Abstract
Recently, the concept of radiomics has emerged from radiation oncology. It is a novel approach for solving the issues of precision medicine and how it can be performed, based on multimodality medical images that are non-invasive, fast and low in cost. Radiomics is the comprehensive analysis of massive numbers of medical images in order to extract a large number of phenotypic features (radiomic biomarkers) reflecting cancer traits, and it explores the associations between the features and patients’ prognoses in order to improve decision-making in precision medicine. Individual patients can be stratified into subtypes based on radiomic biomarkers that contain information about cancer traits that determine the patient’s prognosis. Machine-learning algorithms of AI are boosting the powers of radiomics for prediction of prognoses or factors associated with treatment strategies, such as survival time, recurrence, adverse events, and subtypes. Therefore, radiomic approaches, in combination with AI, may potentially enable practical use of precision medicine in radiation therapy by predicting outcomes and toxicity for individual patients.
Keywords: radiomics, artificial intelligence, precision medicine, radiation therapy, medical images, cancer traits
INTRODUCTION
Artificial intelligence (AI) is based on computational algorithms or systems that can accurately perform high inference from a huge amount of knowledge data [1]. AI was named by John McCarthy et al. at a workshop at Dartmouth College in 1956, but the first and second booms of AI did not instill it either into daily life or into medicine. The current AI boom is the third wave since 2000. Hinton’s group made a breakthrough that almost halved the error rate for object recognition, using a convolutional neural network (CNN) or deep learning (DL), which is an advanced machine-learning technology [2], at the ImageNet Large Scale Visual Recognition Competition 2012 [3]. This event promoted the rapid adoption of DL by the computer vision community. DL architectures are composed of a cascade of multiple (deep) layers of non-linear processing units for learning datasets of input and output images and for extracting image features [3]. Since DL has a greater ability to recognize objects such as verbal and visual object patterns than conventional methods have, the applications of DL have been explored for segmentation of target regions in radiation therapy [4–7].
At around the same time as the third large wave of AI (after 2012), the idea of radiomics emerged from radiation oncology [8, 9] in the form of a novel approach for solving the issues of precision medicine, and how it can be performed based on multimodality medical images in a non-invasive (without biopsy), fast (fast scanning) and low-cost way (no additional examination cost). Radiomics is a new word derived from a combination of ‘radio’, which means radiological images (medical images in a broad sense), and ‘omics’ [10]. The omics are a number of fields of study (genomics, transcriptomics, proteomics and metabolomics) that improve our understanding of tumor biology and clinical management of cancer by comprehensively analyzing the massive amount of information become available on the genome, transcriptome, proteome and metabolome [11]. Precision medicine is a treatment strategy for making decisions about a molecularly targeted agent according to genetic mutations, rather than affected organs. Radiomics is a field that comprehensively analyzes massive numbers of medical images in order to extract a large number of phenotypic features (radiomic biomarkers) reflecting cancer traits, and explores the associations between the features and patients’ prognoses to improve decision-making in precision medicine [12]. Phenotypic features in medical images, which are routinely acquired in clinical practice, can result from the expression of the genotype (the organism’s genetic codes). The medical images are thought to include the internal information (e.g. anatomical, physiological and pathological information) in patients’ specific regions. Radiomics is considered a ‘specialized AI’ for predicting patient prognoses based on medical images. This review paper considers the following issues:
Why is radiomics needed in precision medicine?
The potential of radiomics for avoiding undesirable complications caused by biopsy
The overall procedure of radiomic analysis
What radiomic features reflect
What mathematical models of image features represent
Stratification of individual patients into subtypes using radiomic biomarkers
Radiomcs with AI (machine learning)
Perspectives of radiomics in radiation therapy.
WHY IS RADIOMICS NEEDED IN PRECISION MEDICINE?
