Abstract
Background:
DNA ploidy analysis of cervical intraepithelial neoplasia (CIN) and invasive cervical cancer samples by flow cytometry (FCM) has been established as an aid to prognostic assessment. Liquid based cytology (LBC) increases diagnostic specificity by using ancillary techniques that provide information beyond morphology. The present study was undertaken to assess DNA ploidy in LBC samples as an adjunct for early detection of cervical pre-cancer and cancer.
Methods:
DNA ploidy assessment was performed on LBC samples of 50 cases and 31 controls. Cell pellets were obtained by centrifugation and stained with Telford reagent. At least 20,000 R1 gate (G0-G1) events were acquired on a BD FACSCalibur by using a 575±10 nm filter.
Results:
Mean diploid G1 values were lowered significantly (p<0.01) while diploid S values were significantly elevated (p<0.01) in both high grade squamous intraepithelial lesions (HSILs) and squamous cell carcinomas (SCCs) as compared to controls. Receiver operating curve (ROC) analysis of the diploid G1 value was found to have significant diagnostic potential (AUC=0.682, Z=2.00, p=0.046) for distinction between control and low grade squamous intraepithelial lesion (LSIL) at a cut off value of ≤91.6 with a sensitivity and specificity of 50.0 and 87.1%, respectively.
Conclusions:
ROC analysis of diploid G1 and diploid S values allows discrimination between LSIL and HSIL with sensitivities and specificities of 65 and 100% and 70 and100%, respectively, and between LSIL and SCC cases with values of 71.4 and 100% and 64.3 and 100%, respectively.
Keywords: Liquid Based Cytology, Flowcytometry, Low grade Squamous Intraepithelial Lesion, High grade Squamous
Introduction
Cervical cancer is the second most common cancer in women worldwide, representing global incidence of 5, 26,600 new cases each year and 2, 65,700 death, in that India contributes 67,500 i.e., one-fourth of the world burden (Torre et al., 2015). The malignancy targets an active female population between 30.0-49.9 yrs (Ferlay et al., 2012; WHO, 2012). Mortality in developing countries is unacceptably high and screening program are not in place (Soler et al., 2000). Liquid Based Cytology (LBC) provides the use of ancillary techniques in addition to a good morphology and detection of cytological abnormalities (Monsonego et al., 2001). Cervical epithelial cells fixed in LBC and stained with fluorescent dye can be acquired by flowcytometer for DNA ploidy measurement. Analysis of cells within a cell cycle on flowcytometer produces a histogram which represents DNA ploidy. DNA ploidy assessment by flow cytometric technique could serve as a prognostic factor that allows the estimation of the relative progressive risk into more advanced lesions (Melsheimer et al., 2004). Cells are distributed among three major phases of cell cycle: G0/G1 phase which comprises 85% cells, S phase and G2/M phase which make 15% of the cells in a normal tissue and in majority of low grade or gradually proliferating lesions. DNA aneuploidy in S phase may indicate development of intermediate to late stages of the malignancies (Merkel and McGuire 1990; Ross, 1996). Precancer state results in genetic instability due to unrestricted growth of cancer cells by subsequent mutations in growing pool of proliferating cells. Structural and numerical change may occur in chromosomes resulting in overall change in the DNA content of cells, an event referred to as aneuploidy. DNA ploidy assessment has been established as a prognostic factor in ovarian (Vergote et al., 1993; Kaern et al., 1994) and endometrial cancer (Erba et al., 1989; Evans and Podratz,1996), however in cervical cancer there are conflicting results (Jakobsen, 1984; Willen et al., 1993; Podratz et al., 1993). Molecular basis of aneuploidy remain undefined and divisive (Marx, 2002) with an assumption that mitotic proteins play role in chromosomal instability (Lengauer et al., 1998; Rajagopalan et al., 2003). Another assumption proposes that aneuploidy is itself the cause of genetic instability and cancer (Duesberg et al., 1988; Li et al., 2000). Aneuploid DNA profiles are useful indicators for the biologic aggressiveness of cervical cancer lesions than those of diploid or polyploid profiles (Fu et al., 1989). In this study, we have used light scatter characteristics of cells to evaluate the relationship between DNA ploidy and S phase fraction and correlated with clinicopathological parameters in cervical pre cancer and cancer.
Materials and Methods
Patient samples and procedures
The study sample comprised of 50 cases and 31 controls from the Department of Obstetrics and Gynaecology, Queen Mary’s Hospital, King George’s Medical University and Dr. Ram Manohar Lohia Combined Hospital, Lucknow, India. Samples were collected during cervical screening of women in hospital based setting. All participants signed an informed consent and ethical approval was obtained from Institutional Ethics Committee before recruiting patients. All cases that fulfilled the inclusion criteria that is, females with positive findings on colposcopy and/or visual examination of cervix were recruited for the study. Participants were excluded from the study if they were not willing to participate or cases with malignancy under follow up/ prior therapy. Total of 90 cases were screened of which 81 were included in the study excluding 9 cases with low epithelial cell count.
Cytology
Samples were collected by gynaecologist in Thin prep (TP) vials (Hologic, Inc. USA) containing 20 ml PreservCyt (Thin Prep, Hologic, Boxborough, MA. Cytobrush (RoversR Cervex-BrushR) was simply inserted into the endocervical canal deep enough to allow the shorter bristle to fully contact the ectocervix (keeping the last row of bristles visible). The cytobrush was slowly rotated clockwise to 180 degrees for five times avoiding excessive rotation which would distort the cell. The brush was rinsed into the PreservCyt vial by pushing the broom in the bottom of the vial 10 times, forcing the bristle apart. Smears were prepared on Thinprep 2000 processor (CYTYC Corporation) and cytologic screening was done by conventional Pap staining. All slides were evaluated by a experienced pathologist and diagnosis was made using the Bethesda System (2014). Cases with normal cytology were used as a control for DNA ploidy. Samples were kept at 4°C and FCM was performed within 1-3 weeks of collection.
Flow cytometric analysis
Selection of Cases: Cases in serial showing LSIL, HSIL and SCC as per Bethesda criteria with adequate cells count (100,000 cells) were included for ploidy study.
Sample preparation: 5-10 ml of fixed cells (fixed in PreservCyt solution) were taken in a 15 ml conical centrifuge tube, cell pellet was obtained by centrifugation at 2,000 rpm for 5 min. followed by washing of cell pellet with phosphate buffer saline (PBS, pH-7.4), three times or more depending on debris content in samples. Epithelial cell counting was performed on Neubauer chamber. Minimum epithelial cell count required for staining and acquisition was 1x105 cells.
Staining and acquisition
Cells were stained with Telford reagent (Telford et al., 1991){(EDTA (Fischer Scientific, USA), RNAse A 1mg/ml, (Biochem India), Propidium Iodide 1mg/ml, (Sigma-Aldrich, Miss, USA), Triton X-100, (Sigma-Aldrich, Miss, USA) in PBS)} and incubated in dark at 4°C for 1 hour. Stained cells were filtered by 70µ cell strainer (B.D. Biosciences, USA) and cells were acquired within 1 hour on a flowcytometer (FACSCalibur: Becton, Dickinson and Co.; San Jose, CA) equipped with a 488-nm blue laser. Acquisition and analysis were performed on Cell Quest Pro software (B.D. Biosciences, USA).
At least 20,000 events were acquired in R1 gate drawn around the epithelial cells based on light scatter properties of cells. As shown in Figure 1 (A-B), and Figure 2 (A-B), two dot plot of FSC vs. SSC and FL2-W vs. FL2-A and one histogram of FL-2A vs. Count (Figure 1-C, 2-C) were utilised for acquisition. Pulse processing was used to exclude cell doublets from the analysis. All measurement was performed at low flow rate. The cell cycle profiling of all samples were done in unchanged background using same instrument and by the same observer. Cell populations were properly gated and the number of cells was counted in order to evaluate the eligibility of the specimen for the purpose of precise DNA content analysis. Verification of instrument performance in term of coefficient of variation (CV) and linearity of the fluorescence pulse detectors was checked by DNA QC Particle kit (B.D. Biosciences, Singapore).
Figure 1.
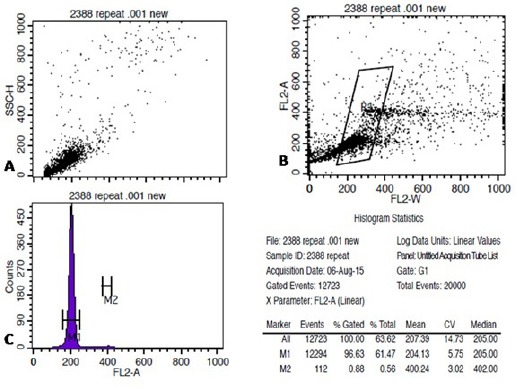
Shows the SAcquisition of Cervical Epithelial Cells on Flowcytometer, Stained with Telford Reagent. A-C shows acquisition of stained cells on FSC vs. SSC, FL2-A vs. FL2-W and FL2-A vs. Count on Cell Quest Pro software(B.D Biosciences, Singapore).
Figure 1.b.

Shows the Analysis of Acquired FCS File on ModFit LT 3.2 (Verity Software House). Based on ModFit analysis case was found to be of Diploid with single G0/G1 peak. Histogram statistics shown on top right.
Figure 2.
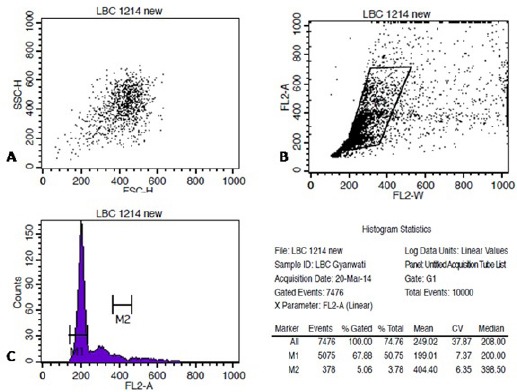
Shows an Aneuploid Case of HSIL on Cytomorphology Acquired on Flowcytometer, Stained with Telford Reagent. A-C shows acquisition of stained cells on FSC vs. SSC, FL2-A vs. FL2-W and FL2-A vs. Count on Cell Quest Pro software (B.D Biosciences, Singapore).
Cell cycle analysis and ploidy
Data obtained was analyzed using the ModFit LT software (DNA Modelling System) version 3.2 (Verity Software House, Inc Topsham, ME). Each histogram was assessed by the estimation of the CV for G0/G1 population. Histogram was considered unsuitable for interpretation if its variation coefficient CV (breadth of the fluorescence signal for normal cell population in G0/G1 phase at the middle of its height) exceeded 9% (Cufer et al., 1997). A sample was considered DNA diploid if, on the histogram, there was a single peak in the G0/G1 phase with DNA content of 2N (Figure 1.b). DNA aneuploidy also known as non diploidy was defined if there was at least one separate second G0/G1 population to the right of the first G0/G1 peak (Figure 2.b). The ploidy was characterized by the DNA index (DI) and the type of DNA content histogram (Diploid: DI= 1, Aneuploid: DI<or >1). For each samples, the percentage distribution were recorded for diploid G1, G2, and diploid S phase, total aneuploid, including aneuploid G1, aneuploid G2, aneuploid S phase, percentage of C.V of diploid population and aneuploid population (if present), DNA index (DI), total aneuploid S phase, total S phase, B.A.D (percentage of background aggregates and debris), debris, aggregates, modeled events, all cycle events, cycle events per channel and Reduced chi-square (RCS-measure of how well the model fitted the data) were estimated.
Figure 2.b.

Shows the Analysis of Acquired FCS File on ModFit LT 3.2 (Verity Software House). Based on ModFit analysis case was found to be Aneuploid on appearance of second G0/G1 population to the right of first G0/G1 peak with DNA index of 1.49.
Statistical analysis
Data were summarized as Mean ± SE (standard error of the mean). Groups were compared by Mann-Whitney U test. Groups were also compared by Kruskal-Wallis (H) analysis of variance (ANOVA) followed by Z test. Diagnostic (sensitivity and specificity) of the cell cycle parameters was done by using the cut off values obtained from receiver operating characteristics (ROC) curve analysis. A two-tailed p<0.05 was considered statistically significant.
Results
DNA ploidy assessment was performed on 50 cases including 10 of Low grade Squamous Intraepithelial Lesion(LSIL), 20 of High grade Squamous Intraepithelial Lesion (HSIL) and 20 of Squamous Cell Carcinoma (SCC) and 31 controls with cervical cytology within normal limits. Analysis was possible in all cases and controls except for 6 cases of SCC where acquisition and analysis was not possible due very low cellularity. The mean (±SD) age of cases was 49.3(±10.58), ranged from 25 to 70 yrs. DNA aneuploidy was observed in 0/10 cases of LSIL, 10/20 cases (50%) of HSIL and 5/14 cases (34.5 %) of SCC. None of controls shows aneuploidy. Diploid LSIL, HSIL and SCC cases had mean CV of 7.18, 7.25 and 7.18 respectively.
Value of cell cycle parameters of Control and Cases: The cell cycle parameters (diploid G1, diploid G2 and diploid S phase) values of control, LSIL, HSIL and SCC are summarized in Table 1. Table 1 shows that the mean diploid G1 value decreases with degree of severity while S Phase Fraction increase with increases in severity. However, diploid G2 value did not show any trend with the severity. Comparing each diploid cell cycle parameter values among four groups, Kruskal-Wallis ANOVA showed significantly (p<0.001) difference in values of both diploid G1 and diploid S phase (Table 1). Furthermore, Z test showed that the mean diploid G1 value was significantly lower (p<0.01) in both HSIL and SCC groups as compared to control group while diploid S value were significantly (p<0.01) higher in both HSIL and SCC groups as compared to control group (Table 2). However, mean diploid G2 value did not differ significantly (p>0.05) among the groups (Table 1 and 2).
Table 1.
Diploid Cell Cycle Parameter Values (Mean ± SE) of Four Groups
| Parameters | Control (n=31) | LSIL (n=10) | HSIL (n=20) | SCC (n=14) | H value | p value |
|---|---|---|---|---|---|---|
| Dip G1 | 95.32 ± 0.52 | 92.64 ± 1.34 | 76.35 ± 4.35 | 73.88 ± 5.82 | 22.60 | <0.001 |
| Dip G2 | 0.20 ± 0.08 | 0.36 ± 0.20 | 0.13 ± 0.10 | 0.89 ± 0.61 | 4.69 | 0.196 |
| Dip S | 4.63 ± 0.47 | 6.98 ± 1.26 | 24.12 ± 4.28 | 25.15 ± 6.01 | 20.44 | <0.001 |
Table 2.
Comparison of Each Diploid Cell Cycle Parameter Value between Groups by Z Test
| Comparisons | Dip G1 | Dip G2 | Dip S | |||
|---|---|---|---|---|---|---|
| Z value | p value | Z value | p value | Z value | p value | |
| Control vs. LSIL | 1.17 | 1.000 | 0.55 | 1.000 | 1.06 | 1.000 |
| Control vs. HSIL | 3.89 | 0.001 | 1.47 | 0.846 | 3.92 | 0.001 |
| Control vs. SCC | 3.87 | 0.001 | 0.41 | 1.000 | 3.41 | 0.004 |
| LSIL vs. HSIL | 1.78 | 0.45 | 1.61 | 0.643 | 1.91 | 0.334 |
| LSIL vs. SCC | 1.98 | 0.287 | 0.81 | 1.000 | 1.72 | 0.511 |
| HSIL vs. SCC | 0.37 | 1.000 | 0.83 | 1.000 | 0.08 | 1.000 |
Assessment of Diploid cell cycle parameter between groups
Comparing the mean value of each cell cycle parameter between two groups, Mann-Whitney U test also showed significantly different and lower (16.8%) diploid G1 value in cases as compared to controls (p<0.001) while significantly different and higher (77.5%) diploid S value in cases as compared to controls (p<0.001). However, mean diploid G2 value not show statistical difference between the two groups, though it was 53.5% higher in cases as compared to control (p=0.423).
Comparison of aneuploid cell cycle parameter between groups
Aneuploidy was found in 50% of HSIL and 34.5% of SCC cases only. The aneuploid cell cycle parameter (An G1, An G2, An S, DI and Total S phase) values are summarized in Table 3. The mean value of An G1, An G2 and DI was higher while An S and Total S phase was lower in SCC as compared to HSIL. Comparing mean value of each aneuploid cell cycle parameter between two groups, Mann-Whitney U test showed significantly different and higher DI value in SCC as compared to HSIL (p=0.027). However, mean value of An G1, An G2, An S and Total S phase was not different (p>0.05).
Table 3.
Aneuploid Cell Cycle Parameter Values (Mean ± SE) of Two Groups
| Parameters | HSIL (n=10) | SCC (n=5) | U Value | p Value |
|---|---|---|---|---|
| An G1 | 67.53 ± 8.81 | 77.40 ± 8.15 | 23 | 0.806 |
| An G2 | 1.95 ± 0.92 | 2.09 ± 0.85 | 20 | 0.54 |
| An S | 30.51 ± 9.04 | 20.49 ± 7.69 | 22 | 0.713 |
| DI | 1.22 ± 0.04 | 1.45 ± 0.12 | 7 | 0.027 |
| Total S phase | 16.04 ± 3.53 | 15.87 ± 11.12 | 15 | 0.221 |
Diagnostic value of cell cycle parameters- Receiver Operating Characteristics (ROC) curve analysis
ROC curve analysis was performed to evaluate diagnostic sensitivity and specificity of diploid cell cycle parameters values to discriminate the cases of LSIL, HSIL and SCC from control is depicted in Table 4.
Table 4.
Diagnostic of Diploid Cell Cycle Parameter Values to Discriminate Cases from Control
| Diagnostic | Parameter | Cutoff value (%) | Sensitivity (95% CI) | Specificity (95% CI) | PPV | NPV | AUC | Z value | p value |
|---|---|---|---|---|---|---|---|---|---|
| Control vs. LSIL | Dip G1 | ≤91.6 | 50 (18.9-81.1) | 87.10 (70.1-96.3) | 55.6 | 84.4 | 0.682 | 2.00 | 0.046 |
| Dip G2 | >0.17 | 50 (18.9-81.1) | 74.19 (55.4-88.1) | 38.5 | 82.1 | 0.566 | 0.62 | 0.538 | |
| Dip S | >7.81 | 50 (18.9-81.1) | 87.10 (70.1-96.3) | 55.6 | 84.4 | 0.673 | 1.66 | 0.098 | |
| Control vs. HSIL | Dip G1 | ≤81.49 | 65 (40.8-84.5) | 100.00 (88.7-100) | 100.0 | 81.6 | 0.792 | 4.69 | <0.001 |
| Dip G2 | ≤0 | 85 (62.1-96.6) | 41.94 (24.6-60.9) | 48.6 | 81.3 | 0.628 | 1.63 | 0.104 | |
| Dip S | >8.93 | 70 (45.7-88.0) | 100.00 (88.7-100) | 100.0 | 83.8 | 0.802 | 4.50 | <0.001 | |
| Control vs. SCC | Dip G1 | ≤90.14 | 78.57 (49.2-95.1) | 96.77 (83.2-99.5) | 91.7 | 90.9 | 0.866 | 6.90 | <0.001 |
| Dip G2 | >2.07 | 14.29 (2.2-42.8) | 100.00 (88.7-100) | 100.0 | 72.1 | 0.464 | 0.39 | 0.701 | |
| Dip S | >8.93 | 71.43 (41.9-91.4) | 100.00 (88.7-100) | 100.0 | 88.6 | 0.811 | 4.08 | <0.001 | |
| Control vs. Cases (LSIL +HSIL + SCC) | Dip G1 | ≤88.48 | 59.09 (43.3-73.7) | 100.00 (88.7-100) | 100.0 | 63.3 | 0.791 | 5.26 | <0.001 |
| Dip G2 | ≤0 | 72.73 (57.2-85.0) | 41.94 (24.6-60.9) | 64.0 | 52.0 | 0.555 | 0.80 | 0.422 | |
| Dip S | >8.93 | 61.36 (45.5-75.6) | 100.00 (88.7-100) | 100 | 64.6 | 0.775 | 5.19 | <0.001 |
The cut off value of diploid G1 at ≤ 91.6 discriminate control vs. LSIL significantly with 50.00% sensitivity (95% CI: 18.9-81.1) and 87.10 specificity (95% CI: 70.1-96.3).
Diploid G1 value at a cut off of ≤ 81.49 showed significant (p< 0.001) diagnostic discrimination between control vs. HSIL (65.00% sensitivity and100%specificity) and control vs. SCC at a cutoff value of ≤ 90.14 (96.77% sensitivity and 78.57% specificity). Diploid G1 value at a cut off of ≤ 88.48 significantly (p< 0.001) discriminate control vs. cases (LSIL + HSIL + SCC) with sensitivity and specificity of 59.09 and 100% respectively (AUC= 0.775). Likewise, diploid S showed significant (p<0.001) diagnostic between control vs. HSIL at a cutoff of >8.93 (65.00% sensitivity and100% specificity) and control vs. SCC at cutoff point of >8.93 (96.77% sensitivity and 78.57% specificity). Diploid S value significantly discriminate control vs. cases (LSIL + HSIL + SCC) with sensitivity and specificity of 59.09 and 100% respectively (AUC= 0.775).
The diagnostic sensitivity and specificity of diploid cell cycle parameter between cases (LSIL, HSIL and SCC) are summarized in Table 5. Diploid G1 and Diploid S values significantly (p<0.05 or p<0.01) discriminates between LSIL vs. HSIL and LSIL vs. SCC. However, none of the Diploid cell cycle parameter could significantly (p>0.05) discriminates between HSIL and SCC cases.
Table 5.
Diagnostic of Diploid Cell Cycle Parameter Values to Discriminate Cases Among Cases
| Diagnostic | Parameter | Cutoff value (%) | Sensitivity (95% CI) | Specificity (95% CI) | +PV | -PV | AUC | Z value | p value |
|---|---|---|---|---|---|---|---|---|---|
| LSIL vs. HSIL | Dip G1 | ≤ 81.49 | 65.00 (40.8-84.5) | 100.00 (69.0-100) | 100.0 | 58.8 | 0.742 | 2.37 | 0.018 |
| Dip G2 | ≤ 0.02 | 90.00 (68.3-98.5) | 50.00 (18.9-81.1) | 78.3 | 71.4 | 0.677 | 1.63 | 0.103 | |
| Dip S | > 12.4 | 70.00 (45.7-88.0) | 100.00 (69.0-100) | 100.0 | 62.5 | 0.78 | 3.3 | 0.001 | |
| LSIL vs. SCC | Dip G1 | ≤ 85.26 | 71.43 (41.9-91.4) | 100.00 (69.0-100) | 100.0 | 71.4 | 0.807 | 3.21 | 0.001 |
| Dip G2 | ≤ 0.17 | 78.57 (49.2-95.1) | 50 (18.9-81.1) | 68.8 | 62.5 | 0.586 | 0.71 | 0.479 | |
| Dip S | > 12.4 | 64.29 (35.2-87.1) | 100.00 (69.0-100.) | 100.0 | 66.7 | 0.75 | 2.50 | 0.012 | |
| HSIL vs. SCC | Dip G1 | > 75.24 | 71.43 (41.9-91.4) | 55 (31.6-76.9) | 52.6 | 73.3 | 0.504 | 0.04 | 0.972 |
| Dip G2 | > 0.02 | 28.57 (8.6-58.1) | 90 (68.3-98.5) | 66.7 | 64.3 | 0.575 | 0.74 | 0.46 | |
| Dip S | ≤ 23.46 | 71.43 (41.9-91.4) | 55 (31.6-76.9) | 52.6 | 73.3 | 0.518 | 0.18 | 0.861 |
Discussion
The current study attempted to assess the analysis of DNA ploidy by flowcytometry on LBC sample as an ancillary test to screen women for preneoplastic lesions. A suspect DNA profile with high S phase value seems to be the clue for developing malignant disease of the uterine cervix. Automation of DNA measurement with flowcytometry through a single cell suspension could be useful to identify patients at risk for developing preneoplastic (LSIL, HSIL) or neoplastic lesions (SCC). More precise discrimination and prediction of malignancy is possible with quantitative analysis of threshold value for cell cycle parameters i.e. G0/G1 (Diploid), Diploid G2, Coefficient of Variation (CV), total S phase and total aneuploidy, aneuploid S phase, DNA index (DI), and RCS as also estimated in all other studies (Chhavi et al., 2010). Cytometric techniques can serve as a marker to detect neoplasia and can provide adjunct information to pathological evaluation for the diagnosis of cervical cancer. DNA aneuploidy along with S Phase Fraction (SPF) analysis can be used to identify the dysplasia and give a predictive value for the malignancy transformation.
In our study diploid G1 and diploid S phase values were significantly higher in HSIL and SCC compared to controls. These results could not significantly differentiate low grade lesions from high grade lesions (LSIL vs. HSIL, LSIL vs. SCC and HSIL vs. SCC). ROC curve analysis of the above cell cycle parameters can more accurately differentiate and predict the malignancy beyond morphology. Though, in our study value of diploid G1 and S phase were most sensitive parameters, with cut off point at ≤91.6, ≤81.49 and ≤90.14 (dip G1) and >8.93 (dip S) significantly differentiate low grade and high grade lesions from controls. Although; diploid S values was not able to differentiate LSIL cases from controls at Cut off point >7.81. Further; ROC analysis also significantly differentiate between LSIL and HSIL by estimation of diploid G1 and diploid S value; this could potentially help us in monitoring the progression of precancerous lesions to cancerous lesions. We have reached the 100% specificity in discrimination of LSIL cases from HSIL when analysed for dip S value, suggesting significance of S phase analysis in LBC of cervical premalignant lesion. Cervical lesions with higher SPF are more likely to persist or progress than normal diploid or with low SPF. Our result could support the hypothesis that higher diploid S value and lower diploid G1 may be associated with progression of cervical carcinoma. Study of Chavi et al. in LBC of cervical cancer cases, found the sensitivity and specificity of diploid G0/G1 to be 96.77% and 100% in discrimination of cases from controls. In contrast to this finding, study by Singh (2000) shows that Aneuploidy was present in 39/79 of mild, 28/36 of moderate, 11/12 of severe dysplasia, 8/57 of ASCUS and in 6/69 controls (Singh et al., 2000). Patient with low S phase fraction often shows better survival compared to the patients with high SPF (Jayat and Ratinaud, 1993). Other studies also supported diploidy as suggestive feature for better survival when compared with aneuploid tumor (Lai et al., 1993).
Many investigators have also examined the prognostic value of cytophotometric DNA content analysis in cervical carcinoma (Fu et al., 1982; Strang et al., 1987a; Strang et al., 1987b; Jelen et al., 1994; Kristensen et al., 1995). However, significance of DNA analysis as a reliable diagnostic and prognostic biomarker in LBC of cervical carcinoma still remains controversial. In invasive cervical carcinoma, DNA ploidy by image cytometry is a prognostic factor to estimate the risk of progression (Antonet al., 1997; Melsheimer et al., 2001; Hornet al., 2002; Melsheimer et al., 2004; Demirel et al., 2013). DNA ploidy analysis of ovarian tumor has been proved as prognostic indicator of disease progression (Demirel et al., 2013). In a study of Endometrial cancer of stage I patients (n=139) by flowcytometry reported that 86% of cases were diploid while 14% were aneuploid. Studies reported aneuploidy in CIN 1 and in normal squamous epithelium by image cytometry on larger sample size (Haroskeet al., 2001).
In our study aneuploidy was found in 50% of HSIL and 35.7 % of SCC cases. This loss of aneuploidy in SCC cases may be due to low count of cervical epithelial cells in our samples due to bleeding at time of sample collection or tumor necrosis. Study carried out by Lage et al. have proven aneuploidy as a predictor of survival and was significantly associated with stage and grade of ovarian cancer. Patient groups with higher SPF seem to die earlier (Lage et al., 1992)
Image cytometry have proved the diagnostic and prognostic value of DNA ploidy; however this technique is relatively slow, may be insensitive for near-diploid aneuploid cell with high coefficient of variation for G0/G1 peaks. Analysis of image cytometry requires a skilled morphologist. Flow cytometry has advantage over image cytometry and provides high-speed, fast results with high number of cells counted. High number of cell counted on flowcytometry enhances sensitivity for near-diploid aneuploid peaks. However, during DNA ploidy analysis by flowcytometry simultaneous morphologic comparison of cells is not possible. The disadvantage of flow cytometry is the requirement for single particles (cells or nuclei) which can lengthen the procedure in order to obtain single –cell suspension for solid tissues or fluid with lot of aggregates/debris.
Cellular DNA content analysis measured by flowcytometry could find prospective benefits on differentially high and low grade CIN, prognostically disease monitoring. Our results could suggest the potential use of SPF analysis as a valuable complement to standard clinical practice. This could be used as companion to cytopathological analysis of precancerous and cancerous lesions.
Acknowledgements
Authors wish to acknowledge and thanks Council of Science & Technology, Uttar Pradesh (CST, UP) for providing grant and Integral University, Lucknow for providing PhD registration to Mr. Sridhar Mishra.
Conflict of interest statement: Authors wish to state that there is no conflict of interest.
References
- Anton M, Nenutil R, Rejthar A, et al. DNA flow cytometry: a predictor of a high-risk group in cervical cancer. Cancer Detect Prev. 1997;21:242–46. [PubMed] [Google Scholar]
- Cufer T, Lamovec J, Bracko M, Lindtner J, US-Krasovec M. Prognostic value of DNA ploidy in breast cancer Stage I-II. Neoplasma. 1997;44:127–32. [PubMed] [Google Scholar]
- Chhavi S, Mona S, Negi MPS, et al. DNA content can improve the detection and prognosis of carcinoma of the cervix. Bio Science Trends. 2010;4:103–9. [PubMed] [Google Scholar]
- Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of Aneuploidy. Proc Natl Acad Sci U S A. 1988;95:13692–697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirel D, Akyürek N, Ramzy I. Diagnostic and prognostic significance of image cytometric DNA ploidy measurement in cytological samples of cervical squamous intraepithelial lesions. Cancer Cytopathol. 2013;24:105–12. doi: 10.1111/cyt.12039. [DOI] [PubMed] [Google Scholar]
- Erba E, Ubezio P, Pepe S, et al. Flow cytometric analysis of DNA content in human ovarian cancers. Br J Cancer. 1989;60:45–50. doi: 10.1038/bjc.1989.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MP, Podratz KC. Endometrial neoplasia: prognostic significance of ploidy status. Clin Obstet Gynecol. 1996;39:696–06. doi: 10.1097/00003081-199609000-00017. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2012;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Fu YS, Cheng L, Huang GI, et al. DNA ploidy analysis of cervical condyloma and intraepithelial neoplasia in specimens obtained by punch biopsy. Anal Quant Cytol Histol. 1989;11:187–95. [PubMed] [Google Scholar]
- Fu YS, Reagan JW, Hsiu JG, Storaasli JP, Wentz WB. Adenocarcinoma and mixed carcinoma of the uterine cervix. I. A. clinicopathological study. Cancer. 1982;49:2560–70. doi: 10.1002/1097-0142(19820615)49:12<2560::aid-cncr2820491225>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Horn LC, Raptis G, Nenning H. DNA cytometric analysis of surgically treated squamous cell cancer of the uterine cervix, stage pT1b1-pT2b. Anal Quant Cytol Histol. 2002;24:23–9. [PubMed] [Google Scholar]
- Haroske G, Baak JP, Danielsen H, et al. Fourth updated ESACP consensus report on diagnostic DNA image cytometry. Anal Cell Pathol. 2001;23:89–5. doi: 10.1155/2001/657642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen A. Ploidy level and short-time prognosis of early cervix cancer. Radiother Oncol. 1984;1:271–75. doi: 10.1016/s0167-8140(84)80010-9. [DOI] [PubMed] [Google Scholar]
- Jayat C, Ratinaud MH. Cell cycle analysis by flowcytometry: Principal and application. Biol cell. 1993;78:15–25. doi: 10.1016/0248-4900(93)90110-z. [DOI] [PubMed] [Google Scholar]
- Jelen I, Valente PT, Gautreaux L, Clark GM. Deoxyribonucleic acid ploidy and S-phase fraction are not significant prognostic factors for patients with cervical cancer. Am J Obstet Gynecol. 1994;171:1511–16. doi: 10.1016/0002-9378(94)90394-8. [DOI] [PubMed] [Google Scholar]
- Kaern J, Trope CG, Kristensen GB, et al. Evaluation of deoxyribonucleic acid ploidy and S-phase fraction as prognostic parameters in advanced epithelial ovarian carcinoma: a prospective study. Am J Obstet Gynecol. 1994;170:479–87. doi: 10.1016/s0002-9378(94)70215-2. [DOI] [PubMed] [Google Scholar]
- Kristensen GB, Kaern J, Abeler VM, et al. No prognostic impact of flow-cytometric measured DNA ploidy and S-phase fraction in cancer of the uterine cervix: A prospective study of 465 patients. Gynecol Oncol. 1995;57:79–5. doi: 10.1006/gyno.1995.1102. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–49. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li R, Sonik A, Reinhard Stindl R, Rasnick D, Duesberg P. Aneuploidy vs. gene mutation hypothesis of cancer: recent study claims mutation but is found to support Aneuploidy. Proc Natl Acad Sci U S A. 2000;97:3236–41. doi: 10.1073/pnas.040529797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CH, Hsueh S, Huang MY, Chang MF, Soong YK. The uses and limitations of DNA flow cytometry in stage IB or II cervical carcinoma. Cancer. 1993;72:3655–62. doi: 10.1002/1097-0142(19931215)72:12<3655::aid-cncr2820721217>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Lage JM, Weinberg DS, Huettner PC, et al. Flow cytometric analysis of nuclear DNA content in ovarian tumors. Association of ploidy with tumor type, histologic grade, and clinical stage. Cancer. 1992;69:2668–75. doi: 10.1002/1097-0142(19920601)69:11<2668::aid-cncr2820691108>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Monsonego J, Autillo-Touati A, Bergeron C, et al. Liquid-based cytology for primary cervical cancer screening: a multi-center study. Br J Cancer. 2001;84:360–66. doi: 10.1054/bjoc.2000.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melsheimer P, Vinokurova S, Wentzensen N, et al. DNA Aneuploidy and integration of human papilloma virus type 16 E6/E7 oncogenic in intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix uteri. Clin Cancer Res. 2004;10:3059–63. doi: 10.1158/1078-0432.ccr-03-0565. [DOI] [PubMed] [Google Scholar]
- Merkel DE, McGuire WL. Ploidy, proliferative activity and prognosis. DNA flow cytometry of solid tumors. Cancer. 1990;65:1194–205. doi: 10.1002/1097-0142(19900301)65:5<1194::aid-cncr2820650528>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Marx J. Debate surges over the origins of genomic defects in cancer. Science. 2002;297:544–46. doi: 10.1126/science.297.5581.544. [DOI] [PubMed] [Google Scholar]
- Melsheimer P, Klaes R, Doeberitz MV, Bastert G. Prospective clinical study comparing DNA flow cytometry and HPV typing as predictive tests for persistence and progression of CIN I/II. Cytometry. 2001;46:166–71. doi: 10.1002/cyto.1101. [DOI] [PubMed] [Google Scholar]
- Melsheimer P, Vinokurova S, Wentzensen N, et al. DNA Aneuploidy and integration of human papillomavirus type 16 E6/E7 oncogenes in intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix uteri. Clin Cancer Res. 2004;10:3059–63. doi: 10.1158/1078-0432.ccr-03-0565. [DOI] [PubMed] [Google Scholar]
- Podratz KC, Wilson TO, Gaffey TA, et al. Deoxyribonucleic acid analysis facilitates the pre-treatment identification of high-risk endometrial cancer patients. Am J Obstet Gynecol. 1993;168:1206–13. doi: 10.1016/0002-9378(93)90370-x. [DOI] [PubMed] [Google Scholar]
- Ross JS. DNA ploidy and cell cycle analysis in cancer diagnosis and prognosis. Oncology. 1996;10:867–87. [PubMed] [Google Scholar]
- Rajagopalan H, Nowak MA, Vogelstein B. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–01. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- Soler ME, Gaffikin L, Blumenthal PD, et al. Cervical cancer screening in developing countries. Prim Care Update Ob Gyns. 2000;7:118–23. doi: 10.1016/s1068-607x(00)00032-9. [DOI] [PubMed] [Google Scholar]
- Strang P, Eklund G, Stendahl U, Frankendal B. S-phase rate as a predictor of early recurrences in carcinoma of the uterine cervix. Anticancer Res. 1987a;7:807–10. [PubMed] [Google Scholar]
- Strang P, Lindgren A, Stendahl U. Blood group antigens in relation to DNA content, S-phase rate and heterogeneity, and their prognostic values in cervical carcinoma. Anticancer Res. 1987b;7:125–8. [PubMed] [Google Scholar]
- Telford WG, King LE, Fraker PJ. Cell Proliferation. 1991;24:447–59. doi: 10.1111/j.1365-2184.1991.tb01173.x. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Vergote IB, Kaern J, Abeler VM, et al. Analysis of prognostic factors in stage I epithelial ovarian carcinoma: importance of degree of differentiation and deoxyribonucleic acid ploidy in predicting relapse. Am J Obstet Gynecol. 1993;169:40–52. doi: 10.1016/0002-9378(93)90129-7. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) Human papillomavirus infection and cervical cancer. 2010. Available at: www.who.int/vaccine_research/diseases/hpv .
- Willen R, Himmelmann A, Langstrom-Einarsson, et al. Prospective malignancy grading, flow cytometry DNA-measurements and adjuvant chemotherapy for invasive squamous cell carcinoma of the uterine cervix. Anticancer Res. 1993;13:1187–96. [PubMed] [Google Scholar]


