Abstract
The archaeon Pyrococcus furiosus grows optimally at 100°C by converting carbohydrates to acetate, carbon dioxide and hydrogen gas (H2), obtaining energy from a respiratory membrane-bound hydrogenase (MBH). This conserves energy by coupling H2 production to oxidation of reduced ferredoxin with generation of a sodium ion gradient. MBH is classified as a Group 4 hydrogenase and is encoded by a 14-gene operon that contains hydrogenase and Na+/H+ antiporter modules. Herein a His-tagged 4-subunit cytoplasmic subcomplex of MBH (C-MBH) was engineered and expressed in P. furiosus by differential transcription of the MBH operon. It was purified under anaerobic conditions by affinity chromatography without detergent. Purified C-MBH had a Fe : Ni ratio of 14 : 1, similar to the predicted value of 13 : 1. The O2 sensitivities of C-MBH and the 14-subunit membrane-bound version were similar (half-lives of ∼15 h in air), but C-MBH was more thermolabile (half-lives at 90°C of 8 and 25 h, respectively). C-MBH evolved H2 with the physiological electron donor, reduced ferredoxin, optimally at 60°C. This is the first report of the engineering and characterization of a soluble catalytically active subcomplex of a membrane-bound respiratory hydrogenase.
Keywords: Hydrogenase, Membrane protein, Differential expression, Archaea
Introduction
Due to limiting fossil fuel availability and a growing need for energy, a major push has occurred recently to generate alternative renewable fuels. Such fuels must be energy efficient as well as carbon neutral, and biohydrogen production meets these criteria (Lee et al., 2010). Hydrogen gas is metabolized by microbes from all three domains of life using the enzyme hydrogenase, which functions to catalyze the reversible reduction of protons to molecular hydrogen (H2) (Vignais and Billoud, 2007). Hydrogenases are classified based on the metal content of their active sites into the [NiFe]-, [FeFe]- and [FeS] cluster free-hydrogenases. The [NiFe]-hydrogenases are ubiquitous in the microbial world and have been extensively studied from numerous mesophilic bacteria (Fontecilla-Camps, 2009; Friedrich et al., 2011; Shafaat et al., 2013). The minimum structure of the [NiFe] hydrogenase is a heterodimer composed of a large and small subunit. The large subunit contains the [NiFe] catalytic site that is coordinated by the sulfur atoms of four cysteine residues organized into two–CxxC– motifs near the N- and C-termini. The small subunit typically contains three iron–sulfur clusters invariably of the [4Fe-4S] type that shuttle electrons between an acceptor/donor for the enzyme and its active site (Vignais and Billoud, 2007). [NiFe]-hydrogenases are classified into four groups based on the phylogeny of their catalytic subunits (Vignais and Billoud, 2007). The majority of crystal structures for [NiFe]-hydrogenases are available for Group 1 hydrogenases (Volbeda et al., 1995, 2013). Recently, the first crystal structure of a Group 3 hydrogenase was determined from Methanothermobacter marburgensis (PDB code: 4OMF) (Vitt et al., 2014).
Group 4 hydrogenases are the least studied of the [NiFe]-hydrogenases. These hydrogenases are defined as the H2-evolving energy-conserving membrane-associated hydrogenases (Vignais and Billoud, 2007). Very little sequence similarity exists between Group 4 hydrogenases and other [NiFe] hydrogenases except for the conserved residues that bind the [NiFe] catalytic site and its proximal [4Fe-4S] cluster, indicating a distinct evolutionary history (Hedderich, 2004, 2005; Schut et al., 2013). Group 4 hydrogenases play an important role in conserving energy by establishing ion gradients across membranes that can be used to generate ATP. These enzymes contain at least six subunits and are much more complex than the characterized dimeric hydrogenases. The simplest members include the 6-subunit ‘energy-conserving’ hydrogenase (Ech) from the archaeon Methanosarcina barkeri, which functions in methanogenesis, and the 7-subunit hydrogenase 3 from Escherichia coli, which oxidizes formate and evolves H2 (Böhm et al., 1990; Sauter et al., 1992; Meuer et al., 1999; Kurkin et al., 2002). Six subunits conserved within the Group 4 hydrogenases are homologous to the catalytic core of the ubiquitous aerobic respiratory complex NADH quinone oxidoreductase or Complex I (NuoBCDIHL) (Hedderich, 2004, 2005; Schut et al., 2013; Marreiros et al., 2013). This conserved homology suggests a close evolutionary history between Group 4 enzymes and Complex I.