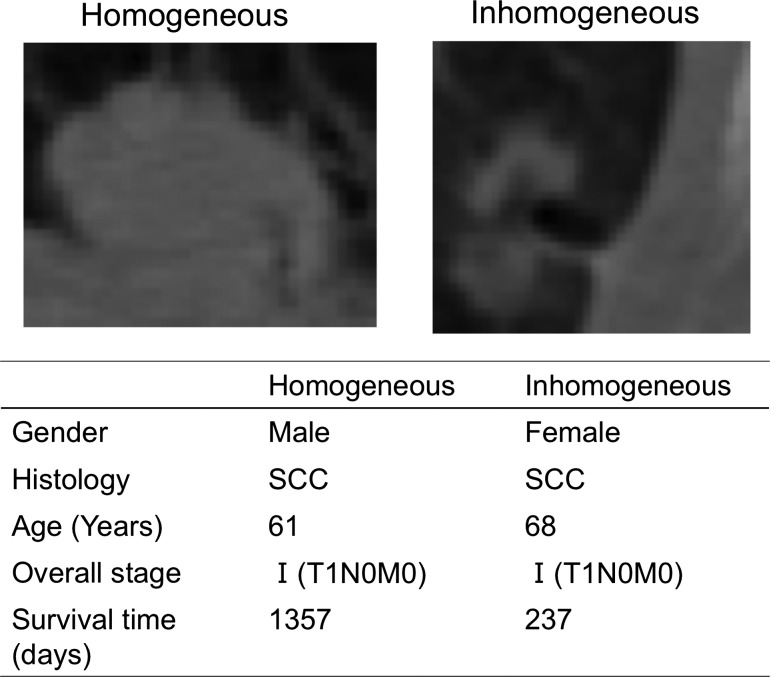
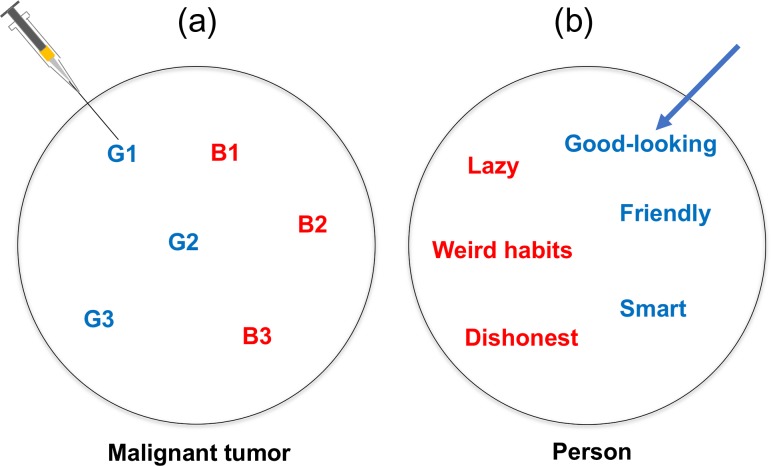
Figure 1 shows two computed tomography (CT) images showing the characteristics of two different tumors of lung cancer patients who received radiation therapy. They appear to differ in appearance in terms of intensity distribution and contour shapes from our subjective points of view, but they have similar histology and patient age. In fact, the two cancer patients had quite different survival times: 3.72 years for the homogeneous cancer (left) and 0.65 years for the inhomogeneous cancer (right), i.e. the patient on the left survived around five times as long as the one on the right. They could belong to different subtypes, from histological and genetic points of view. If the physician of the patient on the right could have predicted the survival time or prognosis prior to the treatment, different strategies might have been chosen. The idea that appropriate treatment strategies are selected according to subtypes is called ‘precision medicine’ [12]. In precision medicine, the subtypes of individual patients need to be identified; generally this is done using ‘wet’ biomarkers [13], i.e. biospecimen-derived biomarkers [14], such as genomic information derived from a part of a tumor obtained in a biopsy [12]. The Food and Drug Administration (FDA) in the USA defines a biomarker as a defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions [15]. Molecular, genomic, histologic, medical imaging, and physiologic characteristics can be considered as examples of biomarkers [16–18]. We now need to determine which biomarkers are more appropriate for the stratification of patients into subtypes that have different prognoses. Measurement of the biomarkers (other than medical imaging biomarkers) could require invasive biopsy or sampling of biospecimens, additional costs, and long examination tests. Furthermore, the biomarker information is limited if the biospecimens are taken from a part of an entire tumor using a single biopsy, because Gerlinger et al. [19] reported that gene-expression signatures of both good and poor prognoses were detected in different locations of the same tumor. Figure 2 describes gene-expression heterogeneity within one tumor, and trait heterogeneity within one person. The imaginary codes G1 to G3 and B1 to B3 represent gene-expression signatures of good and poor prognoses within the one malignant tumor, respectively, in Fig. 2a. A single biopsy of such a heterogeneous tumor could create a misleading estimation of prognosis, based on its genomic information. This situation is similar to that of trait heterogeneity in a person, as shown in Fig. 2b. If you liked a part of your partner, e.g. the positive side (good-looking, friendly, smart) (overestimation) before you lived together, you might be disappointed with the negative side (lazy, weird habits, dishonest) after you lived together.
Fig. 1.
Two computed tomography (CT) images with the characteristics of two different tumors of lung cancer patients who each received radiation therapy. SCC, squamous cell carcinoma.
Fig. 2.
A gene-expression heterogeneity in the same tumor, and trait heterogeneity in the same person.
The issues raised above are drawbacks of precision medicine. Aerts et al. [9] showed the prognostic powers of image features (statistical features and texture features) that have been derived solely from medical (CT) images of lung cancer patients treated with radiation therapy or radiochemotherapy, and the correlations of the image features with gene mutations. Their approach, based on medical images, could overcome the drawbacks of precision medicine, because medical images, which are acquired in a non-invasive, fast, and low-cost way, could obtain the entire information about cancer traits, such as intratumor heterogeneity. The signatures consisting of significant image features associated with patients’ prognoses can be considered ‘dry’ (imaging) biomarkers, and can extensively characterize cancer traits. Many imaging biomarkers based on radiomics have been explored by evaluating several end points to indicate the feasibility of radiomics to estimate overall survival and disease-free survival in radiation therapy [20].
