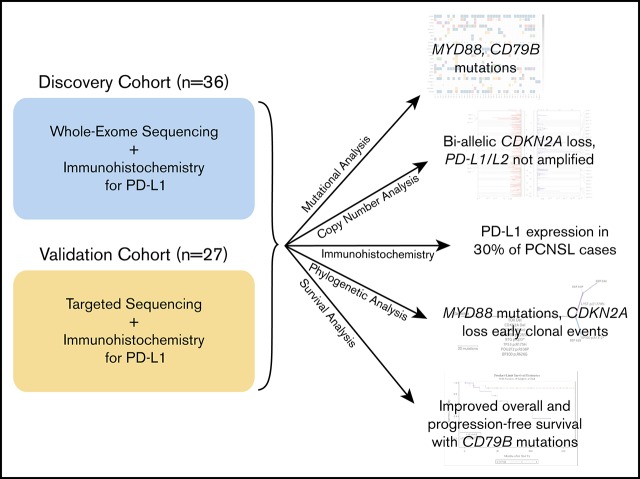
Key Points
MYD88 L265P mutation and CDKN2A loss are early mutational events in PCNSL.
PD-L1 expression was not unregulated in our cohort of PCNSL.
Abstract
The genetic alterations that define primary central nervous system lymphoma (PCNSL) are incompletely elucidated, and the genomic evolution from diagnosis to relapse is poorly understood. We performed whole-exome sequencing (WES) on 36 PCNSL patients and targeted MYD88 sequencing on a validation cohort of 27 PCNSL patients. We also performed WES and phylogenetic analysis of 3 matched newly diagnosed and relapsed tumor specimens and 1 synchronous intracranial and extracranial relapse. Immunohistochemistry (IHC) for programmed death-1 ligand (PD-L1) was performed on 43 patient specimens. Combined WES and targeted sequencing identified MYD88 mutation in 67% (42 of 63) of patients, CDKN2A biallelic loss in 44% (16 of 36), and CD79b mutation in 61% (22 of 36). Copy-number analysis demonstrated frequent regions of copy loss (ie, CDKN2A), with few areas of amplification. CD79b mutations were associated with improved progression-free and overall survival. We did not identify amplification at the PD-1/PD-L1 loci. IHC for PD-L1 revealed membranous expression in 30% (13 of 43) of specimens. Phylogenetic analysis of paired primary and relapsed specimens identified MYD88 mutation and CDKN2A loss as early clonal events. PCNSL is characterized by frequent mutations within the B-cell receptor and NF-κB pathways. The lack of PD-L1 amplifications, along with membranous PD-L1 expression in 30% of our cohort, suggests that PD-1/PD-L1 inhibitors may be useful in a subset of PCNSL. WES of PCNSL provides insight into the genomic landscape and evolution of this rare lymphoma subtype and potentially informs more rational treatment decisions.
Visual Abstract
Introduction
Primary central nervous system lymphoma (PCNSL) is a rare subtype of non-Hodgkin lymphoma, accounting for ∼4% of all newly diagnosed central nervous system (CNS) tumors.1 Although treatment varies, systemic high-dose methotrexate–based chemotherapy remains a foundation of PCNSL therapy. Diffuse-large B-cell lymphoma (DLBCL) constitutes the vast majority of PCNSLs.2-4
PCNSL is characterized by frequent MYD88 L265P activating mutations, biallelic CDKN2A loss, and mutations in CD79B, PIM1, and CARD11.5-8 MYD88 and CD79B are involved in the NF-κB signaling pathway that promotes cell division. MYD88 interacts with toll-like receptors and, in its most commonly mutated form (L265P), leads to increased NF-κB signaling.5,9-11 A higher prevalence of MYD88 L265P mutation in PCNSL and primary testicular lymphoma has been reported compared with DLBCL in all other sites (59.8%, 77.1%, and 16.5%, respectively).7,12-14 These prior studies provide compelling evidence that the presence of MYD88 L265P mutation is a genetic aberration that most commonly occurs in DLBCL within immune-privileged sites (ie, testis and CNS).7 9p24.1 (PD-L1/PD-L2) amplification was also identified in 1 study and may be a mechanism for PCNSL immune evasion.5 However, other studies did not identify changes in PD-L1/PD-L2 copy-number amplification.11
The objective of this study was to perform whole-exome sequencing (WES) of PCNSL samples to identify somatic mutations and copy-number alterations (CNAs) that define this entity and correlate these genetic events with clinical outcomes. It remains unclear whether MYD88 and other previously identified mutations occur as early clonal events in the phylogenetic evolution of PCNSL. To this end, another objective was to obtain paired specimens from patients at disease relapse and employ WES to understand the genomic evolution of PCNSL.
