Abstract
Currently, despite the advances in individualized treatment, breast cancer still remains the deadliest form of cancer in women. Diagnostic, prognostic, and therapy-predictive methods are mainly based on the evaluation of tumor tissue samples and are aimed to improve the overall therapeutic level. Therefore, the exploration of a series of circulating biomarkers, which serve as the information source of tumors and could be obtained by peripheral blood samples, represents a high field of interest. Apart from classical biomarkers, exosomes, which are nanovesicles, are emerging as an accessible and efficient source of cell information. The purpose of this review is to summarize the peculiarities of the presently available breast cancer exosomal biomarkers; the review also provides the prediction of a multitude of potential target genes of exosomal microRNAs using 4 databases.
Keywords: breast cancer, diagnostics, exosome, miRNA, protein
Introduction
Breast cancer (BrCa) remains the most prevalent type of cancer occurring in women worldwide, with no less than 1 000 000 new cases diagnosed each year.1 Although the prognosis of patients with BrCa has improved substantially, approximately one-third of women still die from this disease, mainly from metastasis, especially in the brain.2–4 With the rapid development of modern clinical medicine, early diagnosis of BrCa is crucial to enhance therapeutic outcomes and survivor rate. Mammography is widely used for BrCa screening,5 but it is limited due to the minimum detectable tumor size and the low radiation utilized on some patients.6 Liquid detection, because of its nondestructiveness and low cost, is emerging as a novel method of BrCa diagnosis. Molecular biomarkers, such as the progesterone receptor, estrogen receptor (ER), and human epidermal growth factor receptor (Her2/ErBb2), are used for diagnosis or for predicting outcomes.7,8 However, these biomarkers are not sufficient or reliable to timely screen tumor cells extracted from the peripheral blood of patients with BrCa. Hence, it is imperative to determine more precise biomarkers for diagnosing cancer at an early stage.
Exosomes, secreted by all viable cells, are nanovesicles of approximately 30 to 100 nm in diameter that are produced in the endosomal pathway and play a vital role in paracrine and autocrine cell communication.9 They carry a broad repertoire of cargo, such as proteins, lipids, messenger RNAs (mRNAs), microRNAs (miRNAs), long-noncoding RNA (lncRNA), and DNA.10,11 Furthermore, exosomes broadly exist in physiological or pathological fluids, including plasma, serum, saliva, urine, breast milk, cerebrospinal fluid, and ascites.12 As reported, tumor cells may release more exosomes than normal cells, with a certain number of cancer-specific biomarkers. As they are easily accessible and maintain stability in vitro, exosomes have been regarded as the most promising and fundamental indicators13 for the early diagnosis of patients or for predicting outcomes after different types of treatments with targeting molecules, such as heat shock proteins,14 survivin,15 and miR-221/222.16 These proteins and miRs exist extensively in exosomes and are closely related to the development and metastasis of BrCa.17–19 Indeed, approximately 270 articles on “exosome and BrCa” are found in the PubMed database, most of which were published within the last 5 years. Dissemination of the current knowledge to a broader range of scientific groups or clinical staff can aid in the understanding and utilization of these fascinating nanoparticles. Hence, in this review, we summarize the current knowledge of the diagnostic applications of exosomes in BrCa.
The Biological Activity of Exosomes
Initially, extracellular vehicles (EVs) were discovered by Siekevitz20 as cellular debris after cell damage. In contrast to the prime hypothesis, EVs may contain numerous surface components and cytoplasmic molecules, which originate from their host cells. Although the molecular mechanisms promoting cell-to-cell communication are not fully understood, recent studies have shown that EVs may participate in transferring specific proteins and nucleic acids to recipient cells.21 Exosomes are one kind of cell-derived EVs, with a size of 30 to 120 nm in diameter. The profiles of exosome sare presented in Figure 1. The biological functions of exosomes include antigen presentation, immune regulation, evading apoptosis, drug resistance, and escape from immune surveillance.22,23 As a communication tool, exosomes derived from cancer cells have the potential to transfer a malignant phenotype to normal cells and establish a fertile local and distant microenvironment. As a result, exosomes could be used as a source for biomarkers for predicting metastatic spread, targeted drug delivery, cell free antitumor vaccination, and gene therapy.24,25 In addition, the latest reports have proven that exosomes often correlate with an increase in blood vessel thrombosis by transporting the pro-coagulant receptor, tissue factor (TF).26,27 Fausto28 and colleagues reported that BrCa cell-derived exosomes may induce TF-independent platelet activation and aggregation, as well as TF-dependent plasma clotting and platelet aggregation, by means of thrombin generation.
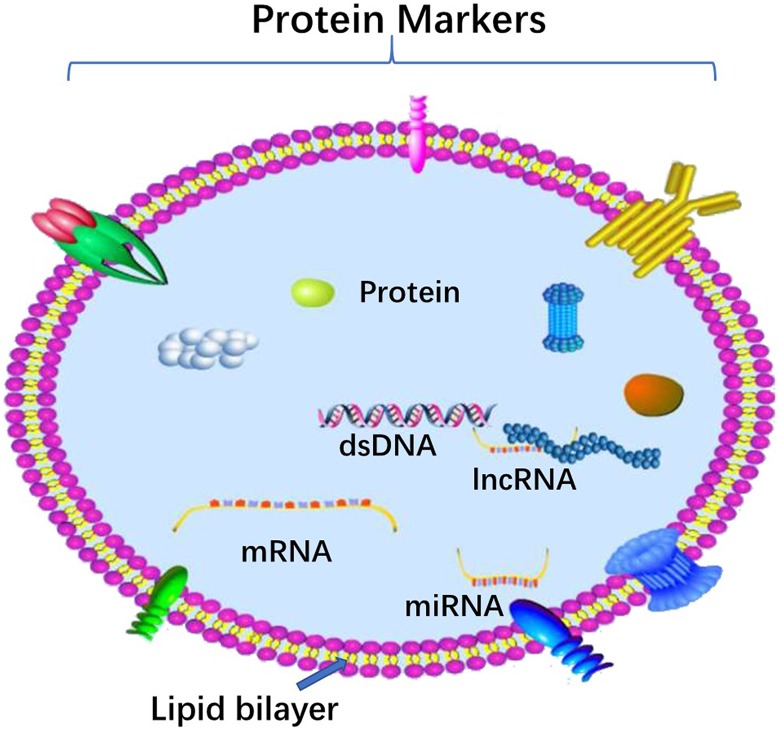
Figure 1.
Schematic Representation of exosomes. The structure and contents of exosomes are demonstrated in this plot. The lipid bilayer membrane of exosomes, sharing similar topology to the original cells, carries typical transmembrane proteins and receptors. In addition, the contents of exosomes can be transferred from the original cell to their target cell, including RNA species (mRNAs, miRNAs, and other types of short RNAs) and a vast array of different proteins.
