Abstract
Lung adenocarcinoma is one of the most common cancers worldwide. However, the molecular mechanisms of lung adenocarcinoma development are still unclear. This study aimed to investigate the expression profiles of anti-lung cancer target genes in different cancer stages and to explore their functions in tumor development. Lung adenocarcinoma transcriptome and clinical data were downloaded from Genomic Data Commons Data Portal, and the anti-lung cancer target genes were retrieved from the Thomson Reuters Integrity database. The results showed that 16 anti-lung target genes were deregulated in all stages. Among these target genes, fibroblast growth factor 22 showed the most important role in transcription regulatory networks. Further analysis revealed that APC, BRIP1, and PTTG1 may regulate fibroblast growth factor 22 and subsequently influence MAPK signaling pathway, Rap1 signaling pathways, and other tumorigenic processes in all stages. Moreover, high fibroblast growth factor 22 expression leads to poor overall survival (hazard ratio = 1.55, P = .019). These findings provide valuable information for the pathological research and treatment of lung adenocarcinoma. Future studies are needed to verify these results.
Keywords: lung adenocarcinoma, anti-lung cancer target genes, gene expression, overall survival, FGF22
Introduction
Lung cancer is one of the most common malignancies in humans and is the main cause of cancer-related death.1 Lung cancer is also an age-related disease. A large US study from 316 682 patients with lung cancer showed that 47% of patients with lung cancer are aged 70 years or older.2 Non-small cell lung cancer (NSCLC), which accounts for approximately 80% of all lung cancer, can be divided into different histological types, including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, of which lung adenocarcinoma (LUAD) is the most common subtype.3 Although significant progress has been made in surgery, chemotherapy, radiation therapy, and molecular targeted therapy in the past few years, the overall 5-year survival rate of patients with lung cancer is still only approximately 15%,4 which is due to the lack of effective early diagnosis methods and the limited efficacy of current therapies. The importance of finding simple and effective biomarkers is not only reflected in diagnosis but also in improving the prognosis of patients with lung cancer.
The American Joint Committee on Cancer (AJCC) lung cancer staging system provides important information for treatment and prognosis. According to the AJCC eighth edition, lung cancer can be divided into 4 stages (stage I-IV) based on various factors, including the size of the tumor, the status of lymph nodes, and metastasis.5 Determining the lung cancer stage is important because it may help determine the best method to understand how far the disease has progressed as well as to contain and eliminate the lung cancer. The treatment of lung cancer depends partly on the stage of the disease, especially for targeted therapy. During lung cancer progression, diverse genetic signatures have been identified to drive this process in genome, transcriptome, and epigenome remodeling.6-8 Therefore, it is necessary to select the right treatment options for patients with different stages.
Currently, many lung cancer risk genes and pathways have been identified, and this information will aid the pathological mechanism research and clinical treatment of lung cancer. Angiogenesis is considered essential for tumor growth and propagation. Vascular endothelial growth factor (VEGF) is a key mediator promoting this process. Recent studies have suggested that all lung cancers aberrantly express VEGF and that LUADs have the highest VEGF expression, which increases tumor aggressiveness and worsens prognosis.9 Through transcriptome network analysis, researchers have found that SPI1, FLI1, FOS, ETS2, EGR1, and PPARG are hub linked to many lung cancer pathways, suggesting that these genes may serve as potential target genes for squamous lung cancer.10 Furthermore, a recent study has shown that gene polymorphisms of some interleukins correlate with prognosis in NSCLC and has identified that IL16 rs7170924 and IL12A rs662959 may act as prognostic factors in patients with NSCLC.11
Thomson Reuters Integrity is a knowledge solution that integrates massive biology, chemistry, and pharmacology data (https://thomsonreutersintegrity.com) and it contains exhaustive therapeutic drug and gene target information for hundreds of human complex diseases. To our knowledge, no study has compared the expression profiles of anti-lung cancer target genes at different tumor stages. Gene expression profiling data sets in diverse stages of lung cancer have been deposited and are available in public databases, such as Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/). In this study, we aimed to identify key anti-lung cancer target genes and to explore the similarities and differences in gene expression change, pathway/biological function damage, and transcriptional regulation abnormalities in different stages of LUAD.