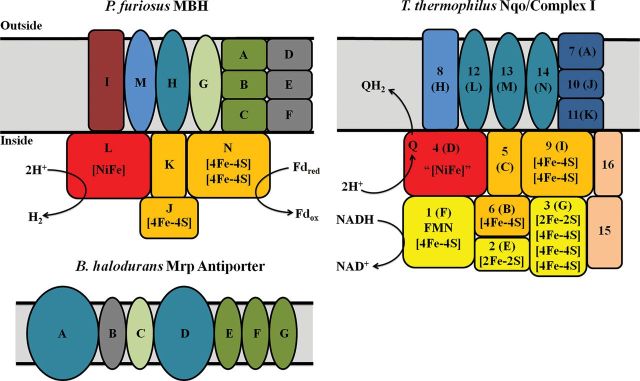
Pyrococcus furiosus is a hyperthermophilic archaeon that grows optimally at 100°C and contains a complex hydrogenase system (Fiala and Stetter, 1986; Schut et al., 2013). It grows by fermenting sugars to acetate, CO2 and H2 and its membrane-bound hydrogenase (MBH) catalyzes H2 production using reduced ferredoxin (Fd) generated from sugar oxidation as the electron donor (Schut et al., 2013). Previous studies of P. furiosus MBH showed that it is encoded by a 14-gene operon (mbhA-N: PF1423-PF1436) (Sapra et al., 2000). Six of the last seven genes in the operon are homologous to those encoding the ‘core’ subunits of Complex I (mbhH,J-N) while another eight subunits (mbhA-H) are homologous to subunits of the Mrp monovalent cation/proton antiporter of some mesophilic bacteria (Fig. 1) (Swartz et al., 2005). The exceptions are mbhI, which does not have homology to subunits of either Complex I or Mrp, and mbhH, which has homology to both (Fig. 1). Mrp catalyzes the efflux of monovalent cations, such as Na+, K+ and Li+ outward in a coupled reaction that transports protons inwards. Of the 14 subunits of MBH, only mbhJKLN are predicted to not encode transmembrane helices (Sapra et al., 2000; Silva et al., 2000). MbhJ and MbhN are proposed to contain one and two [4Fe–4S] clusters, respectively, where MbhJ is the equivalent of the small subunit of the Group 1 dimeric [NiFe]-hydrogenases (Fig. 1). MbhKL are equivalent to the large subunit and contain the [NiFe] active site, with the four Cys residues provided by MbhL.
Fig. 1.
Comparison of homologous subunits between MBH, Complex I and Mrp antiporter. Nqo numbers are given for each T. thermophilus complex I subunit with the correlated Nuo letter in parenthesis. Red subunits represent the catalytic subunits and include mbhL and nuoD. Orange subunits represent the Q module and include mbhJKLN and nuoBCDI. Aqua colored subunits are homologous and include mbhH, nuoLMN, and mrpAD. Blue subunits are homologs to mbhM and include nuoH. Light green subunits are homologous to mbhG and include mrpC. Green subunits are homologous to mbhABC and include mrpEFG. Gray subunits are homologous to mbhDEF and include mrpB. Yellow subunits represent the N module of Complex I and include nuoEFG. MbhI is shown in brown and the dark blue subunits represent nuoAJK. Tan subunits represent two subunits that were observed in the 16-subunit crystal structure of Nqo from T. thermophiles (Efremov et al., 2010). There is no homology between the three complexes for mbhI, nuoAEFGJK or Nqo15–16. Fdred and Fdox represent reduced and oxidized ferredoxin, respectively, and QH2 and Q represent reduced and oxidized ubiquionone, respectively.
The evolutionary linkage between respiratory Complex I and Group 4 [NiFe] hydrogenases has been extensively reviewed (Hedderich, 2004, 2005; Schut et al., 2013). An ‘ancestral group 4 [NiFe] hydrogenase’ has been proposed that evolved into the archaeal, bacterial and eukaryotic (Nuo or Nqo, respectively) Complex I by the shuffling of two distinct modules—Mrp and Mbh. Moreover, another notion of a ‘universal adaptor molecule’ has been proposed to understand the evolution of Nuo or Nqo Complex I and Group 4 hydrogenases (Batista et al., 2013). The universal adaptor molecule is conceived as a set of conserved subunits between Group 4 hydrogenases and Complex I and contains four subunits (nuoB, nuoD, nuoH and nuoL). NuoB and nuoD are analogous to the large and small subunits of the Group 4 hydrogenases (mbhL, mbhJ) while nuoH (mbhM) is believed to function like a membrane anchor that links the hydrogenase module to the membrane, and nuoL (mbhH) is homologous to mrpA and mrpD from the Mrp monovalent cation/proton antiporter (Fig. 1). NuoM and nuoN also show homology to mrpA, mrpD and mbhH (Fig. 1). Close evolutionary relationships have also been observed by analyzing the three-dimensional structure of the hydrophilic domains of the Nqo Complex I of Thermus thermophilus (PDB 2FUG and 3M9S) (Sazanov and Hinchliffe, 2006; Efremov et al., 2010). MBH has significant sequence similarity to the Q-module of Complex I of T. thermophilus, which includes Nqo4, 5, 6 and 9 (Fig. 1) (Hedderich, 2004, 2005). The Q-module has a quinone binding grove in Nqo4, which is where the [NiFe] site is located in the large subunit of MBH (MbhL) and the ‘N2’ [FeS] cluster of Nqo6 is analogous to the proximal [FeS] cluster of small subunit of MBH (MbhJ). Nqo9 shares similarity with MbhN and harbors two [FeS] clusters. Nqo5 is homologous to MbhK and is part of the large subunit.
The best characterized Group 4 hydrogenases are the 6-subunit Ech from M. barkeri and the 14-subunit MBH from P. furiosus. Biochemical studies of Ech have been described but there is no information on its modular structure (Meuer et al., 1999; Kurkin et al., 2002; Hedderich, 2005; Schut et al., 2013), P. furiosus MBH was recently affinity tagged and solubilized using detergent to yield an intact and functional 14-subunit complex and a structural model was obtained based on small angle X-ray scattering (McTernan et al., 2014). To further understand the modular nature of MBH, engineering the 14-gene operon to generate different forms of the enzyme could have major implications. For example, the generation of subcomplexes of MBH could give insight into which subunits are essential for catalytic activity and the generation of a chemical gradient, as well as providing insights into the evolution of the ubiquitous Complex I.