Implications for BrCa Diagnosis
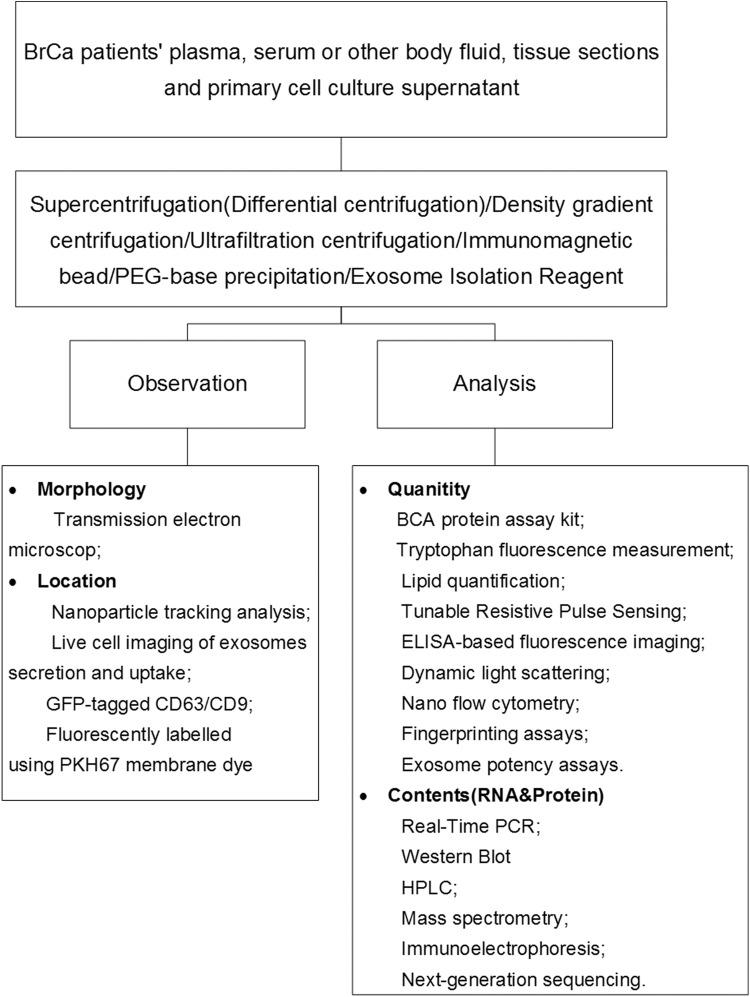
Survival of patients with BrCa depends on the stage at diagnosis, with patients in whom the disease is detected earlier having more favorable outcomes.29 Hence, numerous scientific groups are concentrating on the development of a more efficient and accurate biomarker for BrCa. Apart from classical markers, exosomes are becoming a very accessible source, with extensive information about tumor biological characteristics (proteins and nucleic acids) gathered from liquid biopsy.30 The process of exosome utilization in BrCa diagnosis is shown in Figure 2, which illustrates the use of exosomes in diagnostic and therapeutic applications.
Figure 2.
Exosome application in BrCa diagnosis. This plot demonstrates the process of utilizing exosomes in BrCa diagnosis.
Exosomal Protein in BrCa Diagnosis
Proteins, which are part of the molecular content in exosomes, are heavily dependent on cellular origin. Exosomes possess a series of conservative proteins across species, suggesting valuable, even life-preserving functions.31 For example, almost all exosomes contain cytoplasmic proteins, such as tubulin,32 actin-binding proteins, annexins,33 and Rab proteins,34 such as signal transduction proteins-kinases and heterotrimeric G-proteins.35 Many exosomes also contain heat-shock proteins (Hsp)70 and Hsp9014 and major histocompatibility complex class I molecules.36 Except for a series of normal proteins, the function of exosomes seems to be controlled by cell-specific proteins that reveal the distinctive function of their source cells, such as tumor antigens (gp100, CEA, and Her2)37–39 or death receptors (FasL and TRAIL).40,41 Here, we report the potential functions of exosomes present in BrCa cells that can serve as putative common markers at the early stage. Table 1 lists a detailed brief overview of known BrCa cell-derived exosomal proteins reported by various scientists.
Table 1.
The Exosomal Proteins in Breast Cancer (1).
| Exosomal Protein | Body Fluid | Isolation/Detection Method | Application | Ref. |
|---|---|---|---|---|
| Metalloprotease ADAM10, tetraspanin CD9, HSP 70 and Annexin-1 | Breast cancer cell line culture supernatants, serum, pleural effusions | Western blot | Identification and highly expressed on breast cancer cell derived exosomes | Hernandez et al 42 |
| CD24, tetraspanins and epithelial cell adhesion molecule (EpCam) | Serum, ascites fluid | Ultracentrifugation and sucrose gradient/MACs using beads | Early diagnosis | Rupp et al 43 |
| Developmental endothelial locus-1 (Del-1) | Plasma | ELISA | Diagnosis/monitoring | Moon et al 44 |
| Fibronectin | Plasma | ELISA | Early diagnosis | Moon et al 45 |
| Survivin (Survivn 2B) | Serum | ExoQuick and ELISA | Diagnosis/prognosis | Khan et al.,15 |
| HER-2 | Serum, breast cancer cell line culture supernatants | Ultracentrifugation and Bradford assay | Trastuzumab Resistance and tumor aggressiveness | Ciravolo et al 46 |
| Annexin A2 | Breast cancer cell line culture supernatants | Atomic force microscopy and Western blot | Brain and lung metastasis | Maji et al 47 |
| P-glycoprotein/TrpC5/ABCG2 | MCF-7/S cell culture supernatants | Binding of exosomes to nonmalignant cells | Mlutidrug resistance | Jaiswal et al,48 Matula et al,49 and Ma et al 50 |
| Glutathione S-transferase P1(GSTP-1) | Cell, tissue, and serum | Cell apoptosis and immunofluorescence staining assays | Adriamycin-resistant | Yang et al 51 |
| Ubiquitin carboxyl terminal hydrolase-L1 | MCF-7 cell culture supernatants | Western blot and immunohistochemistry | Adriamycin resistance | Ning et al 52 |
ELISA, enzyme-linked immunosorbent assay.
Evidence from numerous experiments has shown that some of the tumor markers detected in the blood of patients with BrCa are carried by membrane vesicles53; this implies that biomarker research can also benefit from vesicle characterization. Recently, proteomic analysis has become the technique of choice for characterizing exosomes, as a wider knowledge of exosomal protein content can help in understanding the potential roles of exosomes in vivo and in identifying new tumoral markers with diagnostic value.54
Rupp and colleagues43 suggested that both CD24 and EpCAM from ascites and pleural effusions can serve as alternative BrCa-derived exosome markers; these were isolated using magnetic beads and detected by Western blot. Further, in the serum of patients with BrCa, CD24 alone could be detected in the exosomes, but EpCAM was lost; this indicated that exosomal CD24 may serve as a circulating BrCa biomarker. However, CD24 has been implicated in numerous cancer types, including colorectal cancer (CC), so it may serve as a general cancer marker and not a specific BrCa marker.55
BrCa Exosomal Protein as an Early Diagnostic and/or Prognostic Marker
Several exosomal proteins are differentially expressed in certain stages or types of BrCa and may be applied as diagnostic or prognostic markers for general cancer diagnosis. For instance, metalloprotease ADAM10, in addition to tetraspanin CD9, HSP70, and Annexin-1, was specifically expressed in serum/pleural effusion-derived exosomes from patients with BrCa or BrCa cell lines.42 Furthermore, the tetraspanin CD63, a binding partner of integrins and a tumor marker whose expression conversely correlates with cancer metastasis,56–62 and tumor susceptibility gene 101, which is a subunit of the endosomal sorting complex required for transport-1 (ESCRT-1),63 existed exclusively on exosomes.64
Moon and colleagues44,45 have demonstrated that both developmental endothelial Locus-1 (Del-1) and fibronectin on circulating exosomes from the plasma of patients with BrCa could serve as promising biomarkers at the early stage. In addition, Del-1 is also a promising marker to distinguish BrCa from benign breast tumors and noncancerous disease. Khan et al 15 found that the differential expression of exosomal Survivin-2B (proapoptotic protein) in the sera may be used as a diagnostic and/or prognostic marker in patients with early BrCa.