Methods
Data Sources
Lung adenocarcinoma transcriptome and clinical data were downloaded from the GDC Data Portal in September 2017. The data included 533 patients with LUAD and 59 controls, and there were complete transcriptome and clinical data for 513 patients. All samples were RNA sequenced using the Illumina HiSeq 2000 platform (version 2) and were normalized by the fragments per kilobase of exon per million reads mapped method. There were 274, 121, 84 and 26 patients with LUAD at stage I, stage II, stage III, and stage IV, respectively. In order to evaluate and verify the analysis results, we downloaded lung squamous cell carcinoma (LUSC) transcriptome and clinical data from GDC Data Portal and a microarray data set containing 442 patients with LUAD from NCBI-GEO (GSE72094,12 http://ncbi.nlm.nih.gov/geo). Anti-lung cancer target genes were defined as those that had marketed drugs or drugs under development targeting a specific gene. All target genes were searched and downloaded from the Thomson Reuters Integrity Database. In total, we obtained a list of 277 anti-lung cancer target genes.
Differential Expression Gene Analysis
Differential expression gene analysis was performed using R v3.2.2 (https://www.r-project.org/) and Bioconductor Library (http://www.bioconductor.org/). The empirical Bayes algorithm (function “eBayes”) in the limma package13 was used to detect differentially expressed genes between patients with LUAD and controls. A logarithmic transformation (log2) of all gene expression values was performed. Logarithmic transformed fold-change (log2(FC)) of each gene was calculated as the mean expression value in patients minus the mean expression value in controls. Upregulated genes were considered as a log2(FC) ≥1 and a false discovery rate (FDR) adjusted P value ≤.05. Downregulated genes were considered as log2(FC) ≤−1 and a FDR P value ≤.05. The differential expression analysis was performed in the whole cohort and subgroups (stage I, stage II, and stage III).
Transcriptional Regulation Network Analysis
Human transcription factors (TFs) were collected from the Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST) version 2.0 web server (http://www.grnpedia.org/trrust/). TRRUST is a manually curated database of human and mouse transcriptional regulatory networks.14 All human TFs were downloaded from TRRUST, and 751 TFs were mapped to our data. Because the transcriptional regulation information of multiple target genes is not recorded in the database, the coexpression method was used to construct transcriptional regulatory networks of deregulated TFs and deregulated anti-lung cancer genes in each stage. The correlation coefficients of these TFs and target genes were calculated in each stage, and the TF-target pairs with absolute value of correlation coefficients ≥0.5 and FDR P values ≤.05 were selected to construct the transcriptional regulatory network. By definition, the more nodes a target gene contains, the more important its role in the network. Therefore, key anti-lung target genes were screened based on the number of nodes.
Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis
All human reference gene and pathway information was downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/). The KEGG pathway enrichment analysis was performed using a deregulated gene list in each stage. The following formula was used to conduct the enrichment analysis: , where N is the number of all genes in the data; m represents the number of deregulated genes in the data; n is the number of all genes in the enriched KEGG pathway; and k is the number of deregulated genes in the KEGG pathway. A P value ≤.05 was considered significantly enriched.
Survival Analyses
All survival analyses were conducted using the “survival” package in R. Target genes that were deregulated in all stages were screened for survival analysis. Each gene was divided into 2 groups (high and low) according to the median expression value. Kaplan-Meier survival curves were used to show the difference between high and low groups of each target gene with regard to the prognosis of the patient. Cox proportional hazards model was used to explore the association of these target genes and patient prognosis in the whole cohort. A P value ≤.05 was considered significant.