There are limited reports for the successful engineering of ‘minimal’ versions of hydrogenase. This was achieved with the enzyme from a Ralstonia species but involved dissociation of the native complex rather than genetic manipulation (Grzeszik et al., 1997). The first successful example of the engineering of a subcomplex of hydrogenase was accomplished using the genetic system established for P. furiosus in order to make a dimeric version of its heterotetrameric soluble hydrogenase I (SHI) (Hopkins et al., 2011; Lipscomb et al., 2011). Herein, we have successfully engineered a strain of P. furiosus that generates a soluble 4-subunit subcomplex of MBH (C-MBH). Previous attempts to purify the 14-subunit complex led to the purification of subcomplexes of MBH but these were heterogeneous and difficult to characterize (Sapra et al., 2000; Silva et al., 2000). In this study, we have taken advantage of the same genetic system for P. furiosus that has been previously used to make a dimeric version of SHI and to over-express SHI (Hopkins et al., 2011; Lipscomb et al., 2011; Chandrayan et al., 2012). We have engineered a strain of P. furiosus by dividing the native MBH 14-gene operon into two different transcriptional units and have incorporated an affinity tag within the operon. This enabled us to purify a subcomplex of MBH from the cytoplasm (C-MBH) without the use of detergents. This is the first description of the engineering of a MBH to generate a soluble, catalytically active enzyme complex.
Materials and methods
Generation of P. furiosus strains expressing affinity tagged MBH
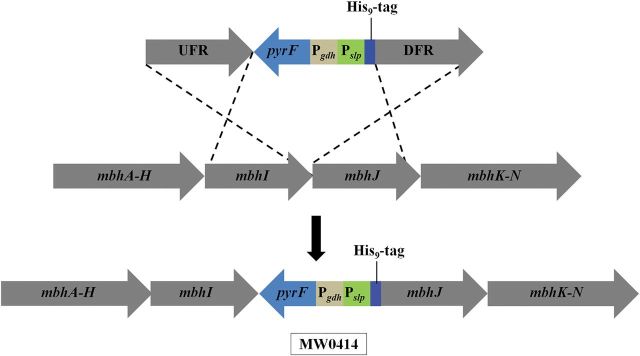
A competent strain of P. furiosus (COM1) was used to manipulate the MBH operon (Lipscomb et al., 2011). A one-step marked knock-in genetic protocol was used in which a polyhistidine (His9) affinity tag was inserted within the operon at the N-terminus of mbhJ (PF1432) yielding strain MW0414 (Fig. 2) (McTernan et al., 2014). The knock-in cassette, which contains the selectable marker and the strong constitutive promoter of the gene encoding the S-layer protein (Pslp) with an in frame His9 tag, were generated by using overlapping PCR (Horton et al., 1989). Prime Star HS polymerase premix (Clonetech, USA) was used to make the knock-in cassette. PyrF was the selectable marker and was placed under the control of the glutamate dehydrogenase promoter (Pgdh). In strain MW0414, the expression of mbhJ and the subsequent five genes (mbhK-N) was under control of Pslp, while the first nine genes are under control of the native MBH promoter (McTernan et al., 2014). Generation of MW0414 required the marker cassette to be placed in front of mbhJ within the MBH operon (Fig. 2). The sequence between mbhI and mbhJ is shown in Supplementary Fig. S1. All transformants were PCR screened for correct insertion and the PCR product was sequenced (Macrogen, MD).
Fig. 2.
The genetic strategy used to insert the His9x tag and to overexpress last five genes (mbhJ-N) of the MBH operon relative the first nine genes (mbhA-I). The tag is inserted at the N-terminus of mbhJ. The abbreviations are: UFR and DFR, upstream and downstream flanking regions (1 kb) of the MBH operon; pyrF, selectable marker; Pgdh and Pslp, promoters for the gene encoding glutamate dehydrogenase and the S-layer protein of P. furiosus, respectively.
Protein expression and purification
All purification steps were carried out anaerobically using a Coy anaerobic chamber (Coy laboratories; MI, USA). Cells were lysed using 50 mM EPPS (4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid; Sigma-Aldrich, USA), pH 8.0, containing 50 μg/ml deoxyribonuclease I (DNase I; Sigma-Aldrich), and 2 mM dithiothreitol (DTT; Inalco, Italy) in a 5 : 1 ratio of buffer to cells in an anaerobic chamber (Coy). After 2 h of incubation at 23°C, cells were passed twice through a French press at a pressure of 1000 psi. Cytoplasmic extract (S100) was prepared by centrifugation in a Beckman-Coulter Optima L-90K ultracentrifuge at 100,000 g for 1 h. Cytoplasmic extract was loaded on a 5-ml FF His-Trap Ni-NTA column (GE Healthcare, USA), which was equilibrated with Ni-NTA buffer A (50 mM Tris–HCl, pH 8.0, containing 400 mM NaCl, 4 mM DTT). The column was washed with Buffer A and the bound protein was eluted with a 20-column volume gradient from 100% Buffer A to 100% Buffer B (Buffer A containing 500 mM imidazole). After the Ni-NTA step, the affinity purified C-MBH was further purified using a Mono-Q column (Bio-Scale Q2 Column) The column was equilibrated in 50 mM Tris, pH 8.0 containing 2 mM sodium dithionite (DT). The bound protein was eluted with a 20 column volume gradient from 100% Buffer A to 100% Buffer B (Buffer A containing 2 M NaCl).