Breast Cancer Metastasis
Currently, the terms “metastatic niche” and “premetastatic niche” are widely accepted to describe regulation at secondary organs at distant sites before cancer cells arrive.65 Tumor cell-derived exosomes, as new critical components of metastasis and premetastatic niches, are able to promote survival and growth of the primary tumor.66 Afterward, they participate in communication with neighboring nontumor cells67 by regulating epithelial to mesenchymal transition,68 inducing local tumor invasion,69 impacting the immune system,70 promoting tumor vascular leakiness and facilitating circulating tumor cell access to distant sites,71,72 which accelerates the progression of the primary tumor. Furthermore, recent discoveries also proved that proteins in exosomes from BrCa cells can promote angiogenesis and organ-specific metastasis. When detected in exosomes, these proteins may imply the presence of tumor metastasis. Maji et al found that Annexin A2(Anx A2)47 in BrCa cell-derived exosomes is an emerging mediator of brain and lung-specific metastasis. Upon delineating the mechanism, it was found that exo-Anx A2 causes macrophage-mediated activation of the p38MAPK, nuclear factor kappa B, and STAT3 pathways and improved secretion of interleukin (IL)′-6 and tumor necrosis factor-α. In addition, Piao73 and colleagues have proven that proliferation of BrCa cell-derived exosomes could create a favorable condition for LN metastatic by stimulating macrophage polarization.
Multidrug Resistance in Exosomal Protein
According to reports, antigens presented in the BrCa exosomes may serve as targets for certain therapeutic antibodies and induce treatment failure by capturing the drugs. Ciravolo and colleagues46 found that exosomes extracted from HER2+ BrCa cell supernatants or the serum of patients with BrCa could bind to trastuzumab. Functional detection revealed that only xenogeneic and autologous HER2+ nanovesicles, but not HER2− ones, inhibited trastuzumab activity in SKBR3 cell proliferation. These findings suggested that HER2-positive exosomes could be a biomarker to indicate trastuzumab resistance and tumor aggressiveness. There are various clinical trials studying circulating HER2 levels as a potential predictor of the trastuzumab response, both as an initial indicator of drug response and as patients progress while on the drug. There was even an Food and Drug Administration-approved HER2 ELISA test developed, which would measure both exo-HER2 and solubilized forms in the extracellular domain. After thorough testing, it was shown that circulating HER2 is not informative, and this product is not used for clinical care.74
The overexpression of P-glycoprotein (P-gp) is associated with the multidrug resistance bioprocess.48 A recent study demonstrated that docetaxel-resistant BrCa cell-derived exosomes transferred P-gp to anticancer drug-sensitive tumor cells; this process is called tunneling nanotube-mediated transportation.49 The sequential research by Ma’s group50 revealed that transient receptor potential channel 5 (TrpC5) in exosomes may be responsible for the acquisition and formation of drug resistance in BrCa cells. At the same time, this transfer upregulates the expression of TrpC5 in recipient cells, thereby increasing the quantities of the drug efflux transporter P-gp by a Ca2+ and transcription factor nuclear factor of activated T-cell isoform c3 (NFATc3)-mediated mechanism.75,76 Currently, a novel multidrug efflux transporter, ATP-binding cassette subfamily G member 2 (ABCG2), is also related to drug resistance, including resistance to mitoxantrone and camptothecin analogs.77
Glutathione S-transferase P1 (GSTP1), a member of the phase II metabolic enzyme family, has been shown to function in detoxifying several anticancer drugs by conjugating them with glutathione. Yang et al 51 have analyzed the exosomal GSTP1 expression in BrCa cells and tissues by apoptosis assays and immunofluorescence staining techniques. Higher GSTP1 expression was observed in the serum of adriamycin-resistant cells and a progressive disease/stable disease group. This study proved for the first time that GSTP1-rich exosomes have the capability to transfer drug resistance and therefore could be helpful in predicting chemoresistance in clinical use.
Adriamycin-resistant human BrCa cells (MCF7/ADM) secrete exosomes carrying UCH-L1 and P-gp proteins into the extracellular microenvironment and are then integrated into adriamycin-sensitive human BrCa cells (MCF7/WT) in a time-dependent manner, thereby transferring the chemoresistance phenotype. Kuan Ning et al 52 have demonstrated that UCH-L1-containing exosomes can transfer chemoresistance to recipient cells, and these exosomes may be useful as noninvasive diagnostic biomarkers for the detection of chemoresistance in BrCa patients, thus achieving more effective and individualized chemotherapy. In conclusion, noninvasive detection of specific exosomal proteins may contribute to the development of new strategies in clinical applications for physicians.
Exosomal miRNA in BrCa Diagnosis
The microRNAs present in BrCa exosomes can serve as a novel ideal biomarker, and their expression pattern is correlated with the degree of tumor malignancy78–81 ( Table 2 ). Lee’s group82 used a nanosized oligonucleotide probe, molecular beacon (MB), to carry out in situ miRNA examination. The results further showed that exosomal miR-21 could be used as a biomarker when compared with MCF-7 exosomes and normal cell-derived exosomes. However, miR-21 as a potential biomarker was widely reported and may not only exist in the exosomes of BrCa.83 Other reports have disclosed that exosomal miR-21 exists in the serum, cerebrospinal fluid, cervicovaginal lavage specimens and peritoneal lavage fluid of CC,93 gastric cancer,94 ovarian cancer,95 pancreatic adenocarcinoma (PC),96 cervical cancer,97 and glioblastoma patients.98
Table 2.
The Exosomal miRNAs and lncRNA in Breast Cancer(1).
