Abstract
We have developed a family of unnatural base pairs (UBPs), exemplified by the pair formed between dNaM and dTPT3, for which pairing is mediated not by complementary hydrogen bonding, but by hydrophobic and packing forces. These UBPs enabled the creation of the first semi-synthetic organisms (SSOs) that store increased genetic information and use it to produce proteins containing non-canonical amino acids. However, retention of the UBPs was poor in some sequence contexts. Here, to optimize the SSO we synthesize two novel benzothiophene-based dNaM analogs, dPTMO and dMTMO, and characterize the corresponding UBPs, dPTMO-dTPT3 and dMTMO-dTPT3. We demonstrate that these UBPs perform similarly to, or slightly worse than dNaM-dTPT3 in vitro. However, in the in vivo environment of an SSO, retention of dMTMO-dTPT3, and especially dPTMO-dTPT3, is significantly higher than that of dNaM-dTPT3. This more optimal in vivo retention results from better replication, as opposed to more efficient import of the requisite unnatural nucleoside triphosphates. Modeling studies suggest that the more optimal replication results from specific internucleobase interactions mediated by the thiophene sulfur atoms. Finally, we show that dMTMO and dPTMO efficiently template the transcription of RNA containing TPT3 and that their improved retention in DNA results in more efficient production of proteins with non-canonical amino acids. This is the first instance of using performance within the SSO as part of the UBP evaluation and optimization process. From a general perspective, the results demonstrate the importance of evaluating synthetic biology “parts” in their in vivo context and further demonstrate the ability of hydrophobic and packing interactions to replace the complementary hydrogen-bonding that underlies the replication of natural base pairs. From a more practical perspective, the identification of dMTMO-dTPT3 and especially dPTMO-dTPT3 represents significant progress towards the development of SSOs with an unrestricted ability to store and retrieve increased information.
Graphical Abstract

INTRODUCTION
The four-letter natural genetic alphabet, conserved throughout nature, is based on the formation of two base pairs via complementary hydrogen-bonding (H-bonding). In the early 1990s, Benner and co-workers reported efforts to develop unnatural base pairs (UBPs) with altered H-bonding topologies,1 and progress with these analogs continues.2 As an alternative approach to developing UBPs, our group3,4 and the Hirao group5 have instead drawn inspiration from Kool and co-workers’ demonstration that H-bonds are not necessary for DNA polymerases to correctly pair a nucleoside triphosphate opposite a template nucleotide6 and have employed shape complementarity and/or hydrophobic and packing forces for pairing. Our efforts have culminated in the development of a class of UBPs, exemplified by dNaM-d5SICS and dNaM-dTPT3 (Figure 1A). DNA containing either of these UBPs is efficiently replicated7,8 and transcribed into RNA.9,10 Most importantly, we have reported that when the corresponding triphosphates are made available within Escherichia coli (via heterologous expression of the nucleoside triphosphate transporter PtNTT211), they can be used to replicate DNA containing the UBP.12,13 Furthermore, the resulting semi-synthetic organism (SSO) can transcribe DNA containing the unnatural nucleotides into mRNA and tRNA, and use the resulting unnatural codon-anticodon pairs to translate proteins with non-canonical amino acids (ncAAs).14 In addition, the forces underlying the pairing of the unnatural nucleotides, as well as their physical properties have been explored by others.15–21
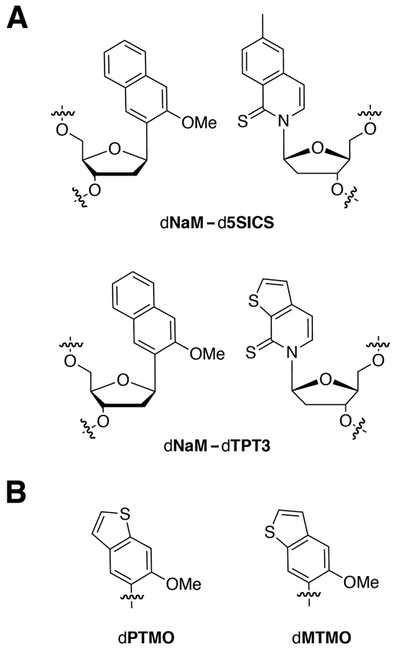
Figure 1.
A) The dNaM-d5SICS and dNaM-dTPT3 UBPs. B) dNaM analogs, dPTMO and dMTMO, sugar and phosphate groups omitted for clarity.
Although UBP retention within the DNA of the SSO is higher with dNaM-dTPT3 than dNaM-d5SICS, it remains sequence-dependent, which limits the extent of the unnatural information the SSO may store.13 To increase retention, we have optimized the SSO by expressing Cas9 and guide RNAs that target and thus cause the degradation of DNA that has lost the UBP.13 In addition, we have determined that the major route to UBP loss is RecA-mediated recombination and that DNA polymerase II facilitates UBP retention in challenging sequences, and thus we have created SSOs wherein the gene encoding polymerase II has been released from its normal repression and/or the gene encoding RecA has been deleted.22 While both the error elimination and avoidance systems increase the range of sequences in which the UBP is efficiently retained, retention remains poor in a handful of sequence contexts, highlighting the need for continued chemical optimization of the UBP.
The discovery of dTPT3 resulted from a survey of d5SICS analogs, the design of which was inspired by a mechanistic model of UBP replication.23 In the model, which was based on both kinetic9 and structural24,25 data, the UBP is synthesized with a unique, mutually induced-fit mechanism. Specifically, pairing of an unnatural triphosphate with its cognate template in the polymerase active site drives the same large conformational change of the polymerase, from an open to a closed conformation, that is induced during the correct pairing of natural nucleotides.26–28 Concomitantly, formation of the closed complex induces the forming UBP to adopt the required Watson-Crick-like structure, as opposed to the cross-strand intercalated structure adopted by the UBP in free duplex DNA. However, after UBP synthesis and polymerase translocation, the UBP is prone to adopt the cross-strand intercalated structure, and deintercalation must occur for DNA synthesis to continue. Thus, efficient replication requires the optimization of each unnatural nucleobase such that intrastrand packing is favored over interstrand packing. The data suggests that the contracted and more polarizable nucleobase of dTPT3 is better able to achieve this delicate balance of packing interactions than is the nucleobase of d5SICS.
