Abstract
Background:
Detection of circulating DNA can be applied for the diagnosis of many malignant neoplasms, including the hepatocellular carcinoma (HCC). The molecular pathogenesis of HCC is complex, involving different genetic and epigenetic alterations, chromosomal aberrations, gene mutations and altered molecular pathways. RASSF1A is a well-established tumor suppressor gene which suffers frequent inactivation due to promoter hypermethylation of CPG islands in multiple tumors including HCC, resulting in the reduction or loss of gene expression.
Objective:
To examine the role of circulating RASSF1A as a non-invasive diagnostic marker for HCC.
Participant and Methods:
A total of 45 HCC patients with a background of HCV infection, 40 cases of HCV infection without tumours and 40 apparently healthy controls were subjected to full history taking, clinical examination, routine laboratory investigations, assessment of serum AFP and detection of circulating hypermethylated RASSF1A gene by methylation-sensitive restriction enzyme digestion and real-time PCR.
Results:
The level of hypermethylated RASSF1A was significantly elevated in the HCC group as compared to the HCV and control groups (p=0.001 for both). Copy number in serum was associated with increased tumor size (p value <0.001). On the other hand, no significant correlation was observed between RASSF1A and AFP (p=0.5). Using ROC curve analysis, the best cut-off for circulating serum RASSF1A to differentiate the HCC group was 8 copies/µl.
Conclusion:
The presence of hypermethylated RASSF1A in serum may be a useful and informative biomarker for HCC diagnosis and might be introduced as a screening method for populations at risk of HCC development.
Keywords: HCC, AFP, RASSF1A gene
Introduction
Hepatocellular carcinoma (HCC) is one of the most common type of liver cancer with high incidence and mortality rate (Kim et al., 2012; El Baz et al., 2013). Many authors study the incidence, prevalence, mortality rate and management of HCC in Egyptian patients (Omran et al., 2015; Khan and Hashim., 2015; Abdelaziz et al., 2015). Hospital-based studies from Egypt have reported an overall increase in the relative frequency of all liver-related cancers in Egypt, from approximately 4% in 1993 to 7.3% in 2003 (El-Zayadi et al., 2005), this may be attributed to the high prevalence of Hepatitis C (HCV) infection in Egypt as previously reported (Lehman et al., 2008; Mohamed et al., 2013). Guerra and his colleagues reported that the Demographic Health Survey (DHS) carried out in Egypt in 2008 for HCV antibody testing revealed high prevalence of HCV infection in Egypt, they reported that the incidence of HCV was 14.7% in the age group 15-59 years and they referred that several factors including to several factors including age, sex, urbanization, poverty, blood transfusion and past history of intravenous injections of anti-schistosomiasis (Guerra et al., 2012).
Epigenetic changes have become established from several years as being one of the most important molecular signatures of human tumors. The discovery of hypermethylation of the CpG islands of certain tumor suppressor genes in cancer links DNA methylation to the classic genetic lesions with the disruption of many cell pathways, from DNA repair to apoptosis, cell cycle and cell adherence.
RASSF1A is one of the most frequently inactivated tumor suppressor genes (TSG) ever identified in human cancer. It has been implicated in the regulation of the cell cycle, apoptosis and genetic instability (Gordon and Baksh, 2011; Agathanggelou et al., 2005). The gene appears to suffer frequent transcriptional inactivation in tumor cells due to aberrant promoter methylation of CpG islands (Dammann et al., 2000; Richter et al., 2009). Hypermethylation in the promoter region is suspected as the main mechanism of silencing observed in many cancers including HCC (Donninger et al., 2007; Van der Weyden et al., 2007).
Current gold standard and most commonly used biomarker for patients at risk for HCC is alpha-fetoprotein (AFP) along with ultrasound every 6 to 12 months both are far from perfect. Serum AFP levels of more than 400ng/mL are considered diagnostic; however, such high values are observed only in a small percentage of patients with HCC. Ultrasound surveillance even performed at every three monthly intervals cannot improve detection of small HCC because of limitations in recall procedures (Farinati et al., 2008). Therefore, there is an urgent need to identify promising tool that could diagnose HCC early or be served as surveillance for patients at risk. This study evaluate the role of circulating hypermethylated RASSF1A as a diagnostic marker for HCC by the use of methylation-sensitive restriction enzyme digestion and real-time PCR detection.