POTENTIAL OF RADIOMICS FOR AVOIDING UNDESIRABLE COMPLICATIONS CAUSED BY BIOPSY
Radiomics can be utilized to avoid undesirable complications caused by biopsy. Regarding brain tumors, craniotomy biopsy or stereotactic biopsy in brain tumors may cause some complications, such as intracerebral haematoma and hemiparesis, depending on tumor locations [21]. Some patients may refuse a needle biopsy to sample tissues from a suspected lung tumor due to concerns about a pneumothorax [22]. In these cases, physicians may decide the treatment policies for patients by reference to the radiomics results without the biopsy. The European Randomized study of Screening for Prostate Cancer (ERSPC) reported that prostate-specific antigen (PSA) screening decreased prostate cancer mortality, but that it increased substantial unnecessary biopsies, which resulted in undesirable complications such as fever, haematuria and haematospermia [23, 24]. In addition, according to Fukugai’s study [25], when using the original biopsy tumor grade rendered by nine different pathologists, the biopsy and prostatectomy results showed weaker agreement. Therefore, the information from radiomics can assist pathologists and radiologists by increasing the accuracy of biopsies or reducing unnecessary biopsies.
OVERALL PROCEDURE FOR RADIOMIC ANALYSIS
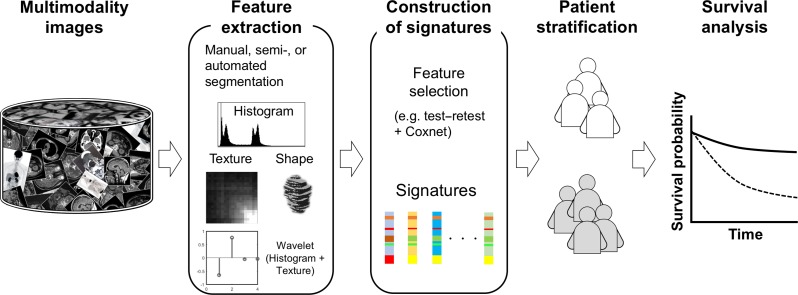
Figure 3 illustrates an overall procedure of radiomic analysis for discovering prognostic signatures that can predict patients’ prognoses. The prognostic signature is a vector including significant features as elements. This procedure, shown in Fig. 3, is similar to what Aerts et al. [9] employed. First, a database including a large number of medical images [such as CT, magnetic resonance (MR) and positron emission tomography (PET) images for specific cancer patients who have received the same treatment] is prepared according to the requirements of radiomic analysis. Second, a large number of image features (e.g. >400), including texture features, are extracted from medical images after manual, semi- or automated segmentation. Third, radiomic features are selected based on their stability (demonstrated e.g. by test–retest [9]) as well as their contributions to the prognoses (e.g. the Coxnet algorithm [26]). Fourth, the patients are stratified into several subtypes of patients by using simple thresholding methods or clustering methods [2, 10]. In general, the threshold values are the medians of image features. Finally, the outcomes of the patients are predicted by performing survival analyses, in which significant features reflecting the prognoses are chosen as signatures. If there are statistically significant differences between the survival curves of two patient subtypes stratified by a radiomic feature, as shown in Fig. 3, the two patient subtypes could have different responses to a same-treatment approach. Therefore, the radiomic features may be associated with patients’ survival times.
Fig. 3.
An overall procedure of radiomic analysis for discovering prognostic signatures that can predict patients’ prognoses.
WHAT RADIOMIC FEATURES REFLECT
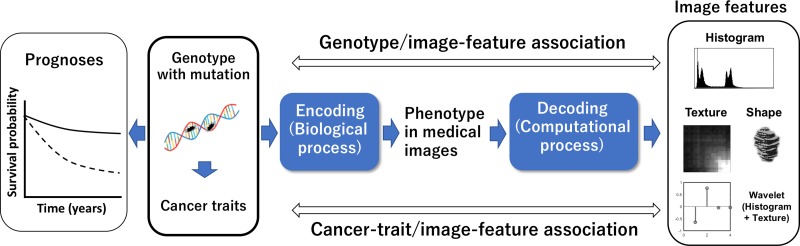
Figure 4 shows radiomic assumptions about associations between prognoses and image features. Genotypes with mutations could determine cancer traits that are associated with the prognoses of patients. On the pathway on the right in Fig. 4, the genotypes are believed to be encoded to the phenotypes expressed in medical images through biological processes. The image features may thus be derived by ‘decoding’ the phenotypes, i.e. medical images [9]. ‘Decoding’ medical images (phenotypes) refers to the computational extraction of image features from medical images using image processing and analysis techniques. Therefore, encoding followed by decoding means that the genotypes having associations with the cancer prognoses might be expressed by the image features, which are denoted as ‘radiomic features’. The image features could reflect cancer traits and prognoses. Furthermore, the authors believe that appropriate mathematical image features can model cancer hallmarks, which include six traits, i.e. sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis [27]. Possible associations between radiomic features and cancer hallmarks are introduced in Section ‘STRATIFICATION OF INDIVIDUAL PATIENTS INTO SUBTYPES USING RADIOMIC BIOMARKERS’.