In the current work, we sought to explore whether a similar approach with the dNaM scaffold could further optimize the UBP. We designed two new nucleobases, dPTMO and dMTMO (Figure 1B), and characterized their ability to be replicated in DNA in vitro, as well as their ability to store and retrieve information in the in vivo environment of an SSO. Neither dPTMO-dTPT3 nor dMTMO-dTPT3 is better replicated in vitro. However, both, especially dPTMO-dTPT3, are better replicated in the SSO, and modeling studies suggest that this results from specific interactions mediated by the thiophene sulfurs. Moreover, we show that both dPTMO and dMTMO efficiently and faithfully direct the transcription of RNA containing TPT3 within the SSO, demonstrating that the more optimal replication of DNA containing the new UBPs results in the more efficient production of proteins containing an ncAA.
RESULTS AND DISCUSSION
The novel nucleobases dPTMO and dMTMO (Figure 1B) were synthesized from their respective bromo- and methoxy- substituted anilines. A Sandmeyer reaction was used to install an iodine, which was followed by an Ullman reaction to produce the arylthiol, which was then converted to the corresponding acetaldehyde diethyl acetals. Friedel-Crafts cyclization then produced the bromo- and methoxy- substituted benzothiophene nucleobases, which were coupled to a disiloxane-protected 2-deoxy-d-ribono-1,4-lactone by lithium-halogen exchange using n-butyllithium, and then reduced with triethylsilane and boron trifluoride diethyl etherate to yield anomeric mixtures of silyl protected nucleoside. Tetra-n-butylammonium fluoride deprotection and subsequent purification via silica gel column chromatography afforded the anomerically pure β-nucleoside of dPTMO, whereas for dMTMO, the reaction resulted in an inseparable anomeric mixture of the free nucleoside. However, re-protection of the 5’-hydroxyl with a 4,4-dimethoxytrityl group allowed for facile separation of the α- and β- anomers via silica gel column chromatography, and deprotection then yielded the pure dMTMO β-nucleoside. In both cases, free nucleosides were converted to the corresponding triphosphates, dPTMOTP and dMTMOTP, as reported previously.23
With dPTMOTP and dMTMOTP in hand, we first explored their insertion opposite dTPT3 using a steady state kinetic assay with the Klenow fragment of E. coli DNA polymerase I (Supporting Information) and a primer-template whose sequence corresponds to one in which dNaM-dTPT3 is only moderately retained in the SSO (Table 1). For comparison, dTTP is inserted opposite dA in this sequence context under the conditions employed with a kcat of 5.1 min−1 and a KM of 0.05 μM, resulting in an efficiency (second order rate constant or kcat/KM) of 1.2 × 108 M−1min−1, while dNaMTP is inserted opposite dTPT3 with a kcat of 10.7 min−1 and a KM of 0.09 μM, resulting in an efficiency of 1.3 × 108 M−1min−1.7 We found that the insertion of dPTMOTP opposite dTPT3 proceeded with a kcat of 19.4 min−1 and a KM of 0.08 μM, resulting in an efficiency of 2.9 × 108 M−1min−1. The insertion of dMTMOTP proceeded with a kcat of 8.5 min−1 and a KM of 0.22 μM, resulting in an efficiency of 4.2 × 107 M−1min−1. Thus, like dNaMTP, dPTMOTP is inserted opposite dTPT3 with an efficiency that is indistinguishable from that of a natural base pair, while dMTMOTP is inserted with a slightly reduced efficiency.
Table 1.
Steady state incorporation kinetic data
| 5’- d(ACAACTTTAACTCACACAATGTA) 3’-d(GAGCTCATGTTGAAATTGAGTGTGTTACAT-Y-TCTAGTGCCGTCTGTTTGTTTTCTTACCTTAG) | ||||
| Y | dXTP | kcat (min−1) | KM (μM) |
kcat/Km (M−1 min−1) |
| A | dTTP | 5.1 ± 0.7 | 0.05 ± 0.02 | 1.2 × 108 |
| TPT3 | dNaMTPa | 10.7 ± 0.4 | 0.09 ± 0.01 | 1.3 × 108 |
| dPTMOTP | 19.4 ± 1.2 | 0.08 ± 0.03 | 2.9 × 108 | |
| dMTMOTP | 8.5 ± 1.7 | 0.22 ± 0.10 | 4.2 × 107 | |
Reproduced from Morris et al.7
To further explore the kinetics of replication, we employed a pre-steady state assay using the same primer-templates (Supporting Information). In this case, reactions were supplemented with the unnatural triphosphate as well as the correct next natural triphosphate, and we characterized the percent of singly and doubly extended primer after 5 s (Table 2). The percent of single-nucleotide extended primers with dNaMTP, dPTMOTP, and dMTMOTP, was 35%, 41%, and 25%, while the percent of doubly-extended primers was 41%, 47%, and 53%. Thus, in agreement with the steady state data, the pre-steady state data suggests that compared to either dNaMTP or dPTMOTP, dMTMOTP is inserted slightly less efficiently; however it also suggests that the resulting dMTMO-dTPT3 UBP is extended slightly more efficiently than either dNaM-dTPT3 or dPTMO-dTPT3.
Table 2.