Gel Filtration chromatography
The molecular weight of C-MBH was determined by analyzing the purified protein on a calibrated Superdex 200 10/300 GL column equilibrated 50 mM Tris pH 8.0, 400 mM NaCl, 2 mM DT. The column was calibrated by using these standards: cytochrome c, bovine serum albumin and thyroglobulin.
Quantitative PCR (Q-PCR) analysis
RNA was isolated using the Absolutely RNA miniprep kit (Agilent technologies, USA). Turbo DNase (Agilent technologies) was used to remove DNA contamination. Complementary DNA (cDNA) was synthesized using a Affinityscript QPCR CDNA synthesis kit (Agilent technologies). CDNA was analyzed using a Brilliant II SYBR green QPCR master mix (Agilent technologies) and measured using a MX3000P instrument (Stratagene, USA). Ct values were normalized to the internal control pyruvate oxidoreductase (pyruvate oxidoreductase, POR, gamma subunit; PF0971).
Other methods
Hydrogenase assays were performed at 80°C and H2 was measured using an Agilent Technologies 6850 gas chromatograph. H2 evolution activity was determined using dithionite-reduced methyl viologen (MV; Sigma-Aldrich) as the electron donor or P. furiosus ferredoxin reduced by P. furiosus POR (Chandrayan et al., 2012). The POR-linked assay contained 100 mM EPPS pH 8.4, 10 mM Na-pyruvate, 0.2 mM coenzyme A, 0.4 mM TPP, 2 mM MgCl2 and 2 mM DTT, POR (30 μg/ml) and ferredoxin (100 μg/ml). The DT-ferredoxin assay contained 100 mM EPPS pH 8.4, 5 mM sodium dithionite and ferredoxin (100 μg/ml). Hydrogen uptake assay was performed by using methyl viologen as an electron acceptor in vials saturated with hydrogen. Oxygen sensitivity assays were carried out by exposing the MBH sample (100 μg/ml) in 50 mM Tris, pH 8.0, containing 400 mM NaCl and 4 mM DTT to air while shaking (30 rpm). Samples were taken at 0, 2, 4, 8, 16 and 32 h to determine residual hydrogenase activity. Thermal stability at 90°C was carried out in the same fashion except that the samples were maintained under anaerobic conditions. The H2 evolution assay at different temperatures was done using 50 μg of each form of MBH (C-MBH, S-MBH and washed membranes, WMs).
Purified C-MBH was analyzed by electrophoresis using 4–20% Tris–glycine NUSEP gels (Bio-Rad, USA). Bands were cut from the sodium dodecyl–polyacrylamide gel electrophoresis (SDS–PAGE), digested with trypsin and were analyzed by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Nickel and iron were measured using an octopole-based ICP-MS (7500ce Agilent Technologies, Tokyo, Japan)), equipped with a MicroMist nebulizer (Cvetkovic et al., 2010). X-band (∼9.6 GHz) electron paramagnetic resonance (EPR) spectroscopy was carried out using a Bruker ESP-300E EPR spectrometer equipped with an ER-4116 dual-mode cavity and an Oxford Instruments ESR-9 flow cryostat.
Temperature studies
MW0414 was grown in a 20-l fermenter at 90°C and switched to 72°C as previously described (Keller et al., 2013). C-MBH was purified as described above.
Results
Construction of P. furiosus to generate C-MBH
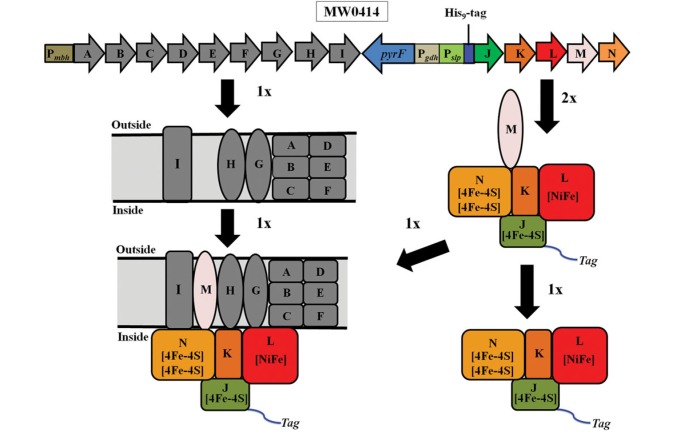
We previously deleted the catalytic subunit from MBH (mbhL) and showed that a functional complex was required for growth (in the absence of elemental sulfur) (Schut et al., 2012). In order to obtain a ‘minimal’ subcomplex of MBH, we decided to engineer the native MBH operon by splitting it into two separate transcriptional units. The first nine genes (mbhA-I) of the operon would be under the control of the native MBH promoter, while the last five genes (mbhJ-N) are under control of the stronger Pslp promoter that controls constitutive expression of the gene encoding the S-layer protein (Fig. 2). At the same time, an affinity-purification tag (His9) was inserted within the operon at the N-terminus of mbhJ (the hydrogenase ‘small’ subunit). We decided to tag the small subunit of MBH as this tag position was effective for the affinity purification of the intact 14-subunit complex (S-MBH) (McTernan et al., 2014). Hence, as shown in Fig. 3, in the engineered strain MW0414, the 14-gene MBH operon is expressed as two transcripts, mbhA-I and mbhJ-N. If the two protein products (MbhA-I and MbhJ-N) were able to combine and give a functional 14-subunit complex, then P. furiosus would be able to grow (Schut et al., 2012). However, due to the relative strengths of the native promoter for MBH (Pmbh) and Pslp, we expected that there would be excess mbhJ-N transcript relative to mbhA-I. Assuming that the transcripts were similarly stable (see below) and were translated with the same efficiency, then MbhJ-N, a potentially soluble form of the enzyme, should be generated in excess of that which combines with MbhA-I to generate functional MBH. Moreover, it should be possible to purify soluble MbhJ-N from the cytoplasmic fraction by its affinity tag (on MbhJ).