Materials and Methods
This study included 85 patients from the inpatient ward and outpatient HCC Clinic of Tropical Medicine Department at Kasr El Ainy Hospital, Cairo University and 40 healthy control subjects, informed consent was taken from all participant enrolled in this study and was approved by the local ethical committee of Clinical and Chemical Pathology Department. Participant were divided into 3 groups as follow:
Group 1: Comprised 45 newly diagnosed HCC patients on top of HCV infection, their age ranged from 38-75 years, 42(93.3%) were males and 3(6.7%) were females.
Group 2: Comprised 40 patients with HCV associated liver disease, their age ranged from 39-67 years. This group comprised 26(65%) males and 14(35%) females.
Group 3: Comprised 40 apparently healthy individuals with no history of chronic hepatitis their age ranged from 21-40 years. This group comprised 38 (95%) males and 2(5%) females.
Diagnosis of HCC was based on imaging techniques following the 2005 diagnostic algorithm (Forner et al., 2008; Sangiovanni et al., 2008).
Patients with hepatitis B virus infection (HBV), other types of tumors or liver metastasis as proved by spiral CT were excluded from this study.
All Groups Were Subjected To:
Full history taking and Clinical examination.
Radiological investigations: Abdominal ultrasonography U/S and spiral CT for detection of hepatic focal lesions, liver cirrhosis, hepatosplenomegaly and presence of ascites.
Complete blood count (CBC), prothrombin time and concentration.
Liver function tests, Kidney function tests.
Special Investigations (HBs Ag, and HCV antibodies, serum AFP).
Molecular study: detection of hypermethylated RASSF1A by real time PCR after enzymatic digestion.
Sample processing
DNA extraction
A 720 µl sterile serum sample was used for DNA extraction for each case using the nucleoSpin® plasma XS supplied by Macherey Nagel (MACHEREY-NAGEL GmbH and Co. KG, Germany), Part number (740900.50). This kit was designed for the efficient isolation of circulating DNA from human blood, plasma or serum. Fragmented DNA as small as 50 – 1000 bp can be purified with high efficiency.
Digestion step
We used a fast digest methylation-sensitive enzyme Bsh1236I (BSTUI) supplied by Thermo Fermentas (subsidiary of Thermo Fisher Scientific Inc., Vilnius Lithuania), Catalog Number (ER0922). The following digestion protocol was used as previously described (Chan et al., 2009), but we apply a modification on their technique, we used 17.5 µL of extracted serum DNA for each sample instead of 35 µL, 5 µL of BSTUI enzyme (50 U) and 2.5 µL of 10 X Buffer R.
All were mixed gently, spinned down for few seconds and incubated at 37 C° for 16 hours in a water bath. Then digested DNA was stored at -20 C°.
Amplification and detection of RASSF1A gene using real time PCR after enzyme digestion
The primer and probe for RASSF1A and β-actin were designated using primer and probe express software (Applied Biosystems), data were illustrated in table (1).
Table 1.
Sequence, Concentration and Part Number of Primer and Probes
| Sequence | Concentration | |
|---|---|---|
| RASSF1A primers | ||
| Forward primer | 5’-AGCCTGAGCTATTGAGCTG-3’ | 40,000 pmol |
| Reversed primer | 5’-CGGCGGATCGGCAAA-3’ | 40,000 pmol |
| RASSF1A probe | 5’FAMTM-CCAACGCGCTGCGCAT(MGBTM)-3’ | 5,000 pmol |
| β -actin primers | ||
| Forward primer | 5’-GCGCCGTTCCGAAAGTT-3’ | 40,000 pmol |
| Reversed primer | 5’-CGGCGGATCGGCAAA-3’ | 40,000 pmol |
| β-actin probe | 5’VICTM-ACCG CCGAGACCGCGTC(MGBTM)-3’ | 5,000 pmol |
We used a multiplex real time PCR assay for detection of RASSF1A (gene of interest) and β-actin (internal control gene) sequences in serum. PCR was carried out in 26.5 μl of total reaction volume containing 12.5 μl of TaqMan universal PCR master mix ll (2X) without UNG, 0.5 μl from each primers of both genes, 0.5 μl from each probe and 11 μl of enzyme digested serum DNA mixture. We performed qPCR using the Applied biosystems StepOne™ detection system (Applied Biosystems, Foster City, CA, USA) under the following conditions: an initiation step for 2 minutes at 50°C was followed by a first denaturation for 10 minutes at 95°C and another step consisting of 50 cycles of 15 seconds at 95°C and 1 minute at 60°C as previously reported (Chang et al., 2009).