Results
Overview of Deregulated Genes and Anti-Lung Cancer Target Genes
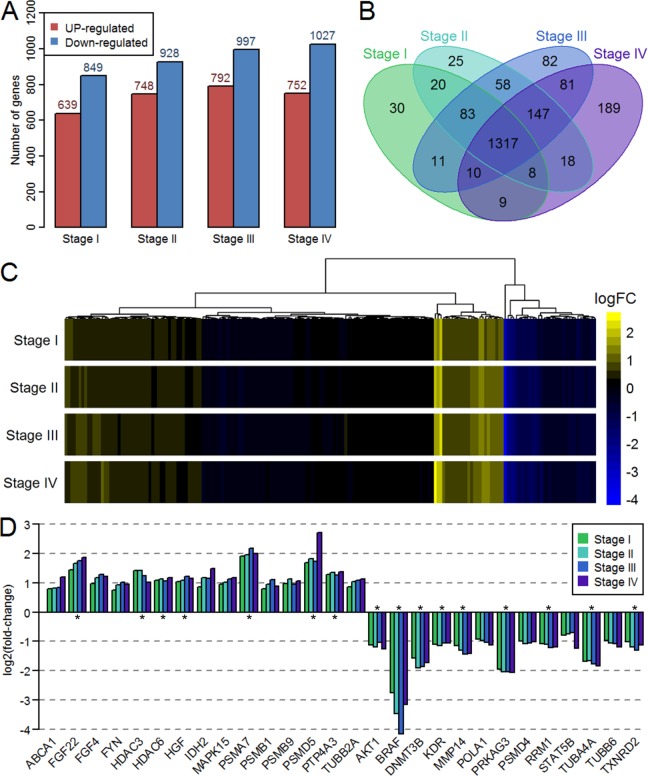
Through differential expression analysis, 1488, 1676, 1789, and 1779 deregulated genes were obtained in stage I, stage II, stage III, and stage IV, respectively. As the disease progressed, the number of differentially expressed genes increased (Figure 1A). A Venn diagram showed that there were 1317 deregulated genes shared by all 4 stages (Figure 1B). Because the number of samples in each stage (274, 121, 84, and 26) vary dramatically, we calculated the deregulated genes in combined stage III and IV samples and then compared to the separate stages analysis. The results showed that more than 85% commonly deregulated genes in separate and combined analysis (Supplementary Figure 1). This suggested that the analyses were unbiased to such sample size variation. The anti-lung target genes were mapped to our data set, and 190 unique target genes were obtained. Figure 1C shows the log2(FC) of these target genes in the 4 stages. There were 16, 23, 25, and 26 deregulated genes in stage I, stage II, stage III, and stage IV, respectively. Moreover, there were 15 upregulated genes and 13 downregulated genes in at least one stage. There were 16 target genes that were deregulated in all stages (Figure 1D).
Figure 1.
Expression profiles of deregulated genes and anti-lung cancer target genes at different stages. A, Number of upregulated and downregulated genes at 4 stages. B, Venn diagram of deregulated genes at 4 stages. C, Logarithmic transformed fold-change of all anti-lung cancer target genes at 4 stages. D, Expression change in anti-lung cancer target genes that were deregulated in at least one stage. The asterisk indicates that the gene is differentially expressed at all stages.
Transcriptional Regulation Networks and Key Target Genes
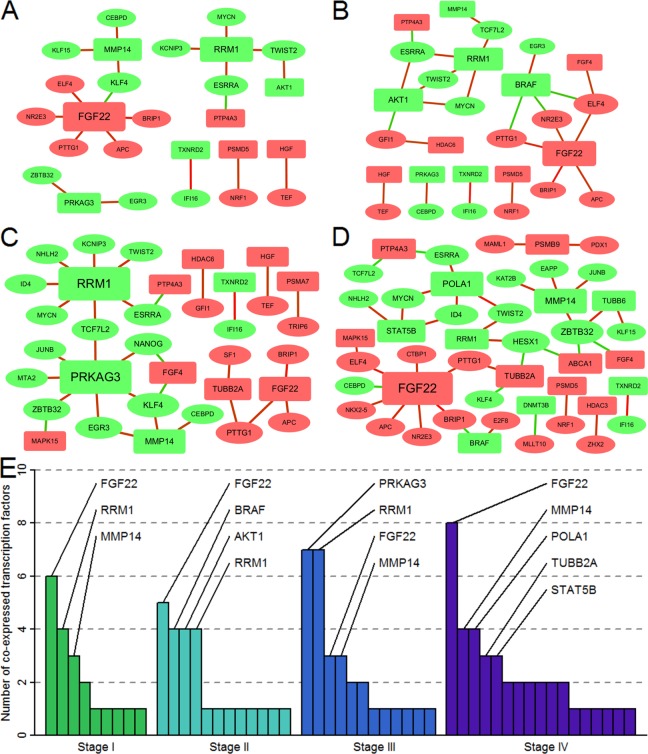
We obtained 49, 53, 58, and 62 significantly deregulated TFs in stage I, stage II, stage III, and stage IV, respectively. The transcription regulatory network of these TFs and target genes is shown in Figure 2. The number of interactions was 20, 25, 30, and 40 in stage I, stage II, stage III, and stage IV, respectively (Figure 3A-D). There were differences in the correlation between TFs and target genes at different stages of the disease. The highest number of TFs coexpressed with fibroblast growth factor 22 (FGF22) was found in stage I, stage II, and stage IV. However, in stage III, PPKAG3 and RRM1 showed the highest number of coexpressed TFs (Figure 3E). According to the number of nodes of target genes in these 4 stages, FGF22 was defined as a key target gene. Furthermore, we found there were 3 TFs that were significantly positive correlated with FGF22 in all stages: APC, BRIP1, and PTTG1.