| Exosomal RNA | Body Fluid | Isolation/Detection Method | Application | Ref. |
|---|---|---|---|---|
| miR-21 | Plasma/MCF-7 cell culture supernatants | In situ nano-sized oligonucleotide probe | Diagnosis | Lee et al,82 and Shi et al 83 |
| miR-101, miR-372 | Serum | Quantitative TaqMan MicroRNA assays and correlated with clinicopathological risk factors | Diagnosis | Eichelser et al 84 |
| miR-373 | Serum | Quantitative TaqMan microRNA assays and correlated with clinicopathological risk factors | Diagnosis of Triple-negative breast cancer | Eichelser et al 84 |
| miR-1246 | Plasma andMDA-MB-231 cells | RT-qPCR | Diagnosis | Hannafon et al,85 and Li et al 86 |
| miR-105 | MCF-10A and MDA-MB-231 cell culture supernatants | RT-qPCR, western blot analysis, and immunofluorescence | Diagnosis | Zhou et al 87 |
| miR-10b | MDA-MB-231 cell culture supernatants | RT-qPCR | Cell invasion | Singh et al 88 |
| miR-338-3p, miR-340-5p, miR-124-3p, miR-29b-3p, miR-20b-5p, miR-17-5p, miR-130a-3p, miR-18a-5p, miR-195-5p, miR-486-5p and miR-93-5p | Human serum and tissues | miRNA PCR array | Prognostic | Aiko et al 89 |
| miR-222/miR-221 | MCF-7 cell culture supernatants | Fluorescence in situ hybridization | Adriamycin-resistant | Yu et al 16 |
| miR-451 | MCF-7 cell culture supernatants | Cell apoptosis and in situ nano-sized oligonucleotide probe | Increase the sensitivity of cell resistance to doxorubicin | Chen et al 90 |
| lncRNA GAS5 | MCF-7 and MDA-MB-231 cell culture supernatants | RT-qPCR | Marker of apoptosis induction | Oguz et al 91 |
| lncRNA MALAT1 | Human specimens and cell lines (MCF-7, MDA-MB-231, and MDA-MB-435 S) | qRT-PCR | Promoted cell proliferation | Zhang et al 92 |
Abbreviation: RT-qPCR, real-time quantitative polymerase chain reaction.
Breast Cancer Exosomal miRNA as a Biomarker
Eichelser et al 84 reported that BrCa-specific miRNAs are expressed in the serum exosomes of patients. They found that the level of exosomal miR-101 and miR-372 was significantly higher in the serum of patients with BrCa than in healthy controls; furthermore, the level of exosomal miR-373 was more elevated in triple-negative patients than in patients with luminal cancers or healthy controls. All the data indicate that exosomal miR-101 and miR-372 could be used as BrCa biomarkers, while exosomal miR-373 is indicative of a triple-negative phenotype.
Hannafon et al 85 showed that certain miRNA species, such as miR-21 and miR-1246, are ubiquitous in human exosomes and are distinctively elevated in patients with BrCa. Furthermore, their results support the concept that circulating exosome microRNA profiles can be used as an important diagnostic tool for BrCa, and the potential mechanism was further discovered by Li et al.86 The exosomal miR-1246 could suppress the expression level of Cyclin-G2 , indicating BrCa progression. In addition, exosome-encapsulated miRNAs could serve as a prognostic marker for metastatic progression in BrCa. Zhou et al 87 demonstrated that the level of serum exosomal miR-105 in BrCa-bearing animals increased swiftly in the premetastatic and metastatic stages; this implies that exosomal miR-105 has the potential to predict or diagnose BrCa when metastasis is developing or has already developed. Additionally, the level of exosomal miRNA-10b88 was higher in TNBC MDA-MB-231 cells, and cell invasion with the noninvasive mammary epithelial cell line HMLE was increased when MDA-MB-231-derived exosomal miRNA-10b was applied to the HMLE cells. In addition, Zhang etal99 have proven that miR19a is transferred via exosomes from astrocytes to mouse and human BrCa cells and mediates adaptive loss of tumor suppressor PTEN in brain metastatic tumor cells.
Recently, Aiko Sueta89 and his group detected 384 miRNAs derived from the exosomes of patients with BrCa and demonstrated that miR-338-3p, miR-340-5p, and miR124-3p were obviously upregulated and miR-29b-3p, miR-20b-5p, miR17-5p, miR-130a-3p, miR-18a-5p, miR-195-5p, miR-486-5p, and miR-93-5p were downregulated rapidly in the recrudescent patients. They provided evidence that these 11 exosomal miRNAs are correlated with BrCa recurrence and may be used as prognostic factors for BrCa prognosis. Several limitations existed in their study, including that there was no reliable endogenous gene for normalization, the period of storage for patient samples was not clear, and they utilized ExoQuick (System Biosciences, Palo Alto, CA, USA) to extract exosomes, which may mix nonexosomal contents with similar size. All of these limitations may cause differences in the miRNA expression pattern between exosomes and tumor tissues.
Multidrug Resistance in Exosomal miRNA
Presently, drug resistance still hinders the treatment of BrCa, causing BrCa to become one of the major deadly cancers in women. It is increasingly reported that exosomal miRNA (including miR-100, miR-222, and miR-30a)100 secreted by drug-resistant BrCa cells may participate in BrCa drug resistance by spreading resistance capacity and altering chemosusceptibility in recipient sensitive cells.
Tamoxifen resistance remains a daunting challenge to the successful treatment of ER+ BrCa. Yu and colleagues16 proved that exosomal miR-221/222 secreted from tamoxifen-resistant BrCa cells could downregulate its target genes of P27 and ERα proteins, which significantly stimulated drug-resistant cell proliferation and colony-forming ability in the presence of tamoxifen. Chen and colleagues90 showed that the disproportionately exported exosomal miRNA-451 from BrCa cells could integrate with mdr1 and downregulate the level of P-gp, leading to lower sensitivity to doxorubicin and poor treatment results.
The Potential Research Strategy of Exosomal miRNAs
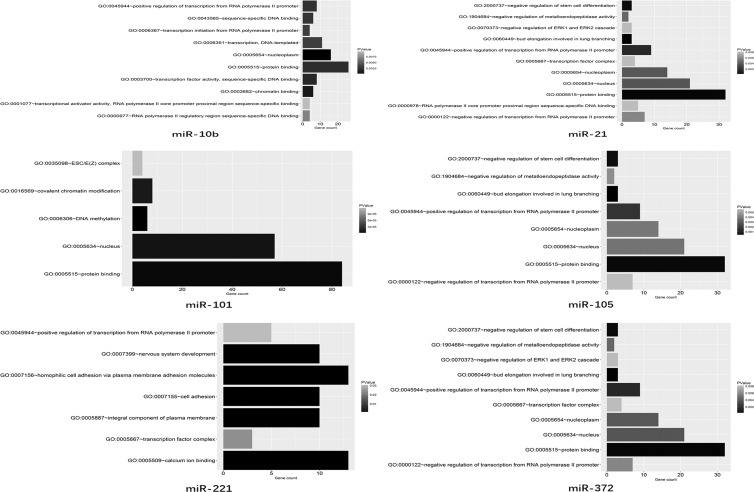
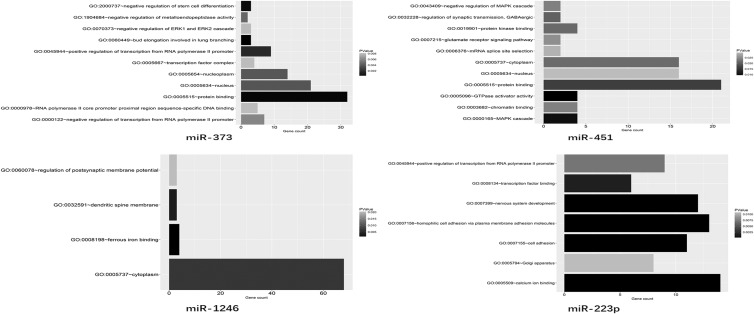
MiRNAs (miRs) are a major class of small noncoding RNAs with lengths varying between 18 and 25 nucleotides. Unlike the evolutionarily conserved long-noncoding RNAs (lncRNAs), miRs are numerous and involved in various genetic networks.101,102 Furthermore, miRs can mediate post-transcriptional gene silencing by binding to target mRNAsvia complementary sequences103 and then modulate mRNA and protein expression among various kinds of cancers.104 Recently, growing evidence has shown that miRscan be encapsulated in secreted exosomes and disseminate through the extracellular fluid to reach remote target cells.105,106 In this review, we utilized the TargetScan, miRanda, miRDB, and PicTar databases to predict targets of the BrCa cell-derived exosomal miRNAs. After screening for potential mRNAs, Gene Ontology enrichment analysis was applied to investigate the major functional categories represented in the aforementioned genes (Figure 3), which would benefit the in-depth research of exosomal miRs in BrCa and help locate the most efficient biomarkers for clinicians.