Pre-steady state kinetic data
| 5’- d(ACAACTTTAACTCACACAATGTA) 3’-d(GAGCTCATGTTGAAATTGAGTGTGTTACAT-TPT3-TCTAGTGCCGTCTGTTTGTTTTCTTACCTTAG) | ||
| dXTP | incorporation (%) | extension (%) |
| dNaMTP | 35 ± 8 | 41 ± 7 |
| dPTMOTP | 41 ± 6 | 47 ± 3 |
| dMTMOTP | 25 ± 4 | 52.8 ± 1.0 |
We next explored the PCR amplification of DNA containing a single UBP. Templates containing the dNaM-dTPT3 UBP were amplified with the natural triphosphates (200 μM each) as well as dTPT3TP and either dNaMTP, dPTMOTP or dMTMOTP (100 μM each) using OneTaq DNA polymerase. Two templates were examined (Table 3): T1, which embeds the UBP within a sequence that is well replicated in the SSO, and T2, which embeds it within a sequence that is poorly replicated in the SSO. DNA was amplified with an extension time of 1 min and monitored by qPCR trace. After maximum amplification was observed, the DNA was purified, and UBP retention was characterized via a second PCR amplification, using a mixture of OneTaq and DeepVent DNA polymerases and a biotinylated analog of dNaMTP followed by a gel mobility shift assay with or without streptavidin to quantify the UBP that was retained in the amplification product (Table 3; Supporting Information). With template T1, retention with dNaMTP was 100%, whereas the retention with dPTMOTP and dMTMOTP was 98% and 80%, respectively. With the more challenging sequence context of T2, retention with dNaMTP was 86%, whereas retention with dPTMOTP and dMTMOTP was 78% and 58%, respectively. Thus, the dNaM-dTPT3 UBP is retained at the highest level, followed closely by dPTMO-dTPT3, and dMTMO-dTPT3 is retained at a somewhat reduced level.
Table 3.
PCR amplification data
| T1: 5’-CTCGAGTACAACTTTAACTCACACAATGTCA-X-TGTCACGGCAGACAAACAAAAGAATGGAATC T2: 5’-CTCGAGTACAACTTTAACTCACACAATGTCC-X-GGTCACGGCAGACAAACAAAAGAATGGAATC | ||
| Sequence | dXTP | retention (%) |
| T1 | dNaMTP | 100 ± 2 |
| dPTMOTP | 98.4 ± 1.0 | |
| dMTMOTP | 80 ± 4 | |
| T2 | dNaMTP | 86 ± 20 |
| dPTMOTP | 78 ± 13 | |
| dMTMOTP | 58 ± 8 | |
While all previous optimization efforts have been based on structure-activity relationship (SAR) data generated in vitro, the availability of the SSO now allows, for first time, the use of in vivo SAR data in the discovery process. Indeed, we have found that in vitro results are not necessarily recapitulated in the in vivo environment,29 the milieu in which the UBPs must ultimately function. Thus, to evaluate the UBPs in the in vivo environment of the SSO, we constructed three derivatives of the pUC19 plasmid containing a single dNaM-dTPT3 UBP within the sequence contexts 1 – 3 (Table 4), in which UBP retention is increasingly difficult.13 Each plasmid was used to transform the SSO (strain YZ313), which was then allowed to recover briefly in media containing dTPT3TP and either dNaMTP, dPTMOTP, or dMTMOTP. After transfer to fresh media containing the same triphosphates and ampicillin (to select for plasmid retention), the SSO was allowed to grow to an OD600 of ~0.7, at which time plasmids were recovered and analyzed for UBP retention as described above for PCR products (Table 4; Supporting Information).
Table 4.
In vivo replication data
| Sequence Contexts 1–4 1: 5’-CTCGAGTACAACTTTAACTCACACAATGTCA-X-TGTCACGGCAGACAAACAAAAGAATGGAATC 2: 5’-CTCGAGTACAACTTTAACTCACACAATGTCC-X-CGTCACGGCAGACAAACAAAAGAATGGAATC 3: 5’-CTCGAGTACAACTTTAACTCACACAATGTCC-X-GGTCACGGCAGACAAACAAAAGAATGGAATC 4: 5’-CTCGAGTACAACTTTAACTCACACAATGTCC-X-AGTCACGGCAGACAAACAAAAGAATGGAATC | ||||||
| SSO | Sequence Context | dXTP | retention (%) | |||
| [dXTP] | ||||||
| 150 μM | 25 μM | 10 μM | 5 μM | |||
| YZ3 | 1 | dNaMTP | 98 ± 4 | 82 ± 15 | 29 ± 11 | 17 ± 21 |
| dPTMOTP | 98 ± 5 | 99 ± 0.5 | 92 ± 6 | 77 ± 22 | ||
| dMTMOTP | 99.0 ± 1.2 | 96 ± 5 | 80 ± 11 | 59 ± 24 | ||
| 2 | dNaMTP | 44 ± 15 | 7 ± 3 | –a | –a | |
| dPTMOTP | 97 ± 10 | 55 ± 14 | –a | –a | ||
| dMTMOTP | 62 ± 22 | 18.7 ± 0.6 | –a | –a | ||
| 3 | dNaMTP | 6 ± 3 | 4.8 ± 1.2 | –a | –a | |
| dPTMOTP | 7 ± 4 | 2 ± 4 | –a | –a | ||
| dMTMOTP | 7.9 ± 0.7 | 5 ± 4 | –a | –a | ||
| ML1 | 3 | dNaMTP | 28 ± 14 | 9 ± 2 | –a | –a |
| dPTMOTP | 60 ± 15 | 19 ± 6 | –a | –a | ||
| dMTMOTP | 25 ± 8 | 10 ± 4 | –a | –a | ||
| 4 | dNaMTP | 26 ± 18 | 11.0 ± 1.4 | –a | –a | |
| dPTMOTP | 41 ± 19 | 21±10 | –a | –a | ||
| dMTMOTP | 22 ± 18 | 12 ± 5 | –a | –as | ||
Not determined
For context 1 and dNaMTP at concentrations of 150 μM, 25 μM, 10 μM, or 5 μM, UBP retentions were 98%, 82%, 29% and 17%, respectively. For dPTMOTP at the same concentrations, retentions were 98%, 99%, 92% and 77%, respectively. For dMTMOTP, retentions were 99%, 96%, 80%, and 59%, respectively. For context 2, with dNaMTP at concentrations of 150 μM and 25 μM, UBP retentions were 44% and 7%, respectively. For dPTMOTP and dMTMOTP at the same concentrations, retentions were 97% and 55%, and 62% and 19%, respectively. Finally, for context 3 and dNaMTP at the same two concentrations, retentions were 6% and 5%, while for dPTMOTP and dMTMOTP, they were 7% and 2% and 8% and 5%, respectively. Thus, in stark contrast to the in vitro results, in all but sequence context 3, retention of both dPTMO-dTPT3 and dMTMO-dTPT3 was significantly higher than that of dNaM-dTPT3.