Methods
WES was performed on tissue from a discovery cohort of 36 patients who were treated at Massachusetts General Hospital for routine care and for whom tissue was available for genetic testing. Patients had known underlying immunodeficiency. WES and phylogenetic reconstruction were performed on paired samples from 4 patients with relapsed PCNSL (including 1 patient’s postmortem specimen). Targeted sequencing of the MYD88 gene was performed on 27 additional PCNSL patients in a validation cohort. WES and targeted sequencing were performed on tumor samples before treatment with chemotherapy except as indicated for the 4 cases of relapsed disease. Characteristics of both cohorts are listed in Table 1.
Table 1.
Cohort characteristics
| Category | ||
|---|---|---|
| Discovery cohort (n = 36) | Pooled discovery and validation cohorts (n = 64) | |
| Mean (±SD) age at first imaging diagnosis, y | 62.07 (±10.01) | 61.27 (±10.75) |
| Sex | ||
| Male | 21 (58) | 35 (55) |
| Female | 15 (42) | 29 (45) |
| Diagnosis | ||
| Symptoms* | ||
| Altered mental status or confusion | 14 (39) | 25 (40) |
| Imbalance, dizziness, or vertigo | 9 (25) | 21 (33) |
| Weakness | 9 (25) | 13 (21) |
| Language difficulties, dysarthria, or aphasia | 10 (28) | 19 (30) |
| Other | 26 (72) | 51 (81) |
| Tumor focality | ||
| Single | 18 (50) | 34 (53) |
| Multiple | 18 (50) | 29 (45) |
| NR | 0 (0) | 1 (2) |
| Tumor location | ||
| Infratentorial | 3 (8) | 6 (9) |
| Supratentorial | 29 (81) | 48 (75) |
| Both | 4 (11) | 8 (13) |
| Midbrain | 0 (0) | 1 (2) |
| NR | 0 (0) | 1 (2) |
| Ocular involvement | ||
| Positive or suspicious | 4 (11) | 7 (10.9) |
| Negative | 31 (86) | 49 (76.6) |
| NR | 1 (3) | 8 (12.5) |
| CSF involvement† | ||
| Positive or suspicious | 8 (22) | 14 (22) |
| Negative | 18 (50) | 27 (42) |
| NR | 10 (28) | 23 (36) |
| Systemic imaging | ||
| Positive or suspicious | 1 (3) | 2 (3) |
| Negative | 35 (97) | 55 (86) |
| NR | 0 (0) | 7 (11) |
| Bone marrow biopsy | ||
| Positive or suspicious | 2 (6) | 2 (3.1) |
| Negative | 10 (28) | 15 (23.4) |
| NR | 24 (67) | 47 (73.4) |
| Testicular ultrasound | ||
| Positive or suspicious | 0 (0) | 0 (0) |
| Negative | 9 (25) | 17 (27) |
| NR | 27 (75) | 47 (73) |
| Therapy | ||
| Initial induction regimen | ||
| Methotrexate, temozolomide, and rituximab | 23 (64) | 41 (64) |
| Methotrexate ± rituximab | 13 (36) | 23 (36) |
| Autologous stem-cell transplantation | ||
| Received | 8 (22) | 19 (30) |
| Did not receive | 28 (78) | 45 (70) |
| Response | ||
| Response to initial therapy | ||
| CR | 30 (83.3) | 57 (89) |
| PR | 3 (8.3) | 4 (6) |
| PD | 3 (8.3) | 3 (5) |
| Relapse | ||
| ≥1 | 14 (39) | 25 (39) |
| First in brain | 10 (71‡) | 15 (60‡) |
| First ocular or systemic | 4 (29‡) | 10 (40‡) |
| Multiple | 5 (14) | 8 (12.5) |
| Follow-up and survival, mo | ||
| Median follow-up | 56.2 | |
| Median PFS | 111 (95% CI, 35 to ∞) | |
| Median OS | NR | |
| Median TTCR | 4 (95% CI, 3-6) |
Data are n (%) unless otherwise indicated.
CI, confidence interval; CR, complete response; CSF, cerebrospinal fluid; NR, not reported/performed; OS, overall survival; PD, progressive or refractory disease; PFS, progression-free survival; PR, partial response; SD, standard deviation; TTCR, time to complete response.
Symptoms not reported for 1 validation patient.
If evaluated within 1 month of diagnosis.
Percentage of patients with specific location of first relapse of all patients who relapsed at least once.
The study was approved by our institutional review board, in accordance with the Declaration of Helsinki. All patients who underwent WES provided written informed consent for use of their tumor tissue, blood, and clinical information. Both a hematopathologist and a neuropathologist confirmed the histologic diagnoses and selected representative formalin-fixed paraffin-embedded (FFPE) samples for WES comprised of at least 40% cancer cells.