Interpretation of Results
The RASSF1A results would be interpreted only if the corresponding β-actin signal is negative. The presence of β-actin signal indicates the presence of incompletely digested unmethylated β-actin sequences, suggesting that the enzyme digestion of the particular sample may be incomplete. Interpretation of results was done by Absolute Quantification using the standard curve method, Slope of the curve was (-3) and the Intercept was (38.67), then we compared unknowns RASSF1A to the curve and extrapolate a value.
Statistical Analysis
Statistical analysis was run on SPSS Statistics 17.0 (SPSS Inc., Chicago, Ill, USA). Quantitative parametric data were presented as mean ± SD while non-parametric data were presented as median and percentiles (25th-75th). Differences between two groups were tested using Mann-Whitney test while differences among several groups were tested using Kruskal-Wallis with Mann-Whitney tests as appropriate. Qualitative data were presented as number (percent) and compared using Chi-square (X2) test. Correlation between different variables were tested using Pearson or Spearman correlation tests as appropriate. Receiver operator characteristic (ROC) curves were constructed for RASSF1A with determination of sensitivity, specificity, positive predictive value “PPV”, Negative Predictive Value” NPV” and accuracy at different cut-off levels. All tests are statistically significant at a P value of < 0.05.
Results
Baseline characteristics of all patients
All cases included in this study (85 cases) were HCV antibodies positive, 75 (88.2%) had cirrhosis and 10 (11.76%) showed no evidence of cirrhosis, out of the 75 cases with cirrhosis 34 (45.3%) cases also suffered from schistosomiasis. The frequency of different clinical data of the studied groups were illustrated in Table 2.
Table 2.
Demonstrate Clinical and Some Laboratory Data of the Studied Groups
| HCC group (45 cases) | HCV group (40 cases) | |
|---|---|---|
| Previous encephalopathy | 3 (6.7%) | 12 (30%) |
| Previous ascites | 24 (53.3%) | 33 (82.5%) |
| Previous variceal bleeding | 9 (20%) | 12 (30%) |
| * MELD score | 11 (9-14.75) | 16.5 (13-18.75) |
| * Serum Bilirubin (mg/dl) | 1.7 (1.2-2.7) | 2.55 (1.4-4.3) |
| ** serum albumin (g/dl) | 2.8 ± 0.5 | 2.4 ± 0.5 |
| * prothrombin concentration | 66.7±11.8 | 51.9±19.9 |
| * Serum creatinine (mg/dl) | 1 (0.8-1.5) | 1.2 (0.9-1.6) |
Data are presented by median (25th-75th percentile),
data are presented by mean±SD
Evaluation of serum methylated RASSF1A as a diagnostic marker for HCC
Promoter methylation of RASSF1A gene was detected in 36 (87.7%) of serum DNA of HCC patients, 25 (62.5%) patients with HCV infection and 2 (5%) out of 40 healthy donors displayed RASSF1A promoter methylation. Quantitation of serum methylation RASSF1A in HCC group revealed a significant elevation when compared to HCV group (61 copies/µl vs. 1.65 copies/µl respectively, p <0.001), data illustrated in Figure 1. The median value of serum methylation RASSF1A in control group was significantly reduced when compared to the other two groups, p value was <0.001 for both.
Figure 1.