Fig. 4.
Radiomics assumptions about associations between prognoses and image features.
WHAT MATHEMATICAL MODELS OF IMAGE FEATURES REPRESENT
In general, the aim of image feature mathematical models is to characterize objects (lesions or anatomical structures in radiomics) that can be recognized in segmented regions, and whose image features are similar to those of objects in the same category, but different or as distinguishable as possible from those of objects in different categories [28]. Objective and quantitative image features would be expected to characterize abnormal lesions or malignant tumors, possibly reflecting the cancer hallmarks in radiomics.
Currently, there are three major types of mathematical feature models, i.e. shape, statistical, and texture features, that are utilized in the radiomics field. Shape features are surface area, surface-area-to-volume ratio, sphericity, spherical disproportion, compactness, and so on [29]. Statistical features obtained from a gray-level histogram represent overall heterogeneity without spatial information in terms of the gray levels within a tumor. Texture features derived from a gray-level co-occurrence matrix (GLCM) [30], gray-level run-length matrix (GLRLM) [31], neighborhood gray-tone difference matrix (NGTDM) [32] and gray-level size zone matrix (GLSZM) [33] represent the local spatial inhomogeneity in terms of gray levels within a tumor. The GLSZM features were employed for characterizing the inhomogeneity of cell nuclei [33].
To obtain the multiresolutional image features, including statistical and texture features, the wavelet transform has been applied for multiresolutionally decomposing multiscale local intensity variations (intratumor inhomogeneity) in an image into several low- and high-frequency components [34]. The multiresolutional decomposition was performed by using a ‘wavelet analysis filter bank’ approach [35] based on a 2D or 3D fast discrete wavelet transformation (fDWT) algorithm.
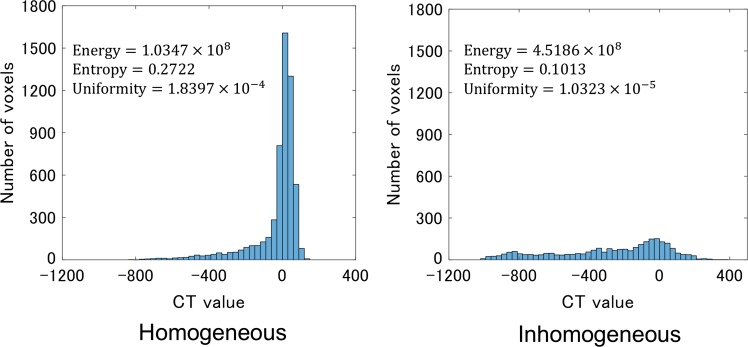
Figure 5 shows CT value histograms of homogeneous and inhomogeneous tumors (Fig. 1) for calculating statistical features. The homogeneous and inhomogeneous tumors have narrow and broad CT value histograms, respectively. The broad histogram with the wide range of CT values indicates tissue inhomogeneity. The statistical features derived from the gray-level histograms are energy, entropy, kurtosis, maximum, mean, mean absolute difference, median, minimum, range, root mean square, skewness, standard deviation, uniformity, and variance, etc. [36].
Fig. 5.
CT value (Hounsfield Unit) histograms of the homogeneous and inhomogeneous tumors (Fig. 1) for calculating statistical features.
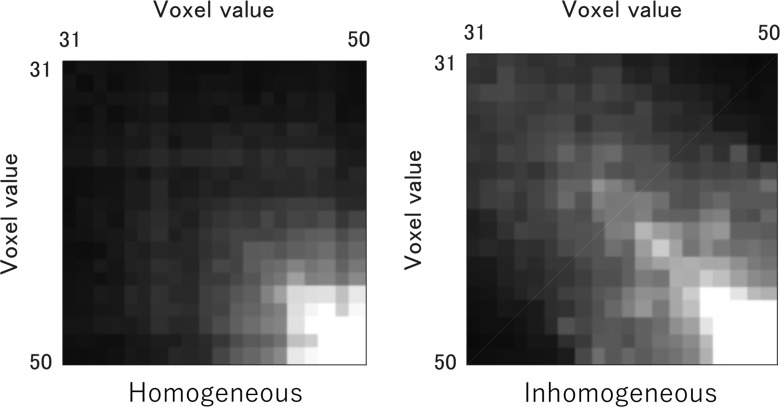
Figure 6 shows GLCMs of homogeneous and inhomogeneous tumors (Fig. 1), which quantify the frequency of all possible combinations of gray-scale values within neighboring voxels. The GLCMs are used for characterization of local tumor inhomogeneity by calculating many features, e.g. autocorrelation, contrast, correlation, cluster prominence, cluster shade, dissimilarity, energy, entropy, homogeneity, maximum probability, sum of squares variance, sum average, sum variance, sum entropy, difference variance, difference entropy, etc. [36]. Glioblastoma (GBM) cases have been classified by using GLCM, which characterized the GBM phenotypes [37].