Sequence data generation and preprocessing
WES was performed using the sequencing platforms at the Broad Institute as previously described.15 A binary sequence alignment file was generated for each sample using the sequencing data processing pipeline known as Picard (http://broadinstitute.github.io/picard/). Picard consists of four steps and has been previously described.16
Cancer genome analysis pipeline.
Analysis, along with quality control, of the WES data was performed using the Broad Institute analysis platform Firehose.17 Somatic single-nucleotide variants (SSNVs), small insertions and deletions (indels), and somatic CNAs were identified as previously described.16 In brief, SSNVs and indels were detected on each tumor-normal pair using the algorithms MuTect and Strelka, respectively.18,19 Potential FFPE artifacts were filtered using a panel of normals filter composed of 374 FFPE samples. Somatic CNAs were estimated by generating a coverage profile for each tumor sample using ReCapSeg, calculating allelic copy ratio of each target segment using AllelicCapseg, and running ABSOLUTE to correct for purity and ploidy of the sample. We then mapped allelic copy ratios to allelic copy numbers via linear transformation.20,21
CCF of SSNVs and indels and phylogenetic analysis.
Cancer cell fraction (CCF) distributions of each SSNV and indel were estimated using ABSOLUTE as previously described.16 Mutation CCFs were clustered across each individual using a Bayesian clustering method, and final clusters were found by sampling from a Dirichlet process using 500 iterations of a Markov chain Monte Carlo sampler as previously described.22,23 Phylogenetic trees were then drawn for each individual using CCF estimates of the final clusters.
Targeted sequencing of MYD88 and DNA purification.
As described, our validation cohort was composed of 27 PCNSL patients who underwent targeted sequencing of MYD88. DNA was purified from FFPE samples. We obtained 1-mm diameter punches from the regions identified by the study pathologists and used the QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer’s protocol. DNA samples were amplified using standard conditions with primers MYD88_ex5_F1 (5′-GTTAACCCTGGGGTTGAAGACT-3′) and MYD88_ex5_R1 (5′-GTGTCTGTGAAGTTGGCATCTC-3′), generating a 374-bp amplicon. The amplified product was used as a template in a seminested polymerase chain reaction with MYD88_ex5_F1 and MYD88_ex5_R2 (5′-GAAGTACATGGACAGGCAGACA-3′), for a final amplicon 291 bp in length. Each sample was amplified and sequenced twice using the Beckman Coulter Genomics Sanger sequencing platform.
Statistical considerations.
We compared the presence of specific comutations (MYD88, IRF4, CARD11, CDKN2A loss, CD79b, PIM1) using McNemar’s test, which evaluates the amount of discordance. A significant P value indicates that for the 2 genes in comparison, the proportions in which 1 is mutated and the other is not mutated are different. Comparison is statistically significant when the false-discovery rate has been controlled at 5%.
OS was defined as the number of months between the date of diagnosis and the date of death resulting from any cause. Follow-up of patients who did not die was censored at the date of last contact. PFS was defined as the number of months between the date of diagnosis and the date of first occurrence of either radiographic disease progression or death resulting from any cause. Follow-up of patients who neither progressed nor died was censored at the date of last contact. Follow-up of patients who did not achieve a CR was censored at the date of last follow-up. Deaths without prior CR were censored events. Note that a competing-risks approach was not used because only 1 patient had a response characterized as progressive disease, and there were no deaths before CR. Demographic variables including age, sex, number of CNS tumors, and tumor location were collected for all patients. Fisher’s exact P values were reported for sex and all demographic analyses; however, a Wilcoxon rank-sum test was used for age. Distributions of OS and PFS were summarized using the Kaplan-Meier method. Median times are accompanied by 95% CIs estimated using log(-log(endpoint)) methodology. Tests of significance were based on the log-rank test.
IHC.