To further explore the utility of the new UBPs, we examined their retention in the optimized SSO ML1,22 wherein the gene encoding RecA has been deleted. We assessed retention in sequence context 3, as well as context 4 (Table 4), as these two contexts are the most challenging for UBP retention in the SSO.13 In context 3, dNaMTP supplied at a concentration of 150 μM or 25 μM resulted in a UBP retention of 28% and 9%, respectively. However, with dPTMOTP at the same concentrations, retention was 60% and 19% respectively, while with dMTMOTP it was 25% and 10%. In sequence 4, dNaMTP at a concentration of 150 μM or 25 μM resulted in a UBP retention of 26% and 11%, respectively. Addition of dPTMOTP at the same concentrations resulted in a retention of 41% and 21%, while the addition of dMTMOTP resulted in a retention of 22% and 12%. Thus, while the optimized SSO ML1 better retains each UBP, its use with the dPTMO-dTPT3 UBP results in a synergistic increase in retention.
The improved retention of dMTMO–dTPT3 and especially dPTMO–dTPT3 in the SSO might result from better PtNTT2–mediated import of the corresponding triphosphates or from better replication within the in vivo environment. To differentiate between these possibilities we characterized the uptake of dNaMTP, dMTMOTP, and dPTMOTP. Briefly, a culture of exponentially growing SSO strain YZ3 (YZ3 and ML1 both deploy the same PtNTT2 transporter system) was treated with varying concentrations of nucleoside triphosphate and incubated for 1 h at 37 °C. Cells were then pelleted and washed before extracting intracellular nucleotides with acidic acetonitrile.30 Shrimp alkaline phosphatase was added to degrade the nucleotides to their corresponding nucleosides, which were then quantified by LC-MS/MS, using external calibration curves for each nucleotide. Initial velocities were plotted against the concentration of triphosphate added to the media, and the resulting curves were fit to the Michaelis-Menten equation to determine the apparent second order rate constants for import (Vmax/KM).31 We found that dNaMTP is imported with a Vmax/KM of 5.37 ± 1.20 × 10−8 nL cell−1 hr−1, while dPTMOTP and dMTMOTP are imported with Vmax/KM values of 7.56 ± 1.9 × 10−8 and 1.37 ± 0.30 × 10−7 nL cell−1 hr−1, respectively. Thus, while dMTMOTP is imported with a 2-fold increased efficiency, dPTMOTP is imported with an efficiency that is virtually identical to dNaMTP, suggesting that its more optimal retention results from improved in vivo replication.
To begin to explore the origins of the improved replication, we modeled the structures of each UBP during replication. Previously, we reported the structure of a pre-insertion complex of KlenTaq DNA polymerase with d5SICSTP paired opposite dNaM in the template (PDB entry 3SV3),24 and the structure of a post-insertion complex in which d5SICS has been incorporated into the primer opposite dNaM (PDB entry 4C8L).25 As mentioned above, in the pre-insertion complex the UBP adopts a Watson-Crick-like structure, while in the post-insertion complex it forms a cross-strand intercalated structure. To generate a model to understand the current results, d5SICSTP was replaced with dTPT3TP and dNaM was retained or replaced with dMTMO or dPTMO. Geometric and electrostatic parameters for each nucleobase were calculated,32 and the initial structures were minimized for 500 cycles of steepest descent with the sander module of AMBER,33 at which point each had converged.
During minimization, only small changes were observed, and all of the nucleotide-polymerase interactions remained virtually unchanged. However, the structures did reveal specific interactions introduced by the thiophene sulfur atoms. In the pre-insertion complex with either dMTMO or dPTMO, the Watson-Crick-like structure is retained. With dPTMO, the sulfur is directed into the developing major groove where it is positioned in an open channel, which in the parent structure contains structural water; with dMTMO, the sulfur is buried by the protein (Figure 2A,B). Thus, the complex with dPTMO may be more stable, and this stability could contribute to its more efficient incorporation as a triphosphate relative to dMTMOTP. A similar stabilizing interaction is obviously absent with dNaMTP, but this may be offset by the increased hydrophobic and packing interactions afforded by its larger aromatic surface area.
Figure 2.
Model of dMTMOTP and dPTMOTP pre-insertion complexes (A and C, respectively) and post-insertion complexes (B and D, respectively). DNA backbone is shown as blue ribbon, oxygen, sulfur, and phosphorous (of the triphosphates in pre-insertion complexes) are colored red, orange, and yellow, respectively. In the pre-insertion complexes, only the UBP is shown, and in the post-insertion complex the UBP and the templating nucleotide are shown.
In the post-insertion complex, the sulfur of both dPTMO and dMTMO are in apparent van der Waals contact with a glycosidic nitrogen. Specifically, with dPTMO the sulfur is positioned 3.6 Å from the glycosidic nitrogen of dTPT3, and with dMTMO, it is positioned 3.4 Å from the glycosidic nitrogen of the templating dG (made possible by the previously described distortion of the template that results from interactions of the dG with the phosphate backbone of the primer terminus) (Figure 2 C,D). Based on the Watson-Crick-like insertion structure, the de-intercalated structures at the same post-insertion position should be free of such destabilizing interactions. Along with the reduced aromatic surface area, this would favor de-intercalation and thus optimize replication.