Figure 3.
Summary of the target gene of miRs. The potential function of the miRs (miR-21, miR-101, miR-105, miR-372, miR-373, miR-451, miR-1246, and miR-222-3p) encapsulated in the BrCa cell-derived exosomes.
Exosomal lncRNA in BrCa Diagnosis
Long-noncoding RNAs, at least 20 bp to 100 kb in length, play an important part in many fundamental cellular processes such as chromatin organization, gene transcription, mRNA turnover, and protein translation, and their deregulation is considered to contribute to carcinogenesis.107,108 In addition, exosomal lncRNAs may participate in the dissemination of cell signals to regulate local cellular microenvironments. Koldemir91 and his group found that growth arrest specific 5 (GAS5), an inhibitor of cell proliferation and promoter of apoptosis, accumulated in exosomes in BrCa cell lines (MCF-7 and MDA-MB-231 cell). In their research, they tracked exosomal GAS5 and assessed the efficacy of therapeutic interventions. The results proved that the accumulation of exosomal GAS5 may represent cell apoptosis and also represent a way to evaluate treatment with radiotherapy and many chemotherapeutic agents. The latest research also found that BrCa cell-derived exosomes could regulate the progression of BrCa by delivering the long-noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1),92 which may indicate that exosomal MALAT1 would be a novel strategy for overcoming BrCa.
Non-BrCa Cell-Derived Exosomes in BrCa Diagnosis
Exosomes are secreted into the extracellular space and efflux into the bloodstream, where they mediate intercellular communication in a very distant way. Hence, non-BrCa cells in this microenvironment will transfer their molecular characteristics to BrCa cells, and this information can serve as a potential source for detecting biomarkers ( Table 3 ). Mei Yang and colleagues109 verified that exosomal miRNAs may be transported from macrophages to BrCa cells under a co-culture system of IL-4-activated macrophages and BrCa cells. Macrophage-derived exosomal miR-223 was significantly elevated in co-cultivation with BrCa cells (SKBR3 and MDA-MB-231 cells) and regulated the invasiveness of BrCa cells.
Table 3.
Non-BrCa Cell Content Regulation.
| Exosomal Contents | Origin | Isolation/Detection Method | Application | Ref. |
|---|---|---|---|---|
| miR-233 | Human macrophages | RT-qPCR | Promotes breast cancer cell invasion | Yang et al 109 |
| The quantity of exosomes | Human mammary epithelial cell line(HMEC B42) | Nanosight LM10 analysis, western blot and PKH labeled | Regulate BrCa cell derived exosomes released | Riches et al 110 |
| ADAM10 | Fibroblast | Antibodies and qRT-PCR primers or primer/probe sets | Breast cancer cell mobilization and proliferation | Shimoda M et al,111,112 |
| miR-21, -378e, and -143 | Cancer-associated fibroblasts | qRT-PCR | Breast cancer epithelial-mesenchymal transition | Donnarumma et al 89 |
| miR-16 | Mesenchymal stem cells | qRT-PCR | Inhibition of angiogenesis | Lee et al 113 |
Abbreviation: RT-qPCR, real-time quantitative polymerase chain reaction.
Andrew Riches et al 110 demonstrated that labeled exosomes from either normal or BrCa cells are incorporated into both normal and cancer cells. Thus, there is a clear interaction of exosomes derived from the extracellular environment with tumor cells and normal cells. At the same time, exosomes from normal mammary epithelial cells markedly inhibit exosome release from BrCa cells. This represents a new regulatory pathway for the feedback control of exosome concentrations. It also seems likely that this is a generic mechanism and may be important for other tumor types.
Evidence suggesting that cancer-associated fibroblast (CAF)-derived exosomes may promote cancer cell motility and metastasis is supported in studies wherein ADAM10-rich exosomes were found to promote the activation of oncogenic signaling.111,112 Numerous studies demonstrated that the CAF-like cell state may determine the role of the tissue inhibitor of metalloproteinases family in the maintenance of the extracellular matrix. It must be emphasized that ADAM10-rich exosomes secreted by human CAFs could enhance cell motility and stimulate cancer cell growth through RhoA and the Notch signaling pathway. Tissue inhibitor of metalloproteinases suppress cancer stroma, while activated fibroblast-secreted exosomes influence tumor progression.
The level of communication between cells is leading to a greater appreciation of the implications of intratumoral heterogeneity and the role of the tumor microenvironment and vasculaturein cancer progression and metastasis.89 Exosomes are emerging as a crucial mediator of cell-to-cell communication in cancer, thus affecting BrCa progression. Three miRs increased evidently in exosomes from CAFs when compared with normal fibroblasts. Donnarumma et al 114 showed that CAF-secreted exosomal ongogenic miR (miR-21, -378e, and -143) could significantly increase the form of mammospheres, improve stem cell and epithelial-mesenchymal transition markers, and promote BrCa progression.
Mesenchymal stem cells (MSCs), which potentially differentiate into multiple cell types, have the capability of migrating to tumor sites and exerting complex effects on tumor progression. The potential mechanisms and roles within the tumor microenvironment have been elucidated by the scientists, such as immune modulation113,115 and regulation of the secreted factors of MSCs. Nevertheless, the paracrine effects of MSC-derived exosomes on the tumor microenvironment are unknown and remain to be explored. Jong-Kuen Lee et al 116 showed that MSC-derived exosomal miR-16 may decrease the expression of vascular endothelial growth factor extensively in tumor cells and subsequently inhibit angiogenesis in vitro and in vivo. Additionally, Koumangoye et al 117 proved that the detachment of adherent cells from various substrata may trigger a rapid and substantial secretion of exosomes, which aggregate on the cell surface and mediate adhesion to various extracellular matrix proteins. In brief, these data indicate that cellular detachment is accompanied by a significant release of exosomes, and at the same time, cellular adhesion and spreading are enhanced by the rapid uptake and disposition of exosomes on the cell surface.