Fig. 3.
Transcription of the engineered MBH operon. (A) Schematic showing the transcribed products of the two different transcriptional units of the engineered MBH operon. The abbreviations are: Pmbh, the native MBH promoter that controls the expression of mbhA-I; pyrF, selectable marker; Pgdh, promoter for the gene encoding glutamate dehydrogenase; Pslp, promoter for the S-layer protein that controls the expression of mbhJ-N. Based on Quantitative PCR analysis, mbhJ-N, under the control of Pslp, were expressed at twice the level of mbhA-I, under the control of the native Pmbh.
Q-PCR analysis
There was no difference in the growth of MW0414 or the parent strain (data not shown), and the specific activity of WMs from COM1 and from MW0414 was the same (∼1.0 U/mg). This confirmed that the native MBH complex in MW0414 was intact and functional despite being synthesized and assembled from two different subcomplexes encoded by two different transcriptional units (Fig. 3). We first used Quantitative PCR (Q-PCR) analysis to determine if the split MBH operon was indeed differentially expressed. The results showed that the mbhJ-N transcript was ∼2-fold higher than that of mbhA-I (Supplementary Fig. S2). Since SHI in the cytoplasm of P. furiosus is a very active enzyme, it was not expected that a soluble form of MBH would be detected by an increase in total cytoplasmic hydrogenase activity, and this proved to be the case. Nevertheless, if an intact soluble form of MBH was produced in the cytoplasm, it should be possible to purify it using its affinity tag.
Affinity purification and characterization of affinity-tagged C-MBH
The cytoplasmic extract of MW0414 was applied to a Ni-NTA column and after washing with buffer a significant amount of hydrogenase activity was eluted with a histidine gradient. Hence, it appeared that a soluble His-tagged subcomplex of MBH had been produced, and this was further purified using a Mono-Q anion exchange column. A total of 21 mg of protein containing 12 units of hydrogenase activity (using methyl viologen as the electron donor) eluted from the Ni-NTA column and 12 mg of protein was obtained from the Mono-Q with no loss of activity.
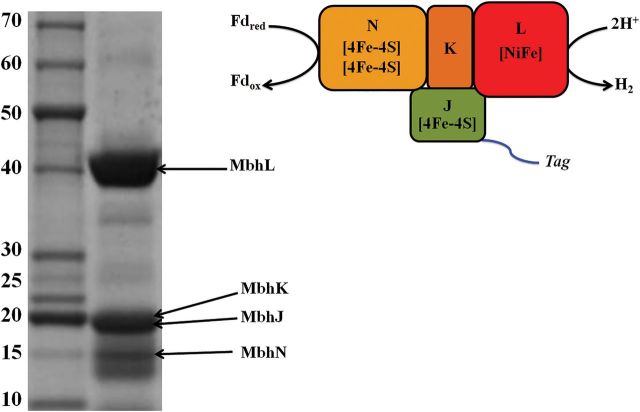
From the molecular analysis (Fig. 4), the second transcript should yield a 5-subunit MbhJ-N complex but MbhM is predicted to be membrane-associated. Analyses by SDS-PAGE revealed that S-MBH was tetrameric (MbhJKLN) rather than pentameric (MbhJ-N) and lacked MbhM. Protein bands corresponding to the calculated molecular weights for all four subunits were evident on the gel. Note that the catalytic subunit, MbhL (calculated MWt of 47,903 Da), is predicted to undergo C-terminal proteolysis during the processing of the [NiFe]-site with the removal of 47 amino acids. This proved to be the case as the band on the gel was consistent with the calculated mass of the mature processed subunit (42,899 Da). The presence of MbhJKLN in C-MBH was confirmed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis from trypsin-digested in-solution samples (Supplementary Table SI). The protein band slightly smaller than MbhN in Fig. 4 could not be identified by this approach.
Fig. 4.
SDS-PAGE of purified C-MBH. Left: MBH subunits identified by LC-MS/MS are labeled with a black arrow. Right: schematic of C-MBH showing location of the affinity-tag. MbhL harbors the [NiFe] active site, MbhK is part of the large subunit, MbhJ harbors one [4Fe4S] cluster and MbhN harbors two [4Fe4S] clusters. Fdred and Fdox represent reduced and oxidized ferredoxin, respectively.