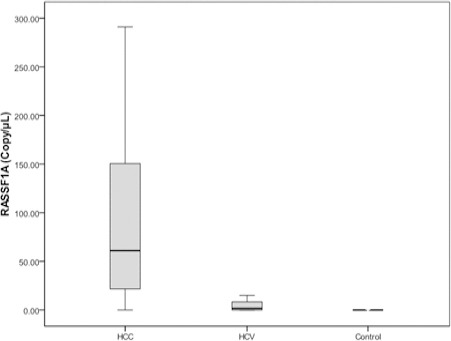
Median Level of Hypermethylated RASSF1A in HCC, Liver Cirrhosis and Control Group
Association between RASSF1A and clinical parameter of HCC group
In HCC group 9 (20%) patients had hematemesis, 3 (6.7%) patient had encephalopathy, 12 (22.7%) had schistosomiasis and 24 (53.3%) out of 45 patients with HCC presented with ascites. Association between serum RASSF1A and different clinical presentation of HCC patients are illustrated in Table 3.
Table 3.
Median Levels of Serum RASSF1A in Relation to Different Clinical Presentation of HCC Patients
| RASSF1A (Copies/µL) | p-value | ||
|---|---|---|---|
| Hematemesis | yes (9 cases) | 47.0 (22.3-69.7) | 0.41 |
| No (36 cases) | 66.2 (19.7-160.5) | ||
| Encephalopathy | yes (3 cases) | 161.0 (0-506) | 0.539 |
| Yes (41 cases) | 65.0 (22.3-154) | ||
| Cirrhosis | No(4 cases) | 71.4 (20.8-160) | 0.34 |
| No(33 cases) | 27.5 (2-138.5) | ||
| Ascites | Yes (24 cases) | 115.0 (26.2-171.5) | 0.001 |
| No (21 cases) | 40.8(20.1-74.1) |
With regard to the HCV group, there was no statistical significant difference on comparing median values of hypermethylated RASSF1A in HCV group regarding the presence or absence of (hematemesis, encephalopathy, bilhareziasis), (p=0.89, 0.61, 0.28) respectively. On the other hand, there was a significant elevation in median values of hypermethylated RASSF1A in HCV patients presented with ascites when compared to patients without ascites (p=0.001).
Effect of cirrhosis on hypermethylated RASSF1A
HCC andHCV groups were further subdivided into two groups (cirrhotic and non-cirrhotic) on the basis of abdominal ultrasound and CT scan. In the HCC group 41(91%) patients were cirrhotic, while 4 (9%) were non-cirrhotic and in HCV group 34 (85 %) patients were cirrhotic while 6 (15%) patients were non–cirrhotic. No significant difference in RASSF1A level was observed when comparing cirrhotic HCC group to non-cirrhotic group (65 copies/µl vs. 27.5 copies/µl respectively, p=0.34). Comparing serum RASSF1A between cirrhotic HCC group and cirrhotic HCV group, a significant elevation was observed in HCC group (65 copies/µl vs. 2.95 copies/µl, p <0.001).
Correlation between tumor size and serum RASSF1A
The median value of tumor size in HCC was 4.2 cm, ranged from 3.2 to 7.3 cm. A significant correlation was found between tumor size and hypermethylated RASSF1A in HCC group (r=0.728 and P<0.001).data is illustrated in figure (2). We further subdivide the HCC group to two groups, the first includes 26 (57.8%) patient with tumor mass < 4 cm in diameter and the second 19 (42.2%) patients with tumor mass diameter > 4 cm. The median level of methylated RASSF1A in the second group was significantly increased when compared to the first group (116 copies/µl vs. 22.3 copies/µl respectively, p <0.001).
Figure 2.
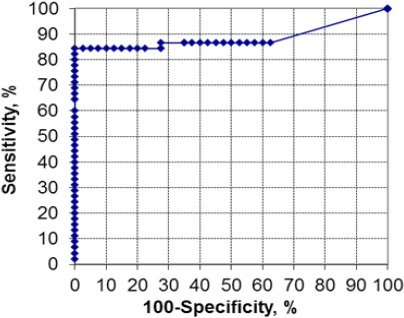
ROC Curve for Hypermethylated RASSF1A
Correlation between biomarkers of Child Turcotte Pugh scoring system, serum AFP and serum RASSF1A
As regard the values of biochemical markers of CTP scoring system in HCV group, a significant correlation was found between methylated RASSF1A and the following parameters (albumin r = -0.45 and p=0.004, bilirubin r =0.463 and p=0.003 and PC, r = -0.54 and p<0.001). In the HCC group, a significant correlation was only found between values of (albumin and bilirubin) and RASSF1A, r = -0.383 and P=0.009, r =0.55 and P<0.001, respectively. The median value of serum AFP in the HCC group was 116 ng/ml, 25th -75th percentile was (22.5-320). No significant correlation was found between AFP and RASSF1A (r= 0.1, p= 0.5).
