu.cn.
Zhida Su, Email: suzhida@smmu.edu.cn.
Supplemental Information
References
- Abad M., Hashimoto H., Zhou H., Morales M.G., Chen B., Bassel-Duby R., Olson E.N. Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Reports. 2017;8:548–560. doi: 10.1016/j.stemcr.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E.R., Sandberg R., Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- Bachoo R.M., Kim R.S., Ligon K.L., Maher E.A., Brennan C., Billings N., Chan S., Li C., Rowitch D.H., Wong W.H. Molecular diversity of astrocytes with implications for neurological disorders. Proc. Natl. Acad. Sci. U S A. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Haim L., Rowitch D.H. Functional diversity of astrocytes in neural circuit regulation. Nat. Rev. Neurosci. 2017;18:31–41. doi: 10.1038/nrn.2016.159. [DOI] [PubMed] [Google Scholar]
- Berninger B., Costa M.R., Koch U., Schroeder T., Sutor B., Grothe B., Gotz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buosi A.S., Matias I., Araujo A.P.B., Batista C., Gomes F.C.A. Heterogeneity in synaptogenic profile of astrocytes from different brain regions. Mol. Neurobiol. 2018;55:751–762. doi: 10.1007/s12035-016-0343-z. [DOI] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboub L.S., Deneen B. Developmental origins of astrocyte heterogeneity: the final frontier of CNS development. Dev. Neurosci. 2012;34:379–388. doi: 10.1159/000343723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis-Donini S., Glowinski J., Prochiantz A. Glial heterogeneity may define the three-dimensional shape of mouse mesencephalic dopaminergic neurones. Nature. 1984;307:641–643. doi: 10.1038/307641a0. [DOI] [PubMed] [Google Scholar]
- Emsley J.G., Macklis J.D. Astroglial heterogeneity closely reflects the neuronal-defined anatomy of the adult murine CNS. Neuron Glia Biol. 2006;2:175–186. doi: 10.1017/S1740925X06000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnie G., Clarke R.B. Mammary stem cells and breast cancer–role of Notch signalling. Stem Cell Rev. 2007;3:169–175. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- Gascon S., Murenu E., Masserdotti G., Ortega F., Russo G.L., Petrik D., Deshpande A., Heinrich C., Karow M., Robertson S.P. Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell. 2016;18:396–409. doi: 10.1016/j.stem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Grande A., Sumiyoshi K., Lopez-Juarez A., Howard J., Sakthivel B., Aronow B., Campbell K., Nakafuku M. Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat. Commun. 2013;4:2373. doi: 10.1038/ncomms3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemian S., O'Rourke C., Phillips J.B., Stromberg I., Af Bjerken S. Embryonic and mature astrocytes exert different effects on neuronal growth in rat ventral mesencephalic slice cultures. Springerplus. 2015;4:558. doi: 10.1186/s40064-015-1362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins K., Joy S., McKay T. Cell signalling pathways underlying induced pluripotent stem cell reprogramming. World J. Stem Cells. 2014;6:620–628. doi: 10.4252/wjsc.v6.i5.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Blum R., Gascon S., Masserdotti G., Tripathi P., Sanchez R., Tiedt S., Schroeder T., Gotz M., Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Yuan Y., Wang D., Su Z. Heterogeneous astrocytes: active players in CNS. Brain Res. Bull. 2016;125:1–18. doi: 10.1016/j.brainresbull.2016.03.017. [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Shimojo H., Sakamoto M., Ohtsuka T., Kageyama R. Genetic visualization of notch signaling in mammalian neurogenesis. Cell Mol. Life Sci. 2013;70:2045–2057. doi: 10.1007/s00018-012-1151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehre K.P., Levy L.M., Ottersen O.P., Storm-Mathisen J., Danbolt N.C. Differential expression of two glial glutamate transporters in the rat brain: quantitative and immunocytochemical observations. J. Neurosci. 1995;15:1835–1853. doi: 10.1523/JNEUROSCI.15-03-01835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Strati K., Ortega S., Canamero M., Blasco M.A., Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li P., Liu K., He Q., Han S., Sun X., Li T., Shen L. Timely inhibition of Notch signaling by DAPT promotes cardiac differentiation of murine pluripotent stem cells. PLoS One. 2014;9:e109588. doi: 10.1371/journal.pone.0109588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Zunder E.R., Ng Y.H., Goronzy I.N., Nolan G.P., Wernig M. Early reprogramming regulators identified by prospective isolation and mass cytometry. Nature. 2015;521:352–356. doi: 10.1038/nature14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson J.P., Goritz C., Tatarishvili J., Dias D.O., Smith E.M., Lindvall O., Kokaia Z., Frisen J. A latent neurogenic program in astrocytes regulated by Notch signaling in the mouse. Science. 2014;346:237–241. doi: 10.1126/science.346.6206.237. [DOI] [PubMed] [Google Scholar]