Purified C-MBH was analyzed using a calibrated S-200 gel filtration column and eluted as a single peak corresponding to a mass of 85 ± 5 kDa (Supplementary Fig. S3). The predicted mass of the MbhJKLN complex calculated from the deduced amino acid sequences is 97 kDa, suggesting that all four subunits are present but that the subcomplex is not globular in shape. To gain a further understanding as to which subunits are associated with C-MBH, we analyzed the purified protein with both inductively coupled plasma mass spectrometry and EPR. Purified C-MBH contained both iron and nickel in a ratio of 14 : 1 (Table I). This result supports the proposed presence of three [4Fe–4S] clusters in the enzyme (Fig. 4), which together with the [NiFe] active site should give a predicted Fe : Ni ratio of 13 : 1. The anaerobically purified protein after reduction with sodium dithionite exhibited a complex rhombic EPR signal at 6 K that could be attributed to multiple [4Fe–4S]1+ clusters (Supplementary Fig. S4). A schematic of purified C-MBH containing four subunits (MbhJKLN) is shown in Fig. 4.
Table I.
Characterization of purified C-MBH
| Property | C-MBH | WM |
|---|---|---|
| DT-Fd H2 evolution activity (U/mg) at 80°C | 0.03 | 0.05 |
| DT-Fd H2 evolution activity (U/mg) at 60°C | 0.06 | 0.01 |
| POR-Fd H2 evolution activity (U/mg) at 80°C | 0.004 | 0.02 |
| H2 evolution : oxidation activity (MV as electron carrier) at 80°C | 13 : 1 | 26 : 1 |
| Half-life (t1/2, h) at 90°C under argon | 8 h | 25 h |
| Half-life (t1/2, h) at 25°C under air | 13 h | 13 h |
| Metal content (Fe : Ni) | 14 : 1 | 19 : 1 |
MBH uses reduced ferredoxin as an electron donor in vivo and the redox protein is proposed to interact with MbhN (Fig. 1) (Schut et al., 2013). C-MBH also evolves H2 using ferredoxin as the electron donor at 80°C, both when reduced by sodium dithionite or by the native pyruvate ferredoxin oxidoreductase (Table I). The ability of the subcomplex to catalyze the physiological reaction is also consistent with all the three [4Fe–4S] clusters being intact and functional in C-MBH. The ratio of H2 evolution to H2 oxidation by C-MBH using methyl viologen as the electron carrier was 13 : 1, which is lower than that measured for the membrane-bound form (26 : 1) but this still demonstrates that this subcomplex version of MBH prefers to evolve hydrogen rather than oxidize it. C-MBH and the WM control were similarly insensitive to inactivation by oxygen, with half-lives under air while shaking of ∼13 h.
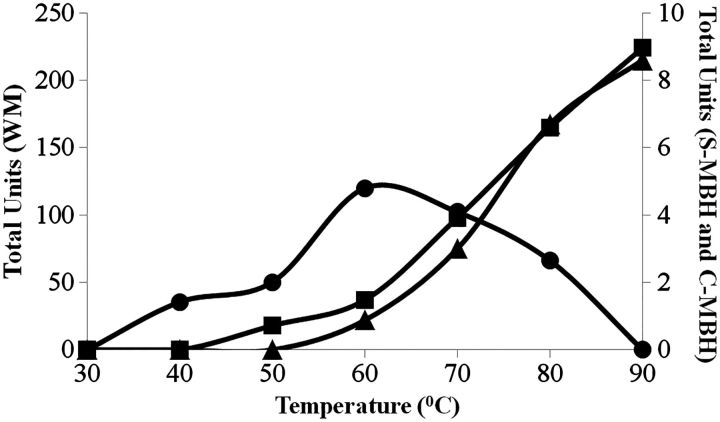
Temperature studies of C-MBH
Purified C-MBH was assayed at different temperatures in order to gain a better understanding of its H2 evolution activity. It was observed that C-MBH had a different temperature profile in comparison to the form of MBH present in WMs or as the solubilized, purified 14-subunit complex (S-MBH) (Fig. 5). With reduced methyl viologen as the electron donor, C-MBH had maximal activity at 60°C and was inactive at 90°C. In comparison the S-MBH and WM forms of MBH were maximally active at or >90°C. Accordingly, H2 evolution by C-MBH using ferredoxin, the physiological electron donor to MBH, was measured at 60°C (Table I), and was higher than the activity measured at 80°C. C-MBH therefore appeared to be less thermostable than MBH in the WM and this proved to be the case during extended incubation at high temperature. Purified C-MBH had a half-life at 90°C of 8 h, which compares with 25 h for MBH in the WM (Table I).
Fig. 5.
Temperature profile for C-MBH activity. The H2 evolution activity using reduced methyl viologen as the electron donor of C-MBH (circular symbols), WM (square symbols) and solubilized and purified MBH (S-MBH, triangles) is shown.