A detailed comparison with the kinetic data is premature, since in each modeled structure, dNaM, dPTMO, or dMTMO is within the template (experimental structures are not available for the other strand context), while the kinetic data was collected with these analogs in, or incorporated into the primer strand. The data is likely more interpretable in terms of the in vivo data, as replication in the SSO obviously requires replication in both strand contexts. While replication in the SSO is mediated by Pol III and Pol II,22 which are not homologous to KlenTaq, the noted interactions are within the primer/template and thus may make polymerase-independent contributions to replication. Thus, the modeling suggests that replication of the dPTMO-dTPT3 UBP is the best of the UBPs examined in the SSO due to the stability of the Watson-Crick-like structure during synthesis and facile de-intercalation once synthesized.
Finally, we explored whether the increased retention in the SSO of the UBPs with dPTMO and dMTMO results in the higher fidelity production of proteins containing ncAAs. To do this, we first integrated dNaM-dTPT3 into the gene encoding superfolder green fluorescent protein (sfGFP), replacing the native 151st codon (TAC) with the unnatural codon AXC (X = NaM). In addition, we integrated the UBP into the gene encoding Methanosarcina mazei tRNAPyl, replacing the anticodon with the unnatural anticodon GYT (Y = TPT3), which is selectively charged with N6-(2-azidoethoxy)-carbonyl-l-lysine (AzK) by the Methanosarcina barkeri pyrrolysyl-tRNA synthetase (PylRS). For comparison, we also constructed the analogous plasmid with the amber stop codon and the corresponding suppressor anticodon. Plasmids bearing both the sfGFP and tRNAPyl genes were then used to transform the SSO strain (YZ3), which also harbored a plasmid encoding PylRS. These transformants were then grown in the presence of dTPT3TP (10 μM) and dNaMTP, dPTMOTP, or dMTMOTP at different concentrations. After growth to an OD600 of 0.4–0.6, the culture medium was supplemented with NaMTP (250 μM), TPT3TP (30 μM), and AzK (10 mM), as well as isopropyl-β-D-thiogalactoside (IPTG; 1 mM) to induce T7 RNAP and tRNAPyl expression. After a brief induction period, expression of sfGFP was initiated by adding anhydrotetracycline (aTc, 100 ng/mL).
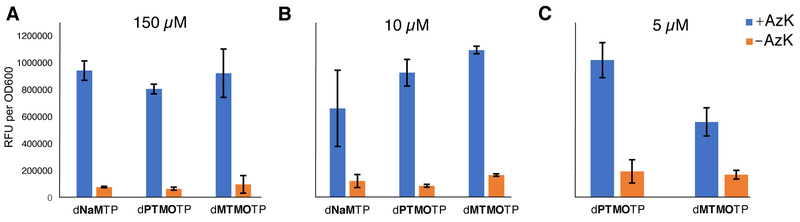
We first explored the level of UBP retention in the sfGFP and tRNAPyl genes as a function of unnatural triphosphate concentration (Table 5). At concentrations of 150 μM, the retentions were 99%, 97% and 92% in the sfGFP gene and 76%, 80% and 82% in the tRNA gene for dNaMTP, dPTMOTP, and dMTMOTP, respectively. At 10 μM the retentions were 73%, 95%, 93% and 71%, 82%, 74%. Finally, while the addition of dNaMTP at a concentration of 5 μM was too low to support cell growth, the addition of dPTMOTP or dMTMOTP at this concentration resulted in 90% or 64% retention in the sfGFP gene and 77% and 71% in the tRNA gene. Thus, as with the sequence contexts examined above, dPTMO-dTPT3 is replicated better than dMTMO-dTPT3, and both are replicated better than the parental dNaM-dTPT3.
Table 5.
UBP retention in the sfGFP and tRNAPyl genes
| Gene | dXTP | retention (%) | ||
|---|---|---|---|---|
| [dXTP] | ||||
| 150 μM | 10 μM | 5 μM | ||
| sfGFP | dNaMTP | 99.2±1.2 | 72.7±1.0 | –a |
| dPTMOTP | 97±2 | 95±3 | 90±5 | |
| dMTMOTP | 92±7 | 93±2 | 64±3 | |
| tRNAPyl | dNaMTP | 76±2 | 71±3 | –a |
| dPTMOTP | 80±2 | 82±6 | 77±3 | |
| dMTMOTP | 82.1±1.1 | 74±3 | 71±5 | |
Not determined
To monitor protein production, culture fluorescence was monitored after induction with IPTG (Figure 3). Cells that had been grown in the presence of dNaMTP, dPTMOTP or dMTMOTP at a concentration of 150 μM, exhibited little fluorescence in the absence of AzK, but significant fluorescence in the presence of AzK, similar to the control amber suppression cells. With cells that had been grown with 10 μM of dNaMTP or dMTMOTP, greater fluorescence was observed in the absence of AzK (relative to the corresponding 150 μM samples). However, fluorescence in the absence of AzK remained low in samples that had been grown in the presence of 10 μM dPTMOTP. Moreover, in the presence of AzK, less fluorescence was observed with cells provided with dNaMTP (again, relative to the corresponding 150 μM samples) than with cells provided with dMTMOTP or dPTMOTP. As mentioned above, cells provided with 5 μM dNaMTP were unable to grow. At this concentration, cells grown with dMTMOTP showed decreased fluorescence in the presence of AzK, while fluorescence in the absence of AzK remained the same (relative to the corresponding 10 μM sample). In contrast, with dPTMOTP, an increase in fluorescence was observed in the absence of AzK, and while in its presence similar levels of fluorescence were observed (relative to that observed with the addition of dPTMOTP at 150 or 10 μM). This data suggests that the higher fidelity replication of DNA containing the dPTMO-dTPT3 UBP results in higher fidelity ncAA incorporation.
Figure 3.
Fluorescence of cells expressing sfGFP with AzK encoded at position 151, in the presence or absence of AzK.