Fig. 6.
GLCMs of the homogeneous and inhomogeneous tumors (Fig. 1), which quantify the frequency of all possible combinations of grayscale values within neighboring voxels.
STRATIFICATION OF INDIVIDUAL PATIENTS INTO SUBTYPES USING RADIOMIC BIOMARKERS
In precision medicine with radiomics, individual patients should be stratified into subtypes using the radiomic biomarkers that contain information on cancer traits (mutation, angiogenesis, metastasis, immune escape) and determine patients’ prognoses (survival time, recurrence, toxicity), according to the various treatment strategies. Liu et al. [38] explored the association between CT-based image features and epidermal growth factor receptor (EGFR) mutation statuses with surgically resected peripheral lung adenocarcinomas. The highest area under the curve (AUC) of 0.709 was achieved in the prediction of the EGFR mutation using a logistic regression model for the combination of clinical factors and image features. The activation of EGFR mutations can be detected by use of image features for the stratification of patients in terms of their responses to tyrosine kinase inhibitors (TKIs) therapy for lung adenocarcinomas. Bak et al. [29] attempted to identify predictive imaging biomarkers that supported genomic alterations and clinical outcomes in patients with lung squamous cell carcinoma (SCC) using a radiomics approach. Mutational profiles for core signaling pathways of lung SCC were stratified into five subtypes: redox stress, apoptosis, proliferation, differentiation, and chromatin remodelers. The range of the gray-level histogram and the right lung volume were significantly associated with alternation of the apoptosis and proliferation pathways, respectively (P < 0.05).
Wang et al. [39] demonstrated that image features (morphological, gray-level, and statistical texture measures) of breast tumors and their surrounding parenchyma on dynamic contrast enhancement (DCE)-MRI could distinguish triple-negative breast cancers from other subtypes with higher accuracy (AUC of 0.878) than when considering the characteristics of the tumor alone, because triple-negative breast cancer would have responses to neither hormonal therapy nor anti-HER2 therapy. Ma et al. [40] investigated whether image features extracted from digital mammography images were associated with molecular subtypes of breast cancer. They showed the AUCs of 0.865 for triple-negative vs non-triple-negative, 0.784 for HER2-enriched vs non-HER2-enriched, and 0.752 for luminal vs non-luminal subtypes.
Yin et al. [41] sought to determine the associations between angiogenesis in renal cell carcinoma and imaging features from PET/MRI. Their study reported significant correlations between radiomic features and tumor microvascular density (MVD) (angiogenesis), and also demonstrated that spatiotemporal features extracted from DCE-MRI had higher correlations with MVD than the textural features extracted from Dixon sequences and 18F-fluoro-2-deoxdeoxyglucose PET (18F-FDG PET). Furthermore, PET/MRI, which took advantages of the combined metabolic and morphological information (radiomic features), had higher correlation with MVD than utilizing PET or MRI alone.
Yang et al. [42] developed and validated a radiomics-based nomogram to predict lymph node metastasis (LNM) in solid lung adenocarcinoma, because LNM of lung cancer was one of the significant factors that were relevant to survival and recurrence. In a validation cohort (n = 53), the AUC of the performance of LNM differentiation was 0.856. Shen et al. [43] constructed a radiomic nomogram for prediction of pre-operation LNM in esophageal cancer. The AUC was 0.771 in a validation cohort (n = 57).
Chen et al. [44] investigated the relationship between programmed cell death ligand 1 (PD-L1) expression and immunohistochemical (IHC) biomarkers or textural features of 18F-FDG PET in patients with head and neck cancer. The p16 (surrogate marker for human papillomavirus (HPV) involvement) and Ki-67 (proliferative marker) staining percentages detected using IHC and several 18F-FDG PET/CT–derived textural features can provide supplemental information to determine tumor PD-L1 expression. Subtypes with tumors with a higher PD-L1 expression may benefit from checkpoint inhibitors.