Immunohistochemistry (IHC) staining for programmed death-1 ligand (PD-L1) was performed on 43 available patient specimens that included both the discovery (n = 22) and validation (n = 21) cohorts. Two patients (PN-059 and PN-079) had both primary and relapsed specimens analyzed. One case underwent IHC of testicular recurrence (PN-078). IHC was performed on FFPE tumor tissue. Specimens were sectioned at 5 μm, mounted onto positively charged glass microscope slides, and baked at 65°C for 60 minutes. Immunostaining was performed on the Leica BOND RX autostainer using the Leica Bond Polymer Refine Detection Kit (Leica Biosystems, Buffalo Grove, IL). Peroxidase block was performed for 5 minutes at room temperature (RT), and antigen retrieval was performed using Epitope Retrieval Solution 2 BOND for 20 minutes at 100°C. Incubation with rabbit monoclonal anti–PD-L1 antibody E1L3N (Cell Signaling Technology, Danvers, MA) was performed at a 1:200 dilution for 15 minutes at RT. Detection was performed through sequential incubations with Post Primary Rabbit Anti-Mouse IgG for 8 minutes at RT, Polymer Anti-Rabbit Poly-HRP-IgG for 8 minutes at RT, and DAB chromogen for 10 minutes at RT, followed by hematoxylin counterstaining for 5 minutes at RT. Stained sections were evaluated and scored by the study neuropathologist. We then scored membranous PD-L1 staining on tumor cells as 1+, 2+, or 3+ expression. Tumor cells with >10% 3+ staining for membranous PD-L1 were considered positive. Prior studies have used between 1% and 5% positivity of all cells in the tumor environment (tumor and nontumor cells), and our staining threshold range was similar to those in prior studies, with an emphasis on characterizing tumor cells with unequivocal membranous expression.5,24-26
Results
WES of PCNSL
We performed WES on a discovery cohort of 36 patients with PCNSL (Figure 1). All samples except 1 (PN-079; obtained at relapse) were obtained from newly diagnosed PCNSL patients. WES was also performed on 4 matched samples obtained at disease relapse. Median follow-up time for our discovery cohort as determined by the inverted Kaplan-Meier method was 56.7 months. Median OS was not reached, and median PFS was 112 months (95% CI, 35 months to ∞) for this cohort.
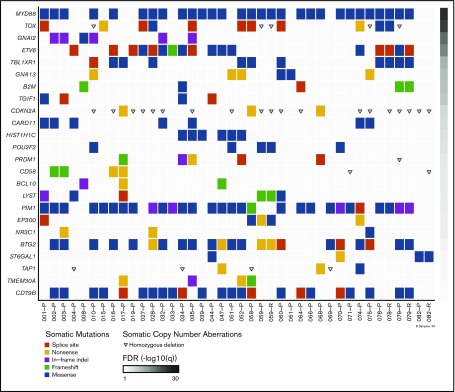
Figure 1.
Somatic mutations in PCNSL. Somatic mutations and copy-number aberrations in PCNSL. The plot includes primary (-P) and relapse samples (-R). The significance of mutations in each gene is shown to the right by a false discovery rate (FDR) q value.
Mean nonsynonymous mutation rate was 2.69/Mb (range, 0.14-6.62/Mb), similar to prior reported mutation rates in PCNSL.5,6,9,11 MYD88 L265P mutation was identified in 26 (72%) of 36 patients. To further assess the frequency of MYD88 mutation, we performed targeted MYD88 sequencing on a validation cohort of 27 additional PCNSL patients. The combined discovery and validation cohort analysis demonstrated MYD88 L265P mutation in 42 (67%) of 63 patients. CARD11 promotes I-κB expression and upregulates the NF-κB pathway. We identified CARD11 missense mutations in 7 (19%) of 36 patients. PIM1 has previously been reported as frequently mutated in PCNSL, and we observed mutations in 27 (75%) of 36 patients.5,11 The PIM1 mutations we identified were most commonly missense (n = 21) but also included in-frame insertion/deletion (n = 4), frameshift (n = 1), or splice site variant (n = 1). CD79b mutation was present in 22 (61%) of 36 of samples. The CD79b mutations were variable, but the most frequent mutation site was Y196, consistent with prior reports.10,27 We also identified frequent mutations in TOX, ETV6, TBL1XR1, and BTG2 (Figure 1).
Using a combined analysis of the discovery and validation cohorts, we did not reach median OS or PFS and did not identify significant survival differences in MYD88-mutant vs wild-type (WT) tumors. Within the discovery cohort, we did not observe a significant effect on OS or PFS for CARD11-, PIM1-, or IRF4-mutant tumors. CD79b mutation was associated with improved PFS, with median PFS of 35 months in CD79b WT tumors (95% CI, 13 months to ∞) vs 112 months in CD79b-mutant tumors (95% CI, 89 months to ∞; P = .02). Similarly, CD79b mutation was also associated with improved OS, with median OS of 105 months in WT tumors (95% CI, 41 months to ∞) vs OS not reached in mutant tumors (P = .03).