We therefore wondered if the yield of purified C-MBH could be improved if the recombinant P. furiosus strain was grown at a temperature lower than 90°C, where the enzyme might be unstable and subject to proteolytic digestion. The MW0414 strain was therefore grown in a 20-liter fermenter at 72°C (Keller et al., 2013). Cytoplasmic extracts were prepared from cells grown at 90°C and at 72°C and C-MBH was purified as described above using two chromatography steps. The yield of C-MBH from the cells grown at the lower temperature was more than twice (9.0 mg vs 4.0 mg) that obtained from cells grown at the higher temperature, indicating that thermal degradation of the enzyme does occur during growth at 90°C. Primers for Q-PCR were designed to bind the first gene under control of either Pmbh (PF1423, mbhA) or Pslp (PF1432, mbhJ) of the MBH operon (Fig. 3). The concentration of RNA was 2-fold higher when generated from the Pslp promoter throughout growth compared with the native MBH promoter and this was independent of the temperature shift from 90 to 72°C (Supplementary Fig. S2).
Discussion
Herein we describe the first successful attempt to engineer a cytoplasmic ‘minimal’ catalytically active subcomplex of a membrane-bound respiratory hydrogenase. We had previously generated a smaller form of the heterotetrameric cytoplasmic SHI from P. furiosus (Hopkins et al., 2011). This was achieved by expressing just two of the four genes that encode the enzyme in a strain in which all four genes had been deleted. However, it was not possible to express simply part of the 14-gene operon encoding MBH since it impossible to obtain a strain of P. furiosus lacking a functional MBH. We therefore engineered the MBH operon by splitting the 14-gene operon into two separate transcriptional units. This approach was risky as it was not known if the protein products of the two different transcripts would be able to find each other and assemble into a functional MBH complex (Fig. 3), which would enable P. furiosus to grow, and if so, would the excess subunits form a functional complex in the cytoplasm. All of this proved to be the case, however, as the over-expressed part of the MBH operon generated a stable cytoplasmic protein with hydrogenase activity, although it contained four (MbhJKLN) rather than the expected five subunits (MbhM was not present). These results show that, surprisingly, splitting and disrupting the MBH operon still produced a functional MBH, and we were able to purify the cytoplasmic subcomplex using the engineered affinity tag.
We also expected to see a 1 : 1 ratio of MBH activity in the membrane (from the 14-subunit MBH) and in the cytoplasmic fraction (from MbhJKLN) based on our Q-PCR data. However, measured at 80°C using the DT-MV H2 evolution assay, the activity of C-MBH was only ∼10% of that of S-MBH purified from the same batch of cells grown at 90°C (McTernan et al., 2014). However, the activity of C-MBH roughly doubled when assayed at 60°C versus 80°C (Fig. 5), so the Ni-NTA purified C-MBH represents ∼20% of the MBH activity present in the membrane in cells grown at 90°C (with no temperature switch). That this was lower than expected is unlikely to be due to the five-subunit version of C-MBH (MbhJ-N) remaining in the membrane (MbhM is predicted to be membrane-associated) since there was no increase in the hydrogenase activity (measured at 80°C) in the membrane. The lower than expected C-MBH activity is more likely due to the thermolability of the subcomplex, and this is supported by the finding that the yield of C-MBH increased >2-fold when cells were grown at 72°C. The amounts of activity and protein associated with purified C-MBH doubled when cells were grown at a lower temperature (90°C then switched to 72°C). Hence, from cells grown at 72°C we are able to purify the equivalent of ∼40% of MBH found in the membranes. Either the remainder is lost due to the low thermal stability of C-MBH or the two transcripts generated from the MBH operon are not translated with the same efficiency.
That purified C-MBH was less stable than S-MBH is not unexpected given that C-MBH lacked 10 partner subunits. What is surprising, however, is that C-MBH retained the very unusual catalytic preference of S-MBH (Table I). In in vitro assays using dyes such as methyl viologen as electron carriers, [NiFe]-hydrogenases preferentially oxidize H2, often by orders of magnitude (Vignais and Billoud, 2007). Pyrococcus furiosus MBH is therefore atypical in that it preferentially catalyzes H2 evolution by a ratio of∼26 : 1 in the standard assays (Table I). It was assumed that this may arise because of the large complex nature of this membrane-bound enzyme where electron transfer is coupled to both H+ and Na+ pumping, which might account for the catalytic preference for the physiological reaction of H2 production. However, the results presented herein show that this is a property of the cytoplasmic component of MBH that is independent of the ion-pumping membrane component. Stability assays of C-MBH showed that it was just as resistant to inactivation by O2 as the native 14-subunit complex, suggesting that the infrastructure of the catalytic subunit is maintained in the cytoplasmic form when it is dissociated from the other ten subunits.
Generation of a cytoplasmic catalytic subcomplex of this respiratory hydrogenase is a significant achievement. In particular, obtaining relatively large amounts of the heterotetrameric soluble complex in the absence of any detergent greatly facilitates structural analyses of this representative of the poorly studied Group 4 hydrogenases, with implications for the evolution of Complex I, and such studies are in progress.
Supplementary data
Funding
This work was supported by a grant (DE-FG05-95ER20175) from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the Department of Energy.
Supplementary Material
Acknowledgements
The authors thank the PAMS Facility at the University of Georgia for the LC-MS/MS analyses. They also thank the Biofermentation and Expression Facility at the University of Georgia for growing P. furiosus.