To directly characterize the protein produced, cells were harvested and lysed 2.5 h after mRNA induction, and the sfGFP produced was isolated and subjected to copper-free click reaction with dibenzocyclooctyl (DBCO) appended to a TAMRA dye, which produces a shift in electrophoretic migration, enabling quantification of the percentage of protein containing the ncAA (Figure 4). Using this assay, we found that virtually all of the control amber suppression cells produced sfGFP containing the ncAA. Cells that were transformed with the unnatural nucleotide-modified plasmid and grown in the presence of 150 μM dNaMTP, dPTMOTP, or dMTMOTP produced sfGFP that showed a 99%, 98%, and 95% shift. When grown in the presence of 10 μM dNaMTP, dPTMOTP, or dMTMOTP, the ncAA content of the protein was found to be 73%, 98% and 94%, respectively. Finally, in the presence of 5 μM unnatural triphosphate, 95% and 69% of protein purified from cells supplemented with dPTMOTP and dMTMOTP, respectively, contained the ncAA (again, as mentioned above, the SSO is unable to grow when supplied with dNaMTP at this concentration). This demonstrates not only that the DNA containing the dMTMO-dTPT3 or dPTMO-dTPT3 UBP is better replicated in the SSO than DNA containing dNaM-dTPT3, but also that both dPTMO and dMTMO can efficiently and faithfully template the transcription of cognate unnatural nucleotides into mRNA and tRNA.
Figure 4.
Western blot analysis of sfGFP with AzK encoded at position 151, purified from cells with AzK content determined by conjugation to TAMRA-DBCO, which results in reduced migration. Values shown below are the percent of the protein shifted, which reveals the percent that was produced with the ncAA.
CONCLUSION
The results of this study demonstrate that the dMTMO–dTPT3 and especially the dPTMO–dTPT3 UBPs are better retained in the DNA of the SSO than the previously most promising UBP, dNaM–dTPT3. The more optimal replication is only manifested within the in vivo environment of the SSO, and not within the in vitro environment employed with the kinetics and PCR assays. It seems likely that this results from differences in recognition by polymerase III and/or polymerase II, which are the polymerases that replicate DNA containing the UBP in vivo,22 relative to the Klenow fragment or OneTaq, the polymerases employed for the in vitro analysis, or from other components of the in vivo replisome (e.g. the β-clamp processivity factor) which are not present in vitro. Regardless, this data emphasizes the importance of using an in vivo environment when evaluating synthetic biology “parts,” as ultimately this is where they must function.
The modeling data suggest that the replication of both dMTMO-dTPT3 and dPTMO-dTPT3 is favored by specific inter-nucleobase interactions mediated by the sulfur of the thiophene moiety that optimize extension of the UBP by favorable packing with the primer nucleobase relative to cross-strand intercalation and packing with template nucleobases. The modeling also predicts that access to a major groove environment, and perhaps molecules of water therein, facilitates the insertion of dPTMOTP opposite dTPT3.
Importantly, the results also demonstrate that both dMTMO and dPTMO efficiently and faithfully template the transcription of mRNA containing TPT3 within the SSO, so that their increased retention during replication results in the higher fidelity production of proteins with the ncAA AzK. Finally, the optimized in vivo replication of dMTMO-dTPT3 and dPTMO-dTPT3 further demonstrates the ability of hydrophobic and packing interactions to replace complementary H-bonding as the force underlying information storage, and these UBPs appear to represent the most promising candidates identified to date. From a practical perspective, the increased range of sequences compatible with retention of dMTMO–dTPT3 and especially dPTMO-dTPT3 increases the potential unnatural information that may be stored in the SSO.
EXPERIMENTAL SECTION
General.
Synthetic details and compound characterizations are provided in Supporting Information. All anhydrous solvents were distilled and/or dried over 4 Å molecular sieves. All other chemical reagents were purchased from Aldrich, unless otherwise noted. 1H and 13C spectra were recorded on a Bruker DPX 400 mHz, Bruker DRX 500 mHz or Bruker AVIII HD 600 mHz NMR instrument. High-resolution mass spectroscopic data were obtained from the core facilities at The Scripps Research Institute.
All bacterial cultures were grown in liquid 2×YT media (casein peptone 16 g/L, yeast extract 10 g/L, NaCl 5 g/L) supplemented with potassium phosphate (50 mM, pH 7). All replication experiments in the SSO were conducted in 96-well microwell plates and translation experiments were conducted in 48-well microwell plates. When needed, antibiotics were used at the following concentrations: 5 μg/mL chloramphenicol, 100 μg/mL ampicillin, 100 μg/mL carbenicillin, 50 μg/mL zeocin. A PerkinElmer EnVision 2103 Multilabel Plate Reader was used to measure cell growth (OD600; 590/20 nm filter), as well as sfGFP fluorescence (485/14-nm excitation filter; 535/25-nm emission filter). Reagents for molecular biology were purchased from New England Biolabs (Ipswich, MA), unless otherwise stated, and were used according to the manufacturer’s protocols. Fully natural primers and oligonucleotides were purchased from Integrated DNA Technologies (San Diego, CA) and oligonucleotides containing unnatural nucleobases dNaM or dTPT3 were synthesized and purified using reverse phase cartridge by Biosearch Technologies (Petaluma, CA), gifted from Synthorx (La Jolla, CA). Plasmids were isolated using a ZR Plasmid Prep Kit (Zymo Research) according to the manufacturer’s recommendations. PCR amplifications were performed using a CFX Connect Real-Time PCR Detection System (Bio-Rad).
Gel-based Kinetics Assays.
Steady-state kinetic reactions7 and presteady-state kinetics reactions34 were performed as reported previously using the primer and template oligonucleotides listed in Table S1. Reaction products were analyzed using denaturing urea PAGE gel electrophoresis, followed by overnight exposure to a phosphor screen (Kodak) and imaging (Typhoon 9410, GE Amersham Biosciences). Image Studio Lite (Li-Cor Biosciences) was used to quantify primer and extended primer gel band intensities for determination of rates of incorporation at each triphosphate concentration. The data was fit to the Michaelis-Menten equation using R Studio (See Figure S1 for a representative plot).35 The reported values for kcat and KM are the averages and standard deviations of three, independent experiments.
PCR Assay.