9p24.1 (PD-L1/PD-L2) CNAs and IHC
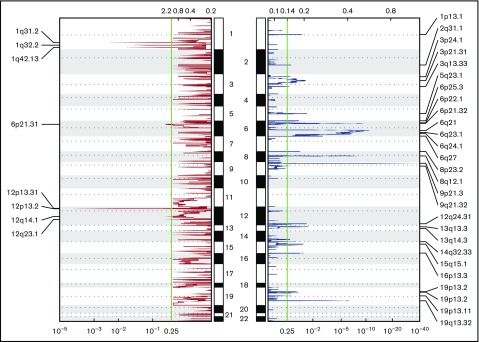
We assessed recurrent CNAs using the Genomic Identification of Significant Targets in Cancer algorithm n all 36 PCNSL cases in the discovery cohort (Figure 2).28 Overall, deletions were the predominant pattern of CNA, with gains being much less frequent (Figure 2). We identified biallelic loss of CDKN2A (9p21.3) in 16 (44%) of 36 patients (Figures 1 and 2). Contrary to other reports, we did not identify copy gain at PD-1 or PD-L1/PD-L2 loci (Figure 2).5 To investigate this further, we performed IHC staining for PD-L1 on 43 PCNSL patient specimens from both the discovery (n = 22) and validation (n = 21) cohorts. We observed tumor cell PD-L1 expression in 13 (30%) of 43 samples and tumor-associated macrophage PD-L1 expression in 16 (37%) of 43 samples. IHC of 1 patient’s matched primary and relapsed tumor specimens did not show PD-L1 expression in either sample. In another patient, no PD-L1 expression was detected by IHC in either the CNS tumor or a simultaneous skin lesion at the time of disease relapse.
Figure 2.
Significant CNAs in the cohort. Chromosomal regions of copy loss indicated in blue. Areas of copy gain indicated in red.
Phylogenetic analysis of PCNSL
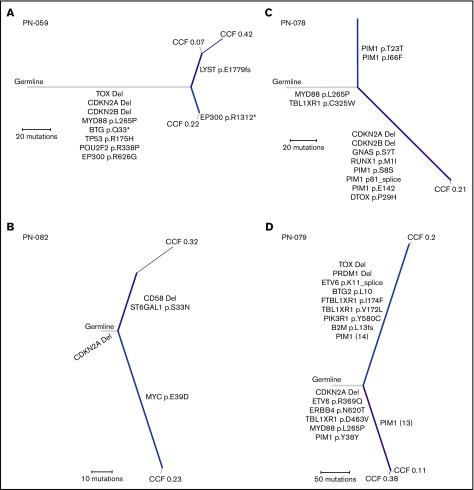
We performed the first WES on matched tumor samples from 4 PCNSL patients, comparing their tumor specimens from the initial pretreatment diagnosis with the specimens at the time of relapse in 3 cases (including one postmortem specimen) and comparing simultaneous CNS and peripheral (skin) relapse specimens in 1 case. Our primary objective was to infer the evolutionary relationship between initial and relapsed specimens both temporally and spatially. In 1 patient, we compared the primary tumor with matched relapsed CNS disease (temporally separated by 413 days) and identified MYD88 L265P mutation and CDKN2A loss as early clonal events with a cancer cell fraction of 1.0 (Figure 3A). Interestingly, in a patient originally diagnosed with PCNSL, with subsequent systemic relapse followed by 2 CNS relapses, MYD88 mutation was not identified, but CDKN2A loss was an early clonal event when comparing the primary tumor with the matched brain relapse specimen obtained at autopsy (temporally separated by 3211 days; Figure 3B). In a separate patient, when comparing disease relapse in the testis with the original CNS sample (temporally separated by 707 days), MYD88 L265P was an early clonal event, as was TBL1XR1 (Figure 3C). CDKN2A loss was identified only in the testicular recurrence. Lastly, in matched samples from a simultaneous brain and skin relapse (temporally separated by 3 days; relapsed 3400 days after primary diagnosis; Figure 3D), we identified MYD88 mutation and CDKN2A loss as early clonal events in both specimens.
Figure 3.
Phylogenetic trees. Phylogenetic trees from 4 different patients with relapsed PCNSL. (A-C) Comparison of the primary tumor with the relapsed specimen: primary and brain relapse (A), primary and systemic relapse (B), and primary and testicular relapse (C). (D) Comparison between simultaneous occurrence of brain and skin lymphoma. The width of each line corresponds to the CCF. Subpopulations (CCF <1) are labeled at the end of each line. The length of the line corresponds to the number of mutations that are present. Relevant mutations are indicated on each branch. Gray, shared; blue, private to primary tumor specimen in the CNS; purple, private to the relapsed or extracranial tumor specimen.
Discussion
Prior studies of WES in PCNSL have shed light on the defining genetic alterations that distinguish PCNSL from systemic DLBCL.5-11,14,29-33 However, given the rarity of PCNSL, study sample sizes tend to be small. Distinguishing features of our study are the relatively large sample size (36 patient cases in the discovery cohort, 27 in the validation cohort), the inclusion of 4 cases with paired, pretreatment tumor specimens with posttreatment relapse specimens to infer the clonal evolution of PCNSL, the association of genetic alterations with clinical outcomes including the observation that CD79b mutation may confer an improved prognosis, and the detailed study of PD-1/PD-L1 amplification and expression.