Diagnostic performance of hypermethylated RASSF1A in discrimination between HCC and HCV
Performance of the hypermethylated RASSF1A gene as a biomarker for distinguishing HCC from HCV infection by real time PCR after enzyme digestion revealed that at a cut off level of 8 copies/µl the sensitivity was 86.7%, specificity was 72.5% PPV was 78%, NPV was 93.8 %, and accuracy was 80% and AUC (95% CI) was 0.889 (0.8-0.97). Data are presented in Figure 2.
Association of serum AFP and serum RASSF1A in the HCC group
We compared the potential diagnostic role of AFP and RASSF1A in HCC group, at an AFP cut off value of 20 ng/ml, 11 (24.4%) patient were <20 ng/ml. From these 11 cases, 8 (72.7%) cases had cut off value 8 copies/µl of serum RASSF1A. Sixteen cases (35%) with HCC at time of diagnosis displayed a value of serum AFP > 200 ng/ml, while 29 (64.5%) cases presented with serum AFP < 200 ng/ml. from the latter group serum RASSF1A at a cut off value of 8 copies/µl could detect 26 (89.6%) of the HCC cases.
Discussion
From 2003 to 2015, many studies have published on the prognostic values of DNA methylation in HCC (Gao et al., 2008; Hernandez-Vargas et al., 2010; Song et al., 2013; Dong et al., 2015). One of the studied genes is RASSF1A, which is a well-known tumour suppressor. RASSF1A inactivation through DNA methylation is one of the most important mechanism known to drive human hepatocarcinogenesis (Nishida et al., 2010). In this study we analysed promoter DNA methylation of RASSF1A in a group of Egyptian HCC patients, HCV chronic carriers and a group of normal healthy individuals. HCC cases in this study were positive for HCV infection and negative for HBV infection. This may be explained by the fact that HCV is more prevalent in Egypt than HBV. Previous Egyptian studies revealed that Egypt is of intermediate endemicity for HBV, having a prevalence of 2–8% (El-Zayadi et al., 2005), while the overall prevalence of anti-HCV antibodies in Egypt was found to be 20% (Arafa et al., 2005). Guo et al., reported that the hepatitis C virus core (HCVc) could up-regulate the methylation status of the RASSF1A promoter (Guo et al., 2011).
Despite the extensive use of methylation-sensitive restriction enzymes in epigenetic research, methylation-sensitive restriction enzyme analysis has not been popular for detecting aberrant DNA methylation in the circulation for the purpose of cancer detection. The reason is probably related to the difficulties in differentiating false-positive results due to incomplete enzyme digestion from the presence of a low concentration of tumor-derived hypermethylated sequences. In this regard Chan and his group introduced a system to check the completeness of enzyme digestion (Chan et al., 2009).
This checking system, we used in our study, consisted of a PCR assay that amplified the β-actin promoter after being subjected to methylation sensitive restriction enzyme digestion. β-actin promoter region has the same number of restriction sites as in the RASSF1A amplicon. Because it was previously shown that the digestion efficiencies for RASSF1A and β-actin sequences are similar (Chan et al., 2006). The presence of a β-actin amplification signal in a sample suggests that the digestion of unmethylated RASSF1A is incomplete. In such cases, the sample results should be excluded. Serum RASSF1A results would be used for further analysis only if the corresponding β-actin signal for the sample was absent (Chan et al., 2009).
We detected a significant elevation in the median level of hypermethylated RASSF1A in sera of HCC patients when compared to HCV and control group, this finding was in agreement with the study done by Chan et al.,(2009) they used the same methodology (methylation sensitive restriction enzyme digestion and real time PCR detection), However in the study of Chan and his group the median RASSF1A concentrations for the HCC patients was lower than that in our study (7.7 x 105 copies/L which is equivalent to 0.77 copies/ µL versus 60 copies/µL. This discrepancy may be attributed to multiple factors:
In our study HCC was developed on top of HCV and not HBV as in the study of Chan and his group.