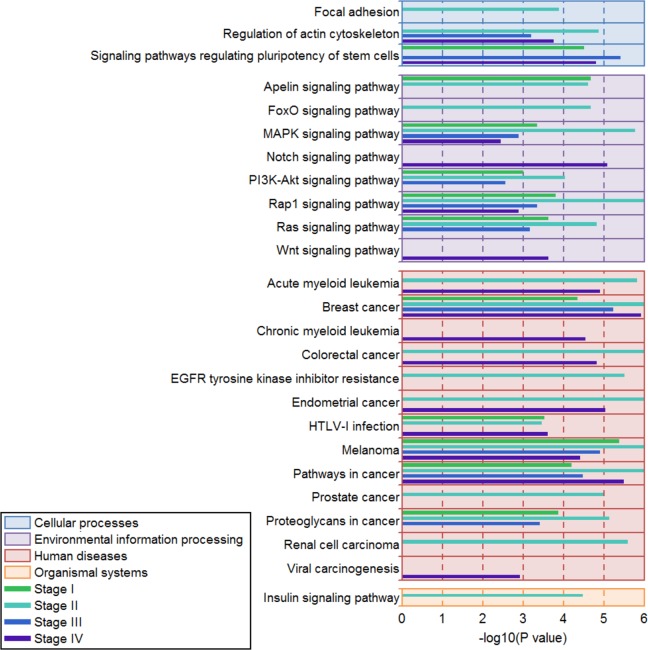
Figure 2.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment in 4 different stages. The figure shows only significantly enriched KEGG pathways and pathways with enriched genes ≥3. Color boxes indicate the type of KEGG pathway. Each stage is represented by a different colored line.
Figure 3.
Transcriptional regulation relationships of anti-lung cancer target genes at different stages. The ellipses represent transcription factors and the rounded rectangles represent target genes. The red and green shapes represent the upregulated and downregulated genes, respectively. The red and green lines indicate the positive correlation and negative correlation, respectively. A, Screened transcriptional regulatory network in stage I. B, Screened transcriptional regulatory network in stage II. C, Screened transcriptional regulatory network in stage III. D, Screened transcriptional regulatory network in stage IV. E, Number of coexpressed transcription factors in anti-lung cancer target genes at different stages.
Enriched KEGG Pathways in the 4 Stages
We used the genes in the transcription regulatory networks to perform KEGG pathway enrichment analysis in all stages. The results showed that multiple signal transduction pathways and carcinogenic pathways were significantly enriched in all stages (Figure 2). There were 11, 20, 10, and 15 significantly enriched KEGG pathways in stage I, stage II, stage III, and stage IV, respectively. According to the pathway information provided by the KEGG website (http://www.kegg.jp/), these pathways were divided into the following 4 categories: cellular processes, environmental information processing, human diseases, and organismal systems. The results showed that 5 pathways were significantly enriched in all stages: MAPK signaling pathway, Rap1 signaling pathway, breast cancer, melanoma, and pathways in cancer. These data suggested that FGF22 and related TFs mainly influence signal transduction function and cancer-related pathways.
Expression and Correlation of FGF22, APC, BRIP1, and PTTG1
The expression of FGF22, APC, BRIP1, and PTTG1 in controls and 4 tumor stages is shown in Figure 4A to D. The results showed that these genes were all upregulated in 4 stages compared to the controls. Linear correlation between FGF22 and other 3 genes is shown in Figure 4E to G. Furthermore, we found that BRIP1 was most correlated with FGF22 (r = 0.886; Figure 4E), followed by PTTG1 (r = 0.765; Figure 4F) and APC (r = 0.713; Figure 4G). Therefore, we speculate that FGF22 was positively regulated by APC, BRIP1, and PTTG1.