With respect to the phylogenetic analyses, 1 patient (Figure 3A) represents the most common clinical scenario in PCNSL: an initial response to treatment followed by relapse confined to the brain. Phylogenetic analysis in this patient demonstrated MYD88 L265P mutation and CDKN2A loss as early clonal events in tumor evolution. In total, 3 of 4 patients with paired specimens had truncal MYD88 mutations and CDKN2A deletions, suggesting that both of these events could be drivers of disease. Our single relapsed patient without MYD88 mutation (Figure 3B) was atypical for PCNSL, given the relapse outside the CNS 6 months after initial diagnosis. Because the tumor genetic profile was atypical for PCNSL (no MYD88, CD79b, or PIM1 alterations) and the patient relapsed systemically shortly after initial diagnosis, it is possible that this patient had systemic germinal center B-cell DLBCL that was not identified on initial extent of disease evaluation. Given the heterogeneity in these clinical scenarios, additional larger studies with matched pretreatment and autopsy tumor samples will be needed to determine whether our results are generalizable.
The efficacy of immune checkpoint blockade in the treatment of PCNSL remains a subject of investigation. There are conflicting findings with regard to copy-number analysis at 9p24.1 (PD-L1) and expression of PD-L1 in PCNSL.5,11 In 1 study, copy gain of PD-L1/PD-L2 was observed in 28 (67%) of 42 samples, whereas we did not identify copy gain of PD-L1/PD-L2 in our cohort.5 We also evaluated membranous PD-L1 expression on tumor cells. Although membranous PD-L1 expression on tumor cells can predict positive response to checkpoint inhibitors in some solid tumors,34-37 the significance of cytoplasmic PD-L1 expression is unknown, and the definition of positive expression in prior reports has been variable.24,25 For example, a prior study reported expression of PD-1/PD-L1 within the tumor microenvironment (including tumor and immune cells) in 90% of PCNSL cases (18 of 20), where positive expression was defined as strong PD-L1 or PD-1 expression at the membrane in at least 5% of cells of any cell type.24 However, in that study, when evaluating only tumor cells, 2 of 20 cases scored positively for PD-L1 membranous expression, more consistent with our findings.24 In another study, a cutoff for positive PD-L1 was defined as membranous or cytoplasmic expression in ≥1% of cells, which resulted in 37.5% (12 of 32) of PCNSL cases expressing PD-L1 within tumor cells.25 The dynamics of PD-1/PD-L1 between tumor and immune cells are complex, and it is unclear whether this information provides a reliable means of associating expression with the clinical efficacy of PD-1/PD-L1 blockade in PCNSL.26,38
We observed that MYD88 L265P and CD79b mutations cooccurred in 50% (18 of 36) of PCNSL cases. Interestingly, MYD88 mutations were not associated with a change in PFS or OS in our cohort, whereas CD79b mutations were associated with improved PFS and OS. In another study by Takano et al,39 MYD88 mutations occurred more frequently in elderly patients (age >65 years) and were associated with worse prognosis, which we did not observe in our study; however, this needs to be investigated in larger cohorts.39 The B-cell antigen receptor (BCR) signaling pathway is dependent on CD79b activity.27 There is downstream convergence between MYD88 activity (NF-κB pathway) and the BCR pathway via MYD88 interaction with IRAK and CD79b with CARD11.27 CD79b normally functions to promote the assembly of the BCR signaling complex.27 This results in I-κB kinase activation and promotion of NF-κB signaling.10,27 CD79b mutation at Y196 in PCNSL has been previously reported.5,10 The frequency of MYD88 L265P and CD79b mutation cooccurrence is frequent and reported to be as high as 100%.5,10,40 Bruton tyrosine kinase (BTK) inhibits NF-κB signaling downstream of the BCR pathway. In systemic activated B-cell DLBCL (genetically similar to PCNSL), inhibition of BTK by the oral BTK inhibitor ibrutinib resulted in a 37% response rate.41 A clinical trial of ibrutinib monotherapy in PCNSL revealed an 83% partial response rate.42 Importantly, prior studies suggest that BTK inhibitor resistance may be mediated through CARD11 mutation.43
Finally, our results indicate that PCNSL is characterized by frequent regions of copy loss. The biallelic deletion of CDKN2A observed in 44% of our cases raises the possibility of investigating the efficacy of CDK4/6 inhibitors in PCNSL patients.