The use of different Extraction kit (Plasma XS, Mechery Nagel) specified for circulating DNA extraction that gives the amplification signal 1 to 2 cycles earlier.
Moreover, we used a modified protocol in which we increased the amount of serum (720 instead of 240 µl) used for extraction. Although the study of Chang et al, used a modified protocol yet the extraction kit was not specified for circulating DNA extraction (QIAamp DNA Mini Kit, Qiagen).
In agreement with our study, Hu et al., reported the expression of hypermethylated RASSF1A in sera and HCC tissue (Hu et al., 2010; Saelee et al., 2010;Huang et al., 2011) and they suggested that RASSF1A methylation should be a potential marker of incipient malignancy in HCC, this finding was in accordance with other investigators (Yeo et al., 2005; Di Gioia et al., 2006; Mohamed et al., 2012; Elshimali et al., 2013; Nishida et al., 2014; Dong et al., 2015). Other authors have reported a high frequency of RASSF1A in the control group (10 %) and they suggest this finding may relate to an underlying undetectable bilharzial liver fibrosis or cirrhosis, as the controls were chosen as those who were negative for hepatitis B and C markers only (Mohamed et al., 2012). Other studies revealed a lower frequency of RASSF1A than our study (Yeo et al., 2005; Zhang et al., 2007), this may be due to the use of a different technique, the authors used the methylation-specific PCR which may degrade up to 96% of DNA without discriminating between methylated and unmethylated DNA, while we used the highly sensitive combination of methylation-sensitive restriction-enzyme digestion which degrade only the unmethylated DNA.
The present study denoted that the serum hypermethylated RASSF1A level was significantly higher in HCC on top of HCV compared to patients with chronic HCV infection without malignancy (p < 0.001). In this context we found that hypermethylated RASSF1A at a cut-off value of 8 copies/ µL could differentiate HCC from patients with HCV with an AUC of 0.889, sensitivity of 86.7%, specificity of 72.5 %, PPV of 78%, NPV of 82, 9% and a predictive accuracy of 80%. This may be of help in early diagnosis of HCC by applying this non-invasive test regularly in those high-risk patients.
The clear difference, our study showed in the median concentrations of circulating hypermethylated RASSF1A in HCC patients in comparison to HCV chronic carriers (> 10 - fold higher in HCC patients than in HCV carriers) (p value< 0,001), suggests that the predominant source of circulating hypermethylated RASSF1A in HCC patients is the tumor itself. This suggestion was supported by Chan and his group when they found a significant reduction in serum RASSF1A in the HCC patients after tumor resection and a significant increase in the circulating concentration of hypermethylated RASSF1A during surveillance programs for HCC from the day of recruitment of hepatitis patients till the day of HCC detection (Chan et al., 2009).
Chronic HCV infection may act as a powerful epi mutagen and may induce methylation of a group of gene including RASSF1A, and may have a role in hepatic carcinogenesis, this was reported by other authors (Lee et al., 2003; Nishida et al., 2008).
This study showed a significant correlation between RASSF1A concentrations in HCC group and tumor size (r = 0.728, p < 0.01), a finding that goes in parallel with a previous study in which the presence of RASSF1A promoter hypermethylation in plasma DNA was found to associate with HCC size of > or =4 cm, p = 0.035 (Yeo et al., 2005). We assume that the significant correlation between tumor size and hypermethylated RASSF1A values may be justified by, and, may also add to the hypothesis that the major source of hypermethylated RASSF1A in HCC patients is the tumor itself. This correlation also may give this marker a prognostic power in follow up of patients after tumor resection and to detect early recurrence.
Considering the relationship between AFP and RASSF1A, our study found the absence of a significant correlation between the serum concentrations of AFP and RASSF1A (r= 0.1, p value = 0.5), these non-significant correlation suggesting that concurrent use of both markers might provide a potentially synergistic set of information. We reported that at serum RASSF1A value 8 copies/µl can pick 89.6% of HCC cases missed by AFP at cut off level 200 ng/ml and 72.7% of HCC cases missed by AFP at cut off level 20 ng/ml.