Figure 4.
Expression and correlation of fibroblast growth factor 22 (FGF22), APC, BRIP1, and PTTG1. A, The expression of FGF22 in controls and 4 tumor stages. B, The expression of APC in controls and 4 tumor stages. C, The expression of BRIP1 in controls and 4 tumor stages. D, The expression of PTTG1 in controls and 4 tumor stages. E, Correlation between FGF22 and APC in whole cohort. F, Correlation between FGF22 and BRIP1 in whole cohort. G, Correlation between FGF22 and PTTG1 in whole cohort. The difference in gene expression between patients and controls was calculated by Student t test. We labeled log2(fold-change) between patients and controls and the statistical significance. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
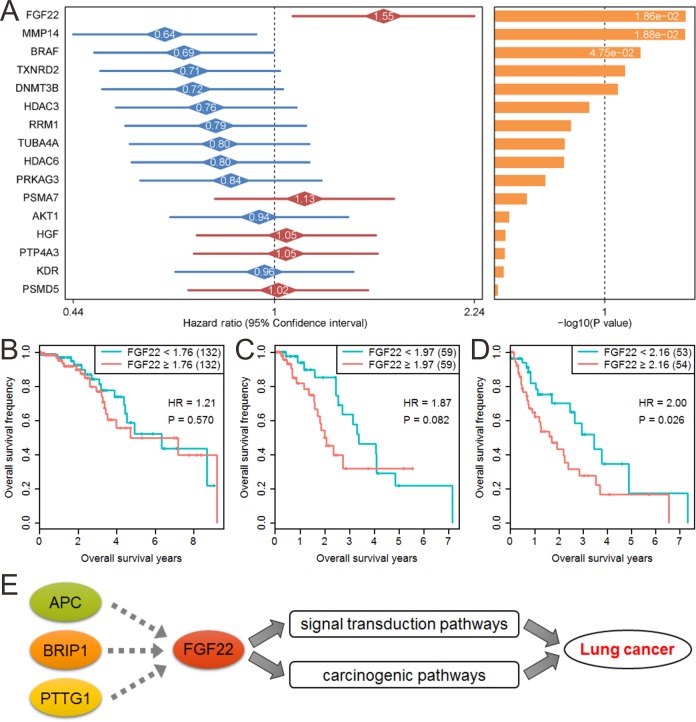
Effect of FGF22 on Prognosis and Its Potential Regulatory Mechanism
The target genes that were deregulated in all stages were used to perform survival analysis. Figure 5A shows the effect of these genes on patients’ overall survival. Among these 16 screened target genes, FGF22 showed the highest significance with survival status (hazard ratio [HR] = 1.55, P = .019). We further analyzed the effect of FGF22 on patient survival in different stages. Due to the smaller number of patients in stage IV, we used stage I (274) versus II (121) versus III + IV (110) for patients’ overall survival evaluation. The results showed that FGF22 has no effect on survival in stage I (HR = 1.21, P = .570; Figure 5B), a boundary significance in stage II (HR = 1.87, P = .082; Figure 5C), and a significant effect in stage III and IV (HR = 2.00, P = .026; Figure 5D). Overall, with the increase of stage, the effect of FGF22 on patients’ prognosis is more significant. We also evaluated the effect of FGF22 on LUSC patients’ overall survival. However, overall analysis or staged analysis all showed no significance (Supplementary Figure 2). Previous analysis showed that FGF22 may positively regulated by APC, BRIP1, and PTTG1. And survival analysis also showed that the high expression of APC, BRIP1, and PTTG1 reduce patients’ overall survival in total patients (Supplementary Figures 3-5). We also used a validation data set containing 442 patients with LUAD to verify the effect of FGF22 and BRIP1 on patients’ prognosis. The results showed that the high expression of FGF22 and PTTG1 reduces patients’ overall survival (HR = 1.59, P = .016 for FGF22; HR = 1.61, P = .013 for PTTG1), and the high expression of BRIP1 also showed boundary significance on patients’ overall survival, whereas the low expression of APC reduces patients’ overall survival (Supplementary Figure