Conclusion
The early detection of metastases and micro metastases using multiple exosomal biomarkers would be advantageous for the patient. The field of exosome detection is characterized by the exploration of novel candidate biomarkers and represents a growing field of research. In addition, a large amount of possible circulating indicators provides a new perspective for surgery-free cancer characterization, while realistically, the application of BrCa tissue-based samples represents now or in the near future an indispensable method for setting up valid genomic-based clinical considerations. More importantly, the quantity of circulating biomarkers is growing almost exponentially, and some of the biomarkers, such as miRs and certain proteins, demonstrate an intriguing ability to distinguish among different cancer subtypes. In BrCa, this profile deserves particular attention because of the high heterogeneity of carcinomas. Although clinical trial studies of exosomes in BrCa are little reported,118 there has been a clinical trial underway for 6 years that has been documented in the web site at https://clinicaltrials.gov (NCT01344109). Other clinical trials have studied the pathogenic and metastatic role of melanoma, colon cancer, lung cancer prostate cancer cell, and pancreatic cancer-derived exosomes (NCT02310451, NCT01294072, NCT03108677, NCT02702856, and NCT02393703), encouraging the use of exosome studies for BrCa. Despite promising results, different kinds of BrCa cell lines or non-neoplastic cell types are known to release different kinds of exosomes, and the high expense of isolation from body fluids has made the clinical application challenging. Additionally, a standard isolation protocol does not exist in the currently available articles. Nevertheless, we still hold that exosomes have opened new avenues in BrCa diagnosis and prognosis due to the minimal invasiveness of sampling and the stability of the bilipid membrane, which makes liquid biopsies repeatable. Additionally, in the development of immunotherapy for high immunogenicity of the most aggressive BrCa subvariants, a specific circulating biomarker might provide more comprehensive information on the immune characteristics of patients with BrCa, which would be extremely useful.
Abbreviations
- ABCG2
ATP-binding cassette subfamily G member 2
- BrCa
breast cancer
- CSF
cerebrospinal fluid
- CAF
cancer-associated fibroblast
- ER
estrogen receptor
- EVs
extracellular vehicles
- ESCRT-1
endosomal sorting complex required for transport-1
- GSTP1
glutathione S-transferase P1
- Hsp
heat-shock proteins
- IL
interleukin
- MB
molecular beacon
- miRNAs
microRNAs
- mRNAs
messenger RNAs
- NFATc3
nuclear factor of activated T-cell isoform c3
- P-gp
P-glycoprotein
- TrpC5
transient receptor potential channel 5.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research work was supported by Doctoral Scientific Research Foundation(201601412 to Yiming Meng) and Natural Science Foundation (20180550488 to Yiming Meng) of Liaoning Province and Excellent Talent Fund of Liaoning Province Cancer Hospital of Yiming Meng.
References
- 1. Cooney MA, Culletonquinn E, Stokes E. Current knowledge of pain after breast cancer treatment: a systematic review. Pain Manag Nurs. 2013;14(2):110–123. [DOI] [PubMed] [Google Scholar]
- 2. Ahmed M, Rubio IT, Klaase JM, Douek M. Surgical treatment of nonpalpable primary invasive and in situ breast cancer. Nat Rev Clin Oncol. 2015;12(2):645–663. [DOI] [PubMed] [Google Scholar]
- 3. Marotti JD, Muller KE, Tafe LJ, Demidenko E, Miller TW. P-Rex1 expression in invasive breast cancer in relation to receptor status and distant metastatic site. Int J Breast Cancer. 2017;2017:4537532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oehrlich NE, Spineli LM, Papendorf F, Park-Simon TW. Clinical outcome of brain metastases differs significantly among breast cancer subtypes. Oncol Lett. 2017;14(1):194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takahashi TA, Lee CI, Johnson KM. Breast cancer screening: does tomosynthesis augment mammography? Cleve Clin J Med. 2017;84(7):522–527. [DOI] [PubMed] [Google Scholar]
- 6. Grimm LJ, Shelby RA, Knippa EE, et al. Patient perceptions of breast cancer risk in imaging-detected low-risk scenarios and thresholds for desired intervention: a multi-institution survey. J Am Coll Radiol. 2018;15(6):911–919. doi:10.1016/j.jacr.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 7. Parise CA, Caggiano V. Risk of mortality of node-negative, ER/PR/HER2 breast cancer subtypes in T1, T2, and T3 tumors. Breast Cancer Res Treat. 2017;165(3):743–750. [DOI] [PubMed] [Google Scholar]
- 8. Wesseling J, Tinterri C, Sapino A, et al. An international study comparing conventional versus mRNA level testing (TargetPrint) for ER, PR, and HER2 status of breast cancer. Virchows Arch. 2016;469(3):297–304. [DOI] [PubMed] [Google Scholar]
- 9. Stahl PD, Barbieri MA. Multivesicular bodies and multivesicular endosomes: the “ins and outs” of endosomal traffic. Sciences Stke signal. Sci STKE. 2002;2002(141):pe32. [DOI] [PubMed] [Google Scholar]
- 10. Javeed N, Mukhopadhyay D. Exosomes and their role in the micro-/macro-environment: a comprehensive review. J Biomed Res. 2017;30(5):386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Human Mol Genet. 2012;21(1):125–134. [DOI] [PubMed] [Google Scholar]
- 12. Aursulesei V, Vasincu D, Timofte D, et al. New mechanisms of vesicles migration. Gen Physiol Biophys. 2016;35(3):287–298. [DOI] [PubMed] [Google Scholar]
- 13. Sun Y, Haglund TA, Rogers AJ, et al. Microfluidics technologies for blood-based cancer liquid biopsies. Anal Chim Acta. 2018;1012:10–29. [DOI] [PubMed] [Google Scholar]
- 14. Michael T, Lasek RJ, Harold G. Axonal maintenance, glia, exosomes, and heat shock proteins. F1000Res. 2016;5:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khan S, Bennit HF, Turay D, et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. J Clin Oncol. 2014;30(suppl 27):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu DD, Wu Y, Zhang XH, et al. Exosomes from adriamycin-resistant breast cancer cells transmit drug resistance partly by delivering miR-222. Tumour Biol. 2016;37(3):3227–3235. [DOI] [PubMed] [Google Scholar]
- 17. Suchorska WM, Lach MS. The role of exosomes in tumor progression and metastasis (Review).Oncol Reports. 2016;35(3):1237–1244. [DOI] [PubMed] [Google Scholar]
- 18. Hwa SY, Thalia N, Cao H, Lee J, Chung J. Emerging roles of exosomes in cancer invasion and metastasis. BMB Reports. 2016;49(1):18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meehan K, Vella LJ. The contribution of tumour-derived exosomes to the hallmarks of cancer. Crit Rev Clin Lab Sci. 2015;53(2):121–131. [DOI] [PubMed] [Google Scholar]
- 20. Siekevitz P. Biological membranes: the dynamics of their organization. Ann Rev Physiol. 1972;34:117–140. [DOI] [PubMed] [Google Scholar]
- 21. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. [DOI] [PubMed] [Google Scholar]
- 22. Groot Kormelink T, Mol S, de Jong EC, Wauben MHM. The role of extracellular vesicles when innate meets adaptive. Semin Immunopathol. 2018;40(5):439–452. doi:10.1007/s00281-018-0681.-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1–11. doi:10.1016/j.pharmthera.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 24. Kooijmans SAA, Schiffelers RM, Zarovni N, Vago R. Modulation of tissue tropism and biological activity of exosomes and other extracellular vesicles: new nanotools for cancer treatment. Pharmacol Res. 2016;111:487–500. [DOI] [PubMed] [Google Scholar]