Cunliffe et al. [45] examined the correlation between the radiologist-defined severity of normal tissue damage following radiation therapy for lung cancer treatment and a set of CT-based texture features. Nineteen features that characterized the severity of radiologic changes from pre-therapy scans were identified. Furthermore, the same group assessed the relationship between radiation dose and changes in a set of mathematical gray-level– and texture-based features and the ability of image features to identify patients who would develop radiation pneumonitis (RP) [46]. This study demonstrated that radiomic features could discriminate between patient subtypes with and without RP.
RADIOMCS WITH MACHINE LEARNING
A number of machine-learning algorithms [2] of AI technologies have been applied in radiomics for prediction or estimation of what will be associated with treatment strategies such as survival time, recurrence, adverse events, and tumor subtypes [47]. The uses of the machine learning, including feature selection methods, are comprehensive in many studies as described in this section, and the most appropriate machine-learning algorithms and feature selection methods are still unknown as we are yet to discover the combinations of significant features with the most prognostic powers.
Leger et al. [48] carried out a comparative study using 12 machine-learning algorithms combined with 11 feature selection methods, and the algorithm performances were assessed to predict loco-regional tumor control and overall survival for patients with head and neck squamous cell carcinoma in a multicenter cohort (n = 213). They found several combinations of machine-learning algorithms and feature selection methods that achieved similar results, e.g. random forest using maximally selected rank statistics had a concordance index of 0.71, and a Coxnet method with least absolute shrinkage and selection operator (LASSO) and elastic-net regularization based on boosting trees (BT) had a concordance index of 0.70 in combination with Spearman feature selection. Using the best performing models, patients were stratified into groups of low and high risk of recurrence. Parmar et al. [49] investigated 12 machine-learning approaches with 14 feature selection methods for radiomics-based prediction of 2-year survival of lung cancer patients treated with radical radiotherapy alone or with chemoradiation therapy. They identified that a Wilcoxon test–based feature selection method (stability = 0.84 ± 0.05, AUC = 0.65 ± 0.02) and a classification random forest method (relative standard deviation = 3.52%, AUC = 0.66 ± 0.03) had the greatest prognostic performances with relatively higher stability against data perturbation. Abdollahi et al. [50] proposed a radiomic analysis approach based on nine machine-learning tools and LASSO [2] for helping in the prediction of hearing loss induced by chemoradiation (cisplatin) therapy for head and neck cancer patients. The LASSO-penalized logistic modeling produced 10 predictive features with the highest performance, as indicated by an AUC of 0.885, for prediction of hearing loss. Shiradkar et al. [51] attempted to identify a radiomic signature derived from pretreatment biparametric MRI (bpMRI) that was predictive of prostate cancer biochemical recurrence after radical prostatectomy/radiotherapy/hormone therapy. The prediction accuracy using a machine learning of support vector machines (SVMs) with a feature selection of joint mutual information (JMI) was 0.84 in AUC. Gabryś et al. [52] investigated whether the machine-learning algorithms [2] (i.e. logistic regression with L1 penalty, logistic regression with L2 penalty, logistic regression with elastic net penalty, k-nearest neighbors, SVM, extra-trees, and gradient tree boosting) with dosiomic, radiomic, and demographic (age and sex) features allowed for xerostomia risk assessment in intensity-modulated radiation therapy (IMRT) for head and neck cancer (hypopharynx/larynx/nasopharynx/oropharynx/other). SVMs and extra-trees were the better performing classifiers compared with the others, whereas the algorithms based on logistic regression were the more appropriate choice for feature selection. Haga et al. [53] evaluated the potential application of radiomics for predicting the histology of early-stage non-small-cell lung cancer (NSCLC) patients, who underwent stereotactic body radiotherapy, by analyzing interobserver variability in tumor delineation. The average AUC for stratification of adenocarcinoma and squamous cell carcinoma subtypes was 0.725 using a naive Bayes model of machine learning.
PERSPECTIVES OF RADIOMICS IN RADIATION THERAPY
Radiomics was able to predict patients’ prognoses based on extensive information about the tumors in a non-invasive, fast, and low-cost way, by stratifying patients into several subtypes such as EGFR and non-EGFR patients, using several imaging biomarkers. AI could be the key to successful radiomics for discovering biomarkers and determining patient stratification. We need to develop mathematical image-feature models of biomarkers to characterize cancer phenotypes or hallmarks and to select appropriate paths [54] in decision-making at each radiation treatment step (diagnosis, treatment planning, treatment execution, and follow-up). AI, including machine learning, may boost the prognostic powers of radiomics. Since many types of machine-learning software are open-sourced and easy-to-use for radiation oncology staff, this third boom in AI could be the practical phase of AI use in radiation oncology. Radiomic approaches may have one practical application in precision medicine by predicting outcomes and toxicity for individual patients in radiation therapy.
ACKNOWLEDGEMENTS
The authors are grateful to all members in the Arimura Laboratory (http://web.shs.kyushu-u.ac.jp/~arimura/), whose comments made enormous contributions to the preparation of this review paper.
CONFLICT OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the contents and writing of the paper.