Oligonucleotides containing dNaM (O4 and O5, see Table S2) were PCR amplified in the presence of SYBR Green (0.5×, Life Technologies), each dNTP (200 μM, GenDEPOT), dTPT3 (100 μM), each unnatural triphosphate of interest (100 μM), and the appropriate forward and reverse primers (1 μM of each P2 and P3, see Table S2) using OneTaq DNA Polymerase (additional details are provided in Supporting Information). Reactions were monitored by qPCR and then processed after maximum amplification (See Figure S2 for representative qPCR amplification curves). The remaining solution was purified by spin-column (DNA Clean and Concentrator-5; Zymo Research) then quantified by absorption at 260 nm using an Infinite M200 Pro Multimode Microplate Reader (Tecan). Percent retention of the UBP was measured using the streptavidin gel shift assay (as described below).
Streptavidin Gel Shift Assay.
PCR products, plasmid minipreps, Golden Gate assembled plasmids (0.5 ng to 2.5 ng), or dNaM-containing oligonucleotides (0.5 fmol) were PCR amplified in the presence of each dNTP (400 μM, GenDEPOT), d5SICSTP (65 μM), dMMO2BioTP (65 μM), and the appropriate forward and reverse primers (1 μM) using a mixture of OneTaq and DeepVent DNA polymerase (additional details are provided in the Supporting Information). Following amplification, 1 μL of each reaction was incubated for 5 min with streptavidin (2.5 μL, 2 μg/mL). Samples with and without streptavidin were mixed with loading buffer, separated on a polyacrylamide TBE gel, stained with SYBR Gold (1×, Thermo Fisher), and imaged using a Molecular Imager Gel Doc XR+ (Bio-Rad) equipped with a 520DF30 filter (Bio-Rad). Percent retention, as previously described and validated,12 can be calculated as a ratio of the signal of the shifted band to the total signal of both shifted and unshifted bands.
In Vivo Replication of the Unnatural Base Pair.
An overnight culture of E. coli strain YZ3 cells grown with chloramphenicol was used to prepare electrocompetent cells for transformation with 2 ng of a Golden Gate pINF plasmid containing the UBP, as described previously13 (see Supporting Information). Cells were transformed by electroporation and then recovered in the presence of dTPT3TP (37.5 μM) and varying concentrations of dNaMTP, dPTMOTP, or dMTMOTP for 1 h at 37 °C. An aliquot of the resulting culture was then used to inoculate growth media with the same triphosphates added and additionally supplemented with ampicillin. The cells were then monitored for growth and collected at an OD600 ~ 0.7 (See Figures S3–S7 for representative growth curves). Plasmids were isolated and then subjected to a streptavidin gel shift assay to determine UBP retention (Supporting Information). A separate aliquot of the culture following electroporation and recovery was used to determine transformation efficiency by plating dilutions onto solid 2×YT media supplemented with chloramphenicol and ampicillin and enumerating colonies after a 16 h incubation at 37 °C.
In Vivo Translation of the Unnatural Base Pair.
E. coli strain YZ3 bearing a pGEX plasmid carrying the sequence of the M. barkeri pyrrolysyl-tRNA synthetase (PylRS), prepared as described previously14 (see Supporting Information), was grown overnight in chloramphenicol and carbenicillin and used to prepare electrocompetent cells for transformation with ~0.5 ng of Golden Gate assembled plasmid containing the UBP embedded within the sfGFP and tRNAPyl genes14 (see Supporting Information). Cells were transformed via electroporation and then recovered in the presence of dNaMTP (150 μM) and dTPT3TP (10 μM) for 1 h at 37 °C. Dilutions of the resulting culture were plated onto solid media supplemented with zeocin, dNaMTP (150 μM), and dTPT3TP (10 μM) and allowed to grow overnight at 37 °C. Single colonies were used to inoculate media supplemented with zeocin, dNaMTP (150 μM), and dTPT3TP (10 μM), and monitored for cell growth. At an OD600 of ~0.7, an aliquot was subjected to plasmid isolation to determine which colonies retained the UBP well, and the remaining culture was stored in glycerol (25% v/v) at −80 °C.
Dilutions of the glycerol stock were then re-plated onto solid media supplemented with zeocin, dNaMTP (150 μM), and dTPT3TP (10 μM) and grown overnight at 37 °C. Single colonies were used to inoculate media supplemented with zeocin, dNaMTP, dPTMOTP, or dMTMOTP (at varying concentrations) and dTPT3TP (10 μM). Cells were monitored for growth and collected at OD600 ~ 0.7. All samples were then simultaneously diluted to OD600 ~ 0.1–0.2 in media supplemented with the same antibiotic and unnatural triphosphates. The remaining culture was pelleted and stored. At OD600 ~ 0.4–0.6, cells were supplemented with NaMTP (250 μM), TPT3TP (30 μM) and AzK (10 mM). From this step forward, cells were shielded from light to minimize photodegradation of AzK. Cells were grown for 20 min at 37 °C before adding IPTG (1 mM) to induce expression of T7 RNAP and tRNAPyl. Cells were then monitored for cell growth and fluorescence. After 1 h, sfGFP expression was induced with anhydrotetracycline (100 ng/mL) and cells were allowed to grow for 2.5 h before harvesting by centrifugation (230 μL for affinity purification of sfGFP and 50 μL for plasmid isolation and streptavidin gel shift assay, see Supporting Information). Cell pellets were then stored at −80 °C until further use.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health GM118178 to F.E.R.), the National Science Foundation Graduate Research Fellowship (DGE-1346837 to A.W.F.), and the Boehringer Ingelheim Fonds PhD Fellowship program (E.C.F.). We thank Synthorx Inc. for providing unnatural triphosphates and oligonucleotides.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Additional Methods, Representative Data, NMR spectra, Supporting Tables and Figures, Primer and Plasmid Sequences, Supporting References.
Notes
The authors declare the following competing financial interests: a patent application has been filed based on the use of UBPs in SSOs and F.E.R. has a financial interest (shares) in Synthorx, Inc., a company that has commercial interests in the UBP.