- 25. Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6(4):287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geddings JE, Mackman N. Tumor-derived tissue factor-positive microparticles and venous thrombosis in cancer patients. Blood. 2013;122(11):1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tehrani M, Friedman TM, Olson JJ, Brat DJ. Intravascular thrombosis in central nervous system malignancies: a potential role in astrocytoma progression to glioblastoma. Brain Pathol. 2008;18(2):164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gomes FG, Sandim V, Almeida VH, et al. Breast-cancer extracellular vesicles induce platelet activation and aggregation by tissue factor-independent and -dependent mechanisms. Thromb Res. 2017;159:24–32. [DOI] [PubMed] [Google Scholar]
- 29. Tsoutsou PG, Vozenin MC, Durham AD, et al. How could breast cancer molecular features contribute to locoregional treatment decision making? Crit Rev Oncol Hematol. 2017;110:43–48. [DOI] [PubMed] [Google Scholar]
- 30. Reclusa P, Sirera R, Araujo A, et al. Exosomes genetic cargo in lung cancer: a truly Pandora’s box. TranslLung Cancer Res. 2016;5(5):483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hartmann A, Muth C, Dabrowski O, Krasemann S, Glatzel M. Exosomes and the prion protein: more than one truth. Front Neurosci. 2017;11:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar D, Manek R, Raghavan V, Wang KK. Protein characterization of extracellular microvesicles/exosomes released from cytotoxin-challenged rat cerebrocortical mixed culture and mouse N2a cells. Mol Neurobiol. 2017;(8):1–13. [DOI] [PubMed] [Google Scholar]
- 33. Vedeler A, Hollã SH, Grindheim AK, Raddum AM. Multiple roles of Annexin A2 in post-transcriptional regulation of gene expression. Curr Protein Pept Sci. 2012;13(4):401–412. [DOI] [PubMed] [Google Scholar]
- 34. An H, Wever OD. Rab27 GTPases distribute extracellular nanomaps for invasive growth and metastasis: implications for prognosis and treatment. Int J Mol Sci. 2013;14(5):9883–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Isola AL, Chen S. Exosomes: the link between GPCR activation and metastatic potential?. Front Genet. 2016;7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pitt JM, André F, Amigorena S, et al. Dendritic cell-derived exosomes for cancer therapy. J Clin Invest. 2016;126(4):1224–1232z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaput N, Schartz NE, André F, et al. Exosomes as potent cell-free peptide-based vaccine. II. Exosomes in CpG adjuvants efficiently prime naive Tc1 lymphocytes leading to tumor rejection. J Immunol. 2004;172(4):2137–2146. [DOI] [PubMed] [Google Scholar]
- 38. Ewaisha R, Gawryletz CD, Anderson KS. Crucial considerations for pipelines to validate circulating biomarkers for breast cancer. Expert RevProteomics. 2015;13(2):201–211. [DOI] [PubMed] [Google Scholar]
- 39. Nd KD, Kulkarni YM, Wu Y, et al. Inferring alterations in cell-to-cell communication in HER2+ breast cancer using secretome profiling of three cell models. Biotechnol Bioeng. 2014;111(9):1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warmoes M, Lam SW, van der Groep P, et al. Secretome proteomics reveals candidate non-invasive biomarkers of BRCA1 deficiency in breast cancer. Oncotarget. 2016;7(39):63537–63548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li A, Zhang T, Zheng M, Liu Y, Chen Z. Exosomal proteins as potential markers of tumor diagnosis. J Hematol Oncol. 2017;10(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galindo-Hernandez O, Villegas-Comonfort S, Candanedo F, et al. Elevated concentration of microvesicles isolated from peripheral blood in breast cancer patients. Arch Med Res. 2013;44(3):208–214. [DOI] [PubMed] [Google Scholar]
- 43. Rupp AK, Rupp C, Keller S, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122(2):437–446. [DOI] [PubMed] [Google Scholar]
- 44. Moon PG, Lee JE, Cho YE, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res. 2016;22(7):1757–1766. [DOI] [PubMed] [Google Scholar]
- 45. Moon PG, Lee JE, Cho YE, et al. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget. 2016;7(26):40189–40199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ciravolo V, Huber V, Ghedini GC, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227(2):658–667. [DOI] [PubMed] [Google Scholar]
- 47. Maji S, Chaudhary P, Akopova I, et al. Exosomal Annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res Mcr. 2017;15(1):93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ritu J, Frederick L, Dalla PV, Grau GE, Bebawy M. Breast cancer-derived microparticles display tissue selectivity in the transfer of resistance proteins to cells. Plos One. 2013;8(4):e61515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matula Z, Németh A, Lőrincz P, et al. The role of extracellular vesicle and tunneling nanotube-mediated intercellular cross-talk between mesenchymal stem cells and human peripheral T cells. Stem Cells Dev. 2016;25(23):1818. [DOI] [PubMed] [Google Scholar]
- 50. Ma X, Chen Z, Hua D, et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc Natl Acad Sci U S A. 2014;111(17):6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang SJ, Wang DD, Jian L, et al. Predictive role of GSTP1-containing exosomes in chemotherapy-resistant breast cancer. Gene. 2017;623:5–14. [DOI] [PubMed] [Google Scholar]
- 52. Ning K, Wang T, Sun X, et al. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J Surgical Oncol. 2017;115(8):932–940. [DOI] [PubMed] [Google Scholar]
- 53. Rivoltini L, Chiodoni C, Squarcina P, et al. TNF-related apoptosis-inducing ligand (TRAIL)-armed exosomes deliver pro-apoptotic signals to tumor site. Clin Cancer Res. 2016;22(14):3499. [DOI] [PubMed] [Google Scholar]
- 54. Chen W, Jiang J, Xia W, Huang J. Tumor-related exosomes contribute to tumor-promoting microenvironment: an immunological perspective. J Immunol Res. 2017;2017:1073947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamkovich SN, Yunusova NV, Stakheeva MN, et al. Isolation and characterization of exosomes from blood plasma of breast cancer and colorectal cancer patients. BiomedKhim. 2017;63(2):165–169. [DOI] [PubMed] [Google Scholar]
- 56. Yang X, Kovalenko OV, Tang W, Claas C, Stipp CS, Hemler ME. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J Cell Biol. 2004;167(6):1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Radford KJ, Thorne RF, Hersey P. Regulation of tumor cell motility and migration by CD63 in a human melanoma cell line. J Immunol. 1997;158(7):3353–3358. [PubMed] [Google Scholar]
- 58. Berditchevski F, Bazzoni G, Hemler ME. Specific association of CD63 with the VLA-3 and VLA-6 integrins. J Biol Chem. 1995;270(30):17784–17790. [DOI] [PubMed] [Google Scholar]
- 59. Kwon MS, Shin SH, Yim SH, et al. CD63 as a biomarker for predicting the clinical outcomes in adenocarcinoma of lung. Lung Cancer. 2007;57(1):46–53. [DOI] [PubMed] [Google Scholar]
- 60. Pols MS, Klumperman J. Trafficking and function of the tetrapanin CD63. Experimental Cell Res. 2009;315(9):1584–1592. [DOI] [PubMed] [Google Scholar]
- 61. Jung KK, Liu XW, Chirco R, Fridman R, Kim HR. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006;25(17):3934–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chirco R, Liu XW, Jung KK, Kim HR. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006;25(1):99–113. [DOI] [PubMed] [Google Scholar]
- 63. Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119(15):3025–3032. [DOI] [PubMed] [Google Scholar]