In conclusion, detection of hypermethylation RASSF1A in serum can be a tool for early screening for HCC. Gene methylation has been implemented to determine tumor specific alteration of circulating RASSF1A as a diagnostic and prognostic marker in cancer patients, still more work needs to be done to establish the role of this marker and to introduce as a routine laboratory investigation for high risk patients.
Compliance with Ethical Standards
Conflict of Interest
The authors (Enas H. Mahmoud, Lamiaa A. Mansour, Maissa El raziky, Amal A. elwahab, Sherif Hamdy, Enas H. El-Sayed and Amr M. Ali) had no conflicts of interest that are directly relevant to the content of this article.
Funding
The authors (Enas H. Mahmoud, Lamiaa A. Mansour, Maissa El raziky, Amal A. elwahab, Sherif Hamdy, Enas H. El-Sayed and Amr M. Ali) didn’t receive any fund for this article.
Ethical approval and informed consent
All procedures performed in this study involving human participants were in accordance with the ethical standards of clinical and chemical pathology research committee. Informed consent was taken from all participants prior to enrolment in this study and the study was approved by the ethical committee of Clinical and Chemical pathology department, Cairo University.
Declaration of Interest
The authors declare no conflict of interest.
References
- Abdelaziz AO, Elbaz TM, Shousha HI, et al. Survival and prognostic factors for hepatocellular carcinoma: an Egyptian multidisciplinary clinic experience. Asian Pac J Cancer Prev. 2015;15:3915–20. doi: 10.7314/apjcp.2014.15.9.3915. [DOI] [PubMed] [Google Scholar]
- Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–08. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- Arafa N, El Hoseiny M, Rekacewicz C, et al. Changing pattern of hepatitis C virus spread in rural areas of Egypt. J Hepatol. 2005;43:418–24. doi: 10.1016/j.jhep.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Chan KC, Lai PB, Mok TS, et al. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem. 2008;54:1528–36. doi: 10.1373/clinchem.2008.104653. [DOI] [PubMed] [Google Scholar]
- Chan KC, Ding C, Gerovassili A. Hypermethylated RASSF1A in maternal plasma: A universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin Chem. 2006;52:2211–18. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, et al. Epigenetic inactivation of a RAS association domain family protein from lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–19. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Di Gioia S, Bianchi P, Destro A, et al. Quantitative evaluation of RASSF1A methylation in the non-lesional, regenerative and neoplastic liver. BMC Cancer. 2006;6:89. doi: 10.1186/1471-2407-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, He H, Zhang W, et al. Combination of serum RASSF1A methylation and AFP is a promising non-invasive biomarker for HCC patient with chronic HBV infection. Diagn Pathol. 2015;4:10. doi: 10.1186/s13000-015-0317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–72. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- Elshimali YI, Khaddour H, Sarkissyan M, et al. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci. 2013;14:9. doi: 10.3390/ijms140918925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Zayadi AR, Badran HM, Barakat EM, et al. Hepatocellular carcinoma in Egypt: a single center study over a decade. World J Gastroenterol. 2005;11:5193–98. doi: 10.3748/wjg.v11.i33.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinati F, Marino D, De Giorgio M, et al. Diagnostic and prognostic role of alpha feto protein in hepatocellular carcinoma both or neither? Am J Gastroenterol. 2006;101:524–32. doi: 10.1111/j.1572-0241.2006.00443.x. [DOI] [PubMed] [Google Scholar]
- Forner A, Vilana R, Ayuso C, et al. Diagnosis of hepatic nodules 20 mm or smaller in cirrhosis: prospective validation of the noninvasive diagnostic criteria for hepatocellular carcinoma. Hepatology. 2008;47:97–04. doi: 10.1002/hep.21966. [DOI] [PubMed] [Google Scholar]
- Gao W, Kondo Y, Shen L, et al. Variable DNA methylation patterns associated with progression of disease in hepatocellular carcinomas. Carcinogenesis. 2008;29:1901–10. doi: 10.1093/carcin/bgn170. [DOI] [PubMed] [Google Scholar]