REFERENCES
- (1).Piccirilli JA; Krauch T; Moroney SE; Benner SA Nature 1990, 343, 33–37. [DOI] [PubMed] [Google Scholar]
- (2).Yang Z; Chen F; Alvarado JB; Benner SA J. Am. Chem. Soc 2011, 133, 15105–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Malyshev DA; Romesberg FE Angew. Chem. Int. Ed. Engl 2015, 54, 11930–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Feldman AW; Romesberg FE Acc. Chem. Res 2018, 51, 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hirao I; Kimoto M Proc. Jpn. Acad. Ser. B Phys. Biol. Sci 2012, 88, 345–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Moran S; Ren RX; Kool ET Proc. Natl. Acad. Sci. USA 1997, 94, 10506–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Morris SE; Feldman AW; Romesberg FE ACS Synth. Biol 2017, 6, 1834–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Malyshev DA; Dhami K; Quach HT; Lavergne T; Ordoukhanian P; Torkamani A; Romesberg FE Proc. Natl. Acad. Sci. USA 2012, 109, 12005–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Seo YJ; Hwang GT; Ordoukhanian P; Romesberg FE J. Am. Chem. Soc 2009, 131, 3246–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lavergne T; Lamichhane R; Malyshev DA; Li Z; Li L; Sperling E; Williamson JR; Millar DP; Romesberg FE ACS Chem. Biol 2016, 11, 1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ast M; Gruber A; Schmitz-Esser S; Neuhaus HE; Kroth PG; Horn M; Haferkamp I Proc. Natl. Acad. Sci. USA 2009, 106, 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Malyshev DA; Dhami K; Lavergne T; Chen T; Dai N; Foster JM; Correa IR Jr.; Romesberg FE Nature 2014, 509, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Zhang Y; Lamb BM; Feldman AW; Zhou AX; Lavergne T; Li L; Romesberg FE Proc. Natl. Acad. Sci. USA 2017, 114, 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhang Y; Ptacin JL; Fischer EC; Aerni HR; Caffaro CE; San Jose K; Feldman AW; Turner CR; Romesberg FE Nature 2017, 551, 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jahiruddin S; Datta A J Phys Chem B 2015, 119, 5839–5845. [DOI] [PubMed] [Google Scholar]
- (16).Galindo-Murillo R; Barroso-Flores J Phys. Chem. Chem. Phys 2017, 19, 10571–10580. [DOI] [PubMed] [Google Scholar]
- (17).Negi I; Kathuria P; Sharma P; Wetmore SD Phys. Chem. Chem. Phys 2017, 19, 16365–16374. [DOI] [PubMed] [Google Scholar]
- (18).Wang Q; Xie XY; Han J; Cui GJ Phys. Chem. B 2017, 121, 10467–10478. [DOI] [PubMed] [Google Scholar]
- (19).Guo WW; Zhang TS; Fang WH; Cui G Phys. Chem. Chem. Phys 2018, 20, 5067–5073. [DOI] [PubMed] [Google Scholar]
- (20).Jahiruddin S; Mandal N; Datta A Chemphyschem 2018, 19, 67–74. [DOI] [PubMed] [Google Scholar]
- (21).Bhattacharyya K; Datta A Chemistry 2017, 23, 11494–11498. [DOI] [PubMed] [Google Scholar]
- (22).Ledbetter MP; Karadeema RJ; Romesberg FE J. Am. Chem. Soc 2018, 140, 758–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Li L; Degardin M; Lavergne T; Malyshev DA; Dhami K; Ordoukhanian P; Romesberg FE J. Am. Chem. Soc 2014, 136, 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Betz K; Malyshev DA; Lavergne T; Welte W; Diederichs K; Dwyer TJ; Ordoukhanian P; Romesberg FE; Marx A Nat. Chem. Biol 2012, 8, 612–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Betz K; Malyshev DA; Lavergne T; Welte W; Diederichs K; Romesberg FE; Marx AJ Am. Chem. Soc 2013, 135, 18637–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Li Y; Korolev S; Waksman G EMBO J. 1998, 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Johnson SJ; Taylor JS; Beese LS Proc. Natl. Acad. Sci. U.S.A 2003, 100, 3895–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Doublie S; Sawaya MR; Ellenberger T Structure 1999, 7, R31–35. [DOI] [PubMed] [Google Scholar]
- (29).Feldman AW; Romesberg FE J. Am. Chem. Soc 2017, 139, 11427–11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Rabinowitz JD; Kimball E Anal. Chem 2007, 79, 6167–6173. [DOI] [PubMed] [Google Scholar]
- (31).Feldman AW; Fischer EC; Ledbetter MP; Liao J-Y; Chaput JC; Romesberg FE J. Am. Chem. Soc 2018, 140, 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Mennucci B; Petersson GA; Nakatsuji H; Caricato M; Li X; Hratchian HP; Izmaylov AF; Bloino J; Zheng G; Sonnenberg JL; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y, Nakai OK,H, Vreven T, Montgomery JA Jr., Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ Gaussian 09, Wallingford CT, 2009. [Google Scholar]
- (33).Case DA; Darden TA; Cheatham III TE; Simmerling CL; Wang J; Duke RE; Luo R; Walker RC; Zhang W; Merz KM; Roberts B; Hayik S; Roitberg A; Seabra G; Swails J; Götz AW; Kolossváry I; Wong KF; Paesani F; Vanicek J; Wolf RM; Liu J; Wu X; Brozell SR; Steinbrecher T; Gohlke H; Cai Q; Ye X; Wang J; Hsieh M-J; Cui G; Roe DR; Mathews DH; Seetin MG; Salomon-Ferrer R; Sagui C; Babin V; Luchko T; Gusarov S; Kovalenko A; Kollman PA AMBER 12 2012, University of California, San Francisco. [Google Scholar]
- (34).Lavergne T; Degardin M; Malyshev DA; Quach HT; Dhami K; Ordoukhanian P; Romesberg FE J. Am. Chem. Soc 2013, 135, 5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).R Core Team R: A language and environment for statistical computing, Vienna, Austria, 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.