In conclusion, PCNSL is a rare subtype of non-Hodgkin lymphoma that is characterized by frequent MYD88 activating mutations as well as CDKN2A biallelic loss. In this study, we show that MYD88 mutation and CDKN2A loss are early clonal events in PCNSL evolution. CARD11 mutations, which may predict resistance to BTK inhibitors, were present in a subset of PCNSL patients. PD-L1 was not amplified in any cases, although expression was detected in 30% of patients; it remains unclear whether the lack of PD-L1 amplification is predictive of a poor response to immune checkpoint blockade, supporting the need for clinical trials.
Acknowledgment
This work was supported by Sidney Kimmel Comprehensive Cancer Core Grant P30CA006973 from the National Institutes of Health, National Cancer Institute.
Authorship
Contribution: P.K.B., T.T.B., and S.C. designed and supervised the research; N.N., M.D.W., and C.M.G. performed the research; M.D.W., C.M.G., M.B., A. Kaplan, M.R.D., I.B., A. Kaneb, J.D., D.R.B., E.B., K.H., B.K., and D.P.C. gathered the biological samples and collected data; N.N., M.D.W., M.L., J.A.F., M.M.-L., M.P.F., and S.F. analyzed and interpreted data; C.M.G., M.L., and A.G.-H. performed statistical analysis; F.J.R and M.H. contributed samples; and N.N., M.D.W., P.K.B., and T.T.B. wrote the manuscript.
Conflict-of-interest disclosure: P.K.B has received research funding from Merck and honorarium from Genentech and consulted for Lilly and Angiochem. T.T.B. has been a pharmaceutical consultant for Merck, NXDC, Amgen, and Proximagen/Upsher, served on a scientific advisory board for Genomicare, been a consultant for Jiahui Health and Champions Biotechnology, and been a contributor for Up to Date. The remaining authors declare no competing financial interests.
Correspondence: Tracy T. Batchelor, 55 Fruit St, Yawkey Center, 9E-9300, Boston, MA 02114; e-mail: tbatchelor@mgh.harvard.edu; and Priscilla K. Brastianos, 55 Fruit St, Yawkey Center, 9E-9300, Boston, MA 02114; e-mail: pbrastianos@mgh.harvard.edu.
References
- 1.Villano JL, Koshy M, Shaikh H, Dolecek TA, McCarthy BJ. Age, gender, and racial differences in incidence and survival in primary CNS lymphoma. Br J Cancer. 2011;105(9):1414-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller DC, Hochberg FH, Harris NL, Gruber ML, Louis DN, Cohen H. Pathology with clinical correlations of primary central nervous system non-Hodgkin’s lymphoma. The Massachusetts General Hospital experience 1958-1989. Cancer. 1994;74(4):1383-1397. [DOI] [PubMed] [Google Scholar]
- 3.Camilleri-Broët S, Martin A, Moreau A, et al. . Primary central nervous system lymphomas in 72 immunocompetent patients: pathologic findings and clinical correlations. Groupe Ouest Est d’étude des Leucénies et Autres Maladies du Sang (GOELAMS). Am J Clin Pathol. 1998;110(5):607-612. [DOI] [PubMed] [Google Scholar]
- 4.Shiels MS, Pfeiffer RM, Besson C, et al. . Trends in primary central nervous system lymphoma incidence and survival in the U.S. Br J Haematol. 2016;174(3):417-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapuy B, Roemer MGM, Stewart C, et al. . Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127(7):869-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukumura K, Kawazu M, Kojima S, et al. . Genomic characterization of primary central nervous system lymphoma. Acta Neuropathol. 2016;131(6):865-875. [DOI] [PubMed] [Google Scholar]
- 7.Lee J-H, Jeong H, Choi J-W, Oh H, Kim Y-S. Clinicopathologic significance of MYD88 L265P mutation in diffuse large B-cell lymphoma: a meta-analysis. Sci Rep. 2017;7(1):1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi JW, Kim Y, Lee JH, Kim YS. MYD88 expression and L265P mutation in diffuse large B-cell lymphoma. Hum Pathol. 2013;44(7):1375-1381. [DOI] [PubMed] [Google Scholar]
- 9.Bruno A, Boisselier B, Labreche K, et al. . Mutational analysis of primary central nervous system lymphoma. Oncotarget. 2014;5(13):5065-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura T, Tateishi K, Niwa T, et al. . Recurrent mutations of CD79B and MYD88 are the hallmark of primary central nervous system lymphomas. Neuropathol Appl Neurobiol. 2016;42(3):279-290. [DOI] [PubMed] [Google Scholar]
- 11.Braggio E, Van Wier S, Ojha J, et al. . Genome-wide analysis uncovers novel recurrent alterations in primary central nervous system lymphomas. Clin Cancer Res. 2015;21(17):3986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheah CY, Wirth A, Seymour JF. Primary testicular lymphoma. Blood. 2014;123(4):486-493. [DOI] [PubMed] [Google Scholar]
- 13.Oishi N, Kondo T, Nakazawa T, et al. . High prevalence of the MYD88 mutation in testicular lymphoma: Immunohistochemical and genetic analyses. Pathol Int. 2015;65(10):528-535. [DOI] [PubMed] [Google Scholar]