FUNDING
There was no funding for this study.
REFERENCES
- 1. Japanese Society for Artificial Intelligence https://www.ai-gakkai.or.jp/about/about-us/jsai_teikan/ (8 September 2018, date last accessed) (in Japanese)
- 2. Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning – Data Mining, Inference, and Prediction. 2nd edn New York:Springer, 2008. [Google Scholar]
- 3. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 4. Liu Y, Stojadinovic S, Hrycushko B et al. . A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS ONE 2017;12:e0185844 10.1371/journal.pone.0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lustberg T, van Soest J, Gooding M et al. . Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol 2018;126:312–7. 10.1016/j.radonc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 6. Men K, Chen X, Zhang Y et al. . Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol 2017;7:315 10.3389/fonc.2017.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cardenas CE, McCarroll RE, Court LE et al. . Deep learning algorithm for auto-delineation of high-risk oropharyngeal clinical target volumes with built-in dice similarity coefficient parameter optimization function. Int J Radiat Oncol Biol Phys 2018;101:468–78. 10.1016/j.ijrobp.2018.01.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lambin P, Rios-Velazquez E, Leijenaar R et al. . Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aerts HJ, Velazquez ER, Leijenaar RT et al. . Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arimura H, Soufi M. A review on radiomics for personalized medicine in cancer treatment. Med Imaging Technol 2018;36:81–9. [Google Scholar]
- 11. Yoo BC, Kim KH, Woo SM et al. (15 August 2017) Clinical multi-omics strategies for the effective cancer management. J Proteomics 2017, pii: S1874-3919(17)30286-5. doi: 10.1016/j.jprot.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 12. Baumann M, Krause M, Overgaard J et al. . Radiation oncology in the era of precision medicine. Nat Rev Cancer 2016;16:234–49. 10.1038/nrc.2016.18. [DOI] [PubMed] [Google Scholar]
- 13. Mobasheri A, Henrotin Y. Biomarkers of (osteo) arthritis. Biomarkers 2015;20:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O’Connor JP, Aboagye EO, Adams JE et al. . Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14:169–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Food and Drug Administration. www.fda.gov (8 September 2018, date last accessed)
- 16. Tanvetyanon T, Creelan BC, Chiappori AA. Current clinical application of genomic and proteomic profiling in non-small-cell lung cancer. Cancer Control 2014;21:32–9. [DOI] [PubMed] [Google Scholar]
- 17. Masuda H, Baggerly KA, Wang Y et al. . Comparison of molecular subtype distribution in triple-negative inflammatory and non-inflammatory breast cancers. Breast Cancer Res 2013;15:R112 10.1186/bcr3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hill DK, Heindl A, Zormpas-Petridis K et al. . Non-invasive prostate cancer characterization with diffusion-weighted MRI: insight from in silico studies of a transgenic mouse model. Front Oncol 2017;7:290 10.3389/fonc.2017.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerlinger M, Rowan AJ, Horswell S et al. . Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peeken JC, Nüsslin F, Combs SE‘Radio-oncomics’: the potential of radiomics in radiation oncology. Strahlenther Onkol 2017;193:767–79. 10.1007/s00066a017-1175a0. [DOI] [PubMed] [Google Scholar]
- 21. Nishihara M, Sasayama T, Kudo H et al. . Morbidity of stereotactic biopsy for intracranial lesions. Kobe J Med Sci 2011;56:E148–53. [PubMed] [Google Scholar]
- 22. Patel JD. Overcoming perceived hurdles in lung cancer screening: the low risk of complications of image-guided transthoracic needle biopsy. J Oncol Pract 2015;11:e360–2. [DOI] [PubMed] [Google Scholar]
- 23. Schröder FH, Hugosson J, Roobol MJ et al. . Screening and prostate cancer mortality: results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet 2014;384:2027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiu PK, Alberts AR, Venderbos LDF et al. . Additional benefit of using a risk-based selection for prostate biopsy: an analysis of biopsy complications in the Rotterdam section of the European Randomized Study of Screening for Prostate Cancer. BJU Int 2017;120:394–400. [DOI] [PubMed] [Google Scholar]
- 25. Fukagai T, Namiki T, Namiki H et al. . Discrepancies between Gleason scores of needle biopsy and radical prostatectomy specimens. Pathol Int 2001;51:364–70. [DOI] [PubMed] [Google Scholar]
- 26. Kickingereder P, Neuberger U, Bonekamp D et al. . Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol 2018;20:848–57. 10.1093/neuonc/nox188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 28. Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd edn.New York, NY: Wiley-Interscience, 2000. [Google Scholar]
- 29. Bak SH, Park H, Lee HY et al. . Imaging genotyping of functional signaling pathways in lung squamous cell carcinoma using a radiomics approach. Sci Rep 2018;8:3284 10.1038/s41598a018-21706a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;3:610–21. [Google Scholar]
- 31. Galloway MM. Texture analysis using gray level run lengths. Comput Graph Image Process 1975;4:172–9. [Google Scholar]
- 32. Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 1989;19:1264–74. 10.1109/21.44046. [DOI] [Google Scholar]
- 33. Thibault G, Fertil B, Navarro C et al. Texture indexes and gray level size zone matrix. Application to cell nuclei classification. In: Proceedings of the 10th International Conference on Pattern Recognition and Information Processing, PRIP 2009, Minsk, Belarus, 2009, pp. 140–5.