References
- Batista A.P., Marreiros B.C., Pereira M.M. Biol. Chem. 2013;394:659–666. doi: 10.1515/hsz-2012-0342. [DOI] [PubMed] [Google Scholar]
- Böhm R., Sauter M., Böck A. Mol. Microbiol. 1990;4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- Chandrayan S.K., McTernan P.M., Hopkins R.C., Sun J., Jenney F.E., Adams M.W.W. J. Biol. Chem. 2012;287:3257–3264. doi: 10.1074/jbc.M111.290916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic A., Menon A.L., Thorgersen M.P., et al. Nature. 2010;466:779–782. doi: 10.1038/nature09265. [DOI] [PubMed] [Google Scholar]
- Efremov R.G., Baradaran R., Sazanov L.A. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- Fiala G., Stetter K. Arch. Microbiol. 1986;145:56–61. [Google Scholar]
- Fontecilla-Camps J.C. Met. Ions Life Sci. 2009;6:151–178. doi: 10.1039/BK9781847559159-00151. [DOI] [PubMed] [Google Scholar]
- Friedrich B., Fritsch J., Lenz O. Curr. Opin. Biotechnol. 2011;22:358–364. doi: 10.1016/j.copbio.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Grzeszik C., Ross K., Schneider K., Reh M., Schlegel H.G. Arch. Microbiol. 1997;167:172–176. [PubMed] [Google Scholar]
- Hedderich R. J. Bioenerg. Biomembr. 2004;36:65–75. doi: 10.1023/b:jobb.0000019599.43969.33. [DOI] [PubMed] [Google Scholar]
- Hedderich R.F.L. J. Mol. Microbiol. Biotechnol. 2005;10:92–104. doi: 10.1159/000091557. [DOI] [PubMed] [Google Scholar]
- Hopkins R.C., Sun J., Jenney F.E., Jr., Chandrayan S.K., McTernan P.M., Adams M.W.W. PLOS One. 2011;6:e26569. doi: 10.1371/journal.pone.0026569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Keller M.W., Schut G.J., Lipscomb G.L., et al. Proc. Natl. Acad. Sci. 2013;110:5840–5845. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkin S., Meuer J., Koch J., Hedderich R., Albracht S.P.J. Eur. J. Biochem. 2002;269:6101–6111. doi: 10.1046/j.1432-1033.2002.03328.x. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Vermaas W.F., Rittmann B.E. Biotechnol. 2010;28:262–271. doi: 10.1016/j.tibtech.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Lipscomb G.L., Stirrett K., Schut G.J., Yang F., Jenney F.E., Jr., Scott R.A., Adams M.W., Westpheling J. Appl. Environ. Microbiol. 2011;77:2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marreiros B.C., Batista A.P., Duarte A.M., Pereira M.M. Biochim. Biophys. Acta. 2013;1827:198–209. doi: 10.1016/j.bbabio.2012.09.012. [DOI] [PubMed] [Google Scholar]
- McTernan P.M., Chandrayan S.K., Wu C.-H., et al. J. Biol. Chem. 2014;289:19364–19372. doi: 10.1074/jbc.M114.567255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuer J., Bartoschek S., Koch J., Kunkel A., Hedderich R. Eur. J. Biochem. 1999;265:325–335. doi: 10.1046/j.1432-1327.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- Sapra R., Verhagen M.F., Adams M.W. J. Bacteriol. 2000;182:3423–3428. doi: 10.1128/jb.182.12.3423-3428.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M., Böhm R., Böck A. Mol. Microbiol. 1992;6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- Sazanov L.A., Hinchliffe P. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- Schut G.J., Nixon W.J., Lipscomb G.L., Scott R.A., Adams M.W.W. Front. Micro. 2012;3:1–6. doi: 10.3389/fmicb.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut G.J., Boyd E.S., Peters J.W., Adams M.W.W. FEMS Microbiol. Rev. 2013;37:182–203. doi: 10.1111/j.1574-6976.2012.00346.x. [DOI] [PubMed] [Google Scholar]
- Shafaat H.S., Rudiger O., Ogata H., Lubitz W. Biochim. Biophys. Acta. 2013;1827:986–1002. doi: 10.1016/j.bbabio.2013.01.015. [DOI] [PubMed] [Google Scholar]
- Silva P., Van den Ban E.C.D., Wassink H., Haaker H., de Castro B., Robb F.T., Hagen W.R. Eur. J. Biochem. 2000;267:6541–6551. doi: 10.1046/j.1432-1327.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- Swartz T.H., Ikewada S., Ishikawa O., Ito M., Krulwich T.A. Extremophiles. 2005;9:345–354. doi: 10.1007/s00792-005-0451-6. [DOI] [PubMed] [Google Scholar]
- Vignais P., Billoud B. Chem. Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- Vitt S., Ma K., Warkentin E., Moll J., Pierik A.J., Shima S., Ermler U. J. Mol. Biol. 2014;426:2813–2826. doi: 10.1016/j.jmb.2014.05.024. [DOI] [PubMed] [Google Scholar]
- Volbeda A., Charon M.H., Piras C., Hatchikian E.C., Frey M., Fontecilla-Camps J.C. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- Volbeda A., Darnault C., Parkin A., Sargent F., Armstrong F.A., Fontecilla-Camps J.C. Structure. 2013;21:184–190. doi: 10.1016/j.str.2012.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.