- 14.Kraan W, Horlings HM, van Keimpema M, et al. . High prevalence of oncogenic MYD88 and CD79B mutations in diffuse large B-cell lymphomas presenting at immune-privileged sites. Blood Cancer J. 2013;3:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger MF, Lawrence MS, Demichelis F, et al. . The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brastianos PK, Nayyar N, Rosebrock D, et al. . Resolving the phylogenetic origin of glioblastoma via multifocal genomic analysis of pre-treatment and treatment-resistant autopsy specimens. NPJ Precis Oncol. 2017;1(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27(18):2601-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cibulskis K, Lawrence MS, Carter SL, et al. . Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811-1817. [DOI] [PubMed] [Google Scholar]
- 20.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557-572. [DOI] [PubMed] [Google Scholar]
- 21.Carter SL, Cibulskis K, Helman E, et al. . Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brastianos PK, Carter SL, Santagata S, et al. . Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5(11):1164-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stachler MD, Taylor-Weiner A, Peng S, et al. . Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47(9):1047-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berghoff AS, Ricken G, Widhalm G, et al. . PD1 (CD279) and PD-L1 (CD274, B7H1) expression in primary central nervous system lymphomas (PCNSL). Clin Neuropathol. 2014;33(1):42-49. [DOI] [PubMed] [Google Scholar]
- 25.Four M, Cacheux V, Tempier A, et al. . PD1 and PDL1 expression in primary central nervous system diffuse large B-cell lymphoma are frequent and expression of PD1 predicts poor survival. Hematol Oncol. 2017;35(4):487-496. [DOI] [PubMed] [Google Scholar]
- 26.Topalian SL, Hodi FS, Brahmer JR, et al. . Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis RE, Ngo VN, Lenz G, et al. . Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulain S, Boyle EM, Tricot S, et al. . Absence of CXCR4 mutations but high incidence of double mutant in CD79A/B and MYD88 in primary central nervous system lymphoma. Br J Haematol. 2015;170(2):285-287. [DOI] [PubMed] [Google Scholar]
- 30.Montesinos-Rongen M, Godlewska E, Brunn A, Wiestler OD, Siebert R, Deckert M. Activating L265P mutations of the MYD88 gene are common in primary central nervous system lymphoma. Acta Neuropathol. 2011;122(6):791-792. [DOI] [PubMed] [Google Scholar]
- 31.Yamada S, Ishida Y, Matsuno A, Yamazaki K. Primary diffuse large B-cell lymphomas of central nervous system exhibit remarkably high prevalence of oncogenic MYD88 and CD79B mutations. Leuk Lymphoma. 2015;56(7):2141-2145. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, Ju H, Kim DH, et al. . CD79B and MYD88 mutations in diffuse large B-cell lymphoma. Hum Pathol. 2014;45(3):556-564. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Aguilar A, Idbaih A, Boisselier B, et al. . Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res. 2012;18(19):5203-5211. [DOI] [PubMed] [Google Scholar]
- 34.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. . Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tawbi HA, Forsyth PA, Algazi A, et al. . Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N Engl J Med. 2018;379(8):722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long GV, Atkinson V, Lo S, et al. . Combination nivolumab and ipilimumab or nivolumab alone in melanoma brain metastases: a multicentre randomised phase 2 study. Lancet Oncol. 2018;19(5):672-681. [DOI] [PubMed] [Google Scholar]
- 37.Gettinger S, Rizvi NA, Chow LQ, et al. . Nivolumab monotherapy for first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(25):2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen BJ, Chapuy B, Ouyang J, et al. . PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res. 2013;19(13):3462-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takano S, Hattori K, Ishikawa E, et al. . MyD88 mutation in elderly predicts poor prognosis in primary central nervous system lymphoma: multi-institutional analysis. World Neurosurg. 2018;112:e69-e73. [DOI] [PubMed] [Google Scholar]
- 40.Ngo VN, Young RM, Schmitz R, et al. . Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson WH, Young RM, Schmitz R, et al. . Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lionakis MS, Dunleavy K, Roschewski M, et al. . Inhibition of B cell receptor signaling by ibrutinib in primary CNS lymphoma. Cancer Cell. 2017;31(6):833-843.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grommes C, Pastore A, Palaskas N, et al. . Ibrutinib unmasks critical role of bruton tyrosine kinase in primary CNS lymphoma. Cancer Discov. 2017;7(9):1018-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]