- 34. Mallat S. A theory for multiresolution signal decomposition: the wavelet representation. IEEE Pattern Anal Mach Intell 1989;11:674–93. 10.1109/34.192463. [DOI] [Google Scholar]
- 35. Strang G, Nguyen T. Wavelets and Filter Banks. Revised edn.Wellesley, MA: Wellesley-Cambridge Press, 1997. [Google Scholar]
- 36. Soufi M, Arimura H, Nakamoto T et al. . Exploration of temporal stability and prognostic power of radiomic features based on electronic portal imaging device images. Phys Med 2018;46:32–44. [DOI] [PubMed] [Google Scholar]
- 37. Chaddad A, Zinn PO, Colen RR Radiomics texture feature extraction for characterizing GBM phenotypes using GLCM. In: Proceedings of International Symposium on Biomedical Imaging 2015;7163822:84–7.
- 38. Liu Y, Kim J, Balagurunathan Y et al. . Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer 2016;17:441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang J, Kato F, Oyama-Manabe N et al. . Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: a pilot radiomics study. PLoS ONE 2015;10:e0143308 10.1371/journal.pone.0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma W, Zhao Y, Ji Y et al. (8 March 2018) Breast cancer molecular subtype prediction by mammographic radiomic features. Acad Radiol 2018, 10.1016/j.acra.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yin Q, Hung SC, Wang L et al. . Associations between tumor vascularity, vascular endothelial growth factor expression and PET/MRI radiomic signatures in primary clear-cell-renal-cell-carcinoma: proof-of-concept study. Sci Rep 2017;7:43356 10.1038/srep43356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang X, Pan X, Liu H et al. . A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J Thorac Dis 2018;10:S807–19. 10.21037/jtd.2018.03.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shen C, Liu Z, Wang Z et al. . Building CT radiomics based nomogram for preoperative esophageal cancer patients lymph node metastasis prediction. Transl Oncol 2018;11:815–24. 10.1016/j.tranon.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen RY, Lin YC, Shen WC et al. . Associations of tumor PD-1 ligands, immunohistochemical studies, and textural features in 18F-FDG PET in squamous cell carcinoma of the head and neck. Sci Rep 2018;8:105 10.1038/s41598a017-18489a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cunliffe AR, Armato SG 3rd, Straus C et al. . Lung texture in serial thoracic CT scans: correlation with radiologist-defined severity of acute changes following radiation therapy. Phys Med Biol 2014;59:5387–98. 10.1088/0031a9155/59/18/5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cunliffe A, Armato SG III, Castillo R et al. . Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol Biol Phys 2015;91:1048–56. 10.1016/j.ijrobp.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Acharya UR, Hagiwara Y, Sudarshan VK et al. . Towards precision medicine: from quantitative imaging to radiomics. J Zhejiang Univ Sci B 2018;19:6–24. 10.1631/jzus.B1700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leger S, Zwanenburg A, Pilz K et al. . A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep 2017;7:13206 10.1038/s41598a017-13448a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parmar C, Grossmann P, Bussink J et al. . Machine learning methods for quantitative radiomic biomarkers. Sci Rep 2015;5:13087 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abdollahi H, Mostafaei S, Cheraghi S et al. . Cochlea CT radiomics predicts chemoradiotherapy induced sensorineural hearing loss in head and neck cancer patients: a machine learning and multi-variable modelling study. Phys Med 2018;45:192–7. 10.1016/j.ejmp.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 51. Shiradkar R, Ghose S, Jambor I et al. (7 May 2018) Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: preliminary findings. J Magn Reson Imaging, 10.1002/jmri.26178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gabryś HS, Buettner F, Sterzing F et al. . Design and selection of machine learning methods using radiomics and dosiomics for normal tissue complication probability modeling of xerostomia. Front Oncol 2018;8:35 10.3389/fonc.2018.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haga A, Wataru T, Shuri A et al. . Classification of early stage non-small cell lung cancers on computed tomographic images into histological types using radiomic features: interobserver delineation variability analysis. Radiol Phys Technol 2017;11:1–9. [DOI] [PubMed] [Google Scholar]
- 54. Lucia F, Visvikis D, Desseroit MC et al. . Prediction of outcome using pretreatment 18F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging 2018;45:768–86. [DOI] [PubMed] [Google Scholar]