- Matyash V., Kettenmann H. Heterogeneity in astrocyte morphology and physiology. Brain Res. Rev. 2010;63:2–10. doi: 10.1016/j.brainresrev.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Krencik R., Ullian E.M., Tsai H.H., Deneen B., Richardson W.D., Barres B.A., Rowitch D.H. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Zang T., Zou Y., Fang S., Smith D.K., Bachoo R., Zhang C.L. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim N.A., Goldman S.A., Nedergaard M. Heterogeneity of astrocytic form and function. Methods Mol. Biol. 2012;814:23–45. doi: 10.1007/978-1-61779-452-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda H. A review of functional heterogeneity among astrocytes and the CS56-specific antibody-mediated detection of a subpopulation of astrocytes in adult brains. Anat. Sci. Int. 2018;93:161–168. doi: 10.1007/s12565-017-0420-z. [DOI] [PubMed] [Google Scholar]
- Petersen P.H., Zou K., Hwang J.K., Jan Y.N., Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P., Romanov R.A., Spigolon G., Masini D., Martin-Montanez E., Toledo E.M., La Manno G., Feyder M., Pifl C., Ng Y.H. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson's disease model. Nat. Biotechnol. 2017;35:444–452. doi: 10.1038/nbt.3835. [DOI] [PubMed] [Google Scholar]
- Rusnakova V., Honsa P., Dzamba D., Stahlberg A., Kubista M., Anderova M. Heterogeneity of astrocytes: from development to injury –– single cell gene expression. PLoS One. 2013;8:e69734. doi: 10.1371/journal.pone.0069734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanbeck R., Martini S., Bernoth K., Just U. The Notch signaling pathway: molecular basis of cell context dependency. Eur. J. Cell Biol. 2011;90:572–581. doi: 10.1016/j.ejcb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Shan Z.Y., Liu F., Lei L., Li Q.M., Jin L.H., Wu Y.S., Li X., Shen J.L. Generation of dorsal spinal cord GABAergic neurons from mouse embryonic stem cells. Cell Reprogram. 2011;13:85–91. doi: 10.1089/cell.2010.0055. [DOI] [PubMed] [Google Scholar]
- Su Z., Niu W., Liu M.L., Zou Y., Zhang C.L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Zang T., Liu M.L., Wang L.L., Niu W., Zhang C.L. Reprogramming the fate of human glioma cells to impede brain tumor development. Cell Death Dis. 2014;5:e1463. doi: 10.1038/cddis.2014.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Hu X., Wang D., Yuan Y., Qin S., Tan Z., Gu Y., Huang X., He C., Su Z. Establishment and characterization of primary astrocyte culture from adult mouse brain. Brain Res. Bull. 2017;132:10–19. doi: 10.1016/j.brainresbull.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tsai H.H., Li H., Fuentealba L.C., Molofsky A.V., Taveira-Marques R., Zhuang H., Tenney A., Murnen A.T., Fancy S.P., Merkle F. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337:358–362. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zang C., Liu X.S., Aster J.C. The role of Notch receptors in transcriptional regulation. J. Cell Physiol. 2015;230:982–988. doi: 10.1002/jcp.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.L., Su Z., Tai W., Zou Y., Xu X.M., Zhang C.L. The p53 pathway controls SOX2-mediated reprogramming in the adult mouse spinal cord. Cell Rep. 2016;17:891–903. doi: 10.1016/j.celrep.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson U., Bushong E.A., Price D.L., Smarr B.L., Phung V., Terada M., Ellisman M.H., Pekny M. Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proc. Natl. Acad. Sci. U S A. 2006;103:17513–17518. doi: 10.1073/pnas.0602841103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Rawls J., Molkentin J.D., Black B.L., Olson E.N. Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C. Mol. Cell. Biol. 1999;19:2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T.J., Bevan M.J. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- Zhang R., Engler A., Taylor V. Notch: an interactive player in neurogenesis and disease. Cell Tissue Res. 2018;371:73–89. doi: 10.1007/s00441-017-2641-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Barres B.A. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr. Opin. Neurobiol. 2010;20:588–594. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Zhu Y.B., Gao W., Zhang Y., Jia F., Zhang H.L., Liu Y.Z., Sun X.F., Yin Y., Yin D.M. Astrocyte-derived phosphatidic acid promotes dendritic branching. Sci. Rep. 2016;6:21096. doi: 10.1038/srep21096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zine A., Aubert A., Qiu J., Therianos S., Guillemot F., Kageyama R., de Ribaupierre F. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J. Neurosci. 2001;21:4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.