- 64. Menck K, Klemm F, Gross JC, Pukrop T, Wenzel D, Binder C. Induction and transport of Wnt 5a during macrophage-induced malignant invasion is mediated by two types of extracellular vesicles. Oncotarget. 2013;4(11):2057–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30(6):836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maia J, Caja S, Strano Moraes MC, et al. Exosome-based cell-cell communication in the tumor microenvironment. Front Cell DevBiol. 2018;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tauro BJ, Mathias RA, Greening DW, et al. Oncogenic H-ras reprograms Madin-Darby canine kidney (MDCK) cell-derived exosomal proteins following epithelial-mesenchymal transition. Mol Cell Proteomics. 2013;12(8):2148–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Casadei L, Calore F, Creighton CJ, et al. Exosome-Derived miR-25-3p and miR-92a-3p Stimulate Liposarcoma Progression. Cancer Res. 2017;77(14):3846–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boyiadzis M. The emerging roles of tumor-derived exosomes in hematological malignancies. Leukemia. 2017;31(6):1259–1268. [DOI] [PubMed] [Google Scholar]
- 71. Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2001;21(2):139–146. [DOI] [PubMed] [Google Scholar]
- 73. Yin JP, Kim HS, Hwang EH, Woo J, Zhang M, Moon WK. Breast cancer cell-derived exosomes and macrophage polarization are associated with lymph node metastasis. Oncotarget. 2018;9(7):7398–7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tchou J, Lam L, Li YR, Edwards C, Ky B, Zhang H. Monitoring serum HER2 levels in breast cancer patients. Springerplus. 2015;4:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dong Y, Pan Q, Jiang L, et al. Tumor endothelial expression of P-glycoprotein upon microvesicular transfer of TrpC5 derived from adriamycin-resistant breast cancer cells. Biochem Biophys Res Commun. 2014;446(1):85–90. [DOI] [PubMed] [Google Scholar]
- 76. Xin M, Yanfei C, Dongxu H, et al. Transient receptor potential channel TRPC5 is essential for P-glycoprotein induction in drug-resistant cancer cells. Proc Natl Acad Sci USA. 2012;109(40):16282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kong JN, He Q, Wang G, et al. Guggulsterone and bexarotene induce secretion of exosome-associated breast cancer resistance protein and reduce doxorubicin resistance in MDA-MB-231 cells. Int J Cancer. 2015;137(7):1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tetta C, Ghigo E, Silengo L, Deregibus MC, Camussi G. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine. 2013;44(1):11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yan S, Han B, Gao S, et al. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget. 2017;8(36):60149–60158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Weidle UH, Dickopf S, Hintermair C, Kollmorgen G, Birzele F, Brinkmann U. The role of micro rnas in breast cancer metastasis: preclinical validation and potential therapeutic targets. Cancer Genomics Proteomics. 2018;15(1):17–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. He Y, Deng F, Yang S, et al. Exosomal microRNA: a novel biomarker for breast cancer. Biomarkers Med. 2018;12(2):177–188. [DOI] [PubMed] [Google Scholar]
- 82. Ji HL, Kim JA, Min HK, Kang JY, Rhee WJ. In situ, single step detection of exosome microRNA using molecular beacon. Biomaterials. 2015;54:116–125. [DOI] [PubMed] [Google Scholar]
- 83. Shi J. Considering exosomal miR-21 as a biomarker for cancer. Ochiya T, Takahashi R, eds. J Clin Med. 2016;5(4):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Eichelser C, Stückrath I, Müller V, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5(20):9650–9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li XJ, Ren ZJ, Tang JH, Yu Q. Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem. 2017;44(5):1741–1748. [DOI] [PubMed] [Google Scholar]
- 87. Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer. 2014;13(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sueta A, Yamamoto Y, Tomiguchi M, Takeshita T, Yamamoto-Ibusuki M, Iwase H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget. 2017;8(41):69934–69944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen WX, Zhong SL, Ji MH, et al. MicroRNAs delivered by extracellular vesicles: an emerging resistance mechanism for breast cancer. Tumor Biol. 2014;35(4):2883–2892. [DOI] [PubMed] [Google Scholar]
- 91. Koldemir O, Özgür E, Gezer U. Accumulation of GAS5 in exosomes is a marker of apoptosis induction. Biomed Rep. 2017;6(3):358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang P, Zhou H, Lu K, Lu Y, Wang Y, Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9(4):e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tokuhisa M, Ichikawa Y, Kosaka N, et al. Exosomal miRNAs from peritoneum lavage fluid as potential prognostic biomarkers of peritoneal metastasis in gastric cancer. Plos One. 2015;10(7):e0130472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. [DOI] [PubMed] [Google Scholar]
- 96. Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu J, Sun H, Wang X, et al. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int J Mol Sci. 2014;15(1):758–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Akers JC, Ramakrishnan V, Kim R, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastoma biomarker development. Plos One. 2013;8(10):e78115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang L, Zhang S, Yao J, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen WX, Liu XM, Lv MM, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of MicroRNAs. Plos One. 2014;9(4):e95240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signaling. 2010;3(107):ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gomes AQ, Nolasco S, Soares H. Non-Coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14(8):16010–16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Croston TL, Lemons AR, Beezhold DH, Green BJ. MicroRNA Regulation of Host Immune Responses following Fungal Exposure. Front Immunol. 2018;9:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lucas K, Raikhel AS. Insect MicroRNAs: biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol. 2013;43(1):24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14(7):14240–14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Translational Med. 2015;13:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. Embo J. 2012;31(3):522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. [DOI] [PubMed] [Google Scholar]
- 109. Yang M, Chen J, Su F, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells—a new regulatory pathway. EurJ Cancer. 2014;50(5):1025–1034. [DOI] [PubMed] [Google Scholar]
- 111. Shimoda M, Jackson HW, Khokha R. Tumor suppression by stromal TIMPs. Mol Cell Oncol. 2016;3(3):e975082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shimoda M, Principe S, Jackson HW, et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat Cell Biol. 2014;16(9):889–901. [DOI] [PubMed] [Google Scholar]
- 113. Liao Y, Lei J, Liu M, et al. Mesenchymal Stromal Cells Mitigate Experimental Colitis via Insulin-like Growth Factor Binding Protein 7-mediated Immunosuppression. Mol Ther. 2016;24(10):1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Donnarumma E, Fiore D, Nappa M, et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget. 2017;8(12):19592–19608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kasoju N, Wang H, Zhang B, et al. Transcriptomics of human multipotent mesenchymal stromal cells: Retrospective analysis and future prospects. Biotechnol Adv. 2017;35(4):407–418. [DOI] [PubMed] [Google Scholar]
- 116. Lee JK, Park SR, Jung BK, et al. Exosomes Derived from Mesenchymal Stem Cells Suppress Angiogenesis by Down-Regulating VEGF Expression in Breast Cancer Cells. Plos One. 2013;8(12):e84256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Koumangoye RB, Sakwe AM, Goodwin JS, Patel T, Ochieng J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. Plos One. 2011;6(9):e24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Domenyuk V, Zhong Z, Stark A, et al. Plasma exosome profiling of cancer patients by a next generation systems biology. Sci Rep. 2017;7:42741. [DOI] [PMC free article] [PubMed] [Google Scholar]