- Gordon M, Baksh S. RASSF1A Not a prototypical Ras effector. Small GTPases. 2011;2:148–57. doi: 10.4161/sgtp.2.3.16286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra J, Garenne M, Mohamed MK, et al. HCV burden of infection in Egypt: results from a nationwide. J Viral Hepat. 2012;19:560–67. doi: 10.1111/j.1365-2893.2011.01576.x. [DOI] [PubMed] [Google Scholar]
- Guo C, Zhang X, Pfeifer GP. The tumor suppressor RASSF1A prevents dephosphorylation of the mammalian STE20-like kinases MST1 and MST2. J Biol Chem. 2011;286:6253–61. doi: 10.1074/jbc.M110.178210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vargas H, Lambert MP, le Calvez-Kelm F, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS One. 2010;5:e9749. doi: 10.1371/journal.pone.0009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Chen G, Yu H, et al. Clinicopathological significance of RASSF1A reduced expression and hypermethylation in hepatocellular carcinoma. Hepatol Int. 2010;4:423–32. doi: 10.1007/s12072-010-9164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZH, Hu Y, Hua D, et al. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol. 2011;91:702–07. doi: 10.1016/j.yexmp.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Khan G, Hashim MJ. Burden of virus-associated liver cancer in the Arab world 1990-2010. Asian Pac J Cancer Prev. 2015;16:265–70. doi: 10.7314/apjcp.2015.16.1.265. [DOI] [PubMed] [Google Scholar]
- Kim dY, Han KH. Epidemiology and surveillance of hepatocellular carcinoma. Liver Cancer. 2012;1:2–14. doi: 10.1159/000339016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee HJ, Kim JH, et al. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371–78. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman EM, Soliman AS, Ismail K, et al. Patterns of hepatocellular carcinoma incidence in Egypt from a population-based Cancer registry. Hepatol Res. 2008;38:465–73. doi: 10.1111/j.1872-034X.2007.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NA, Swify EM, Amin NF, et al. Is serum level of methylated RASSF1A valuable in diagnosing hepatocellular carcinoma in patients with chronic viral hepatitis C? Arab J Gastroenterol. 2012;13:111–5. doi: 10.1016/j.ajg.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Mohamoud YA, Mumtaz GR, Riome S, et al. The epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesis. BMC Infect Dis. 2013;13:288. doi: 10.1186/1471-2334-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N. Impact of hepatitis virus and aging on DNA methylation in human hepatocarcinogenesis. Histol Histopathol. 2010;25:647–54. doi: 10.14670/HH-25.647. [DOI] [PubMed] [Google Scholar]
- Nishida N, Chishina H, Arizumi T, et al. Identification of epigenetically inactivated genes in human hepatocellular carcinoma by integrative analyses of methylation profiling and pharmacological unmasking. Dig Dis. 2014;32:740–46. doi: 10.1159/000368015. [DOI] [PubMed] [Google Scholar]
- Nishida N, Nagasaka T, Nishimura T, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–18. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran DA, Awad AH, Mabrouk MA, et al. Application of data mining techniques to explore predictors of HCC in Egyptian patients with HCV-related chronic liver disease. Asian Pac J Cancer Prev. 2015;16:381–5. doi: 10.7314/apjcp.2015.16.1.381. [DOI] [PubMed] [Google Scholar]
- Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer;from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–28. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Saelee P, Wongkham S, Chariyalertsak S. RASSF1A promoter hypermethylation as a prognostic marker for hepatocellular carcinoma. Asian Pac J Cancer Prev. 2010;11:1677–81. [PubMed] [Google Scholar]
- Sangiovanni A, Manini MA, Iavarone M, et al. The diagnostic and economic impact of contrast imaging techniques in the diagnosis of small hepatocellular carcinoma in cirrhosis. Gut. 2008;59:638–44. doi: 10.1136/gut.2009.187286. [DOI] [PubMed] [Google Scholar]
- Song MA, Tiirikainen M, Kwee S, et al. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8:e55761. doi: 10.1371/journal.pone.0055761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo W, Wong N, Wong WL, et al. High frequency of promoter hypermethylation of RASSF1A in tumor and serum of patients with hepatocellular carcinoma. Liver Int. 2005;25:266–72. doi: 10.1111/j.1478-3231.2005.01084.x. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Wu HC, Shen J, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter hypermethylation in serum DNA. Clin Cancer Res. 2007;13:2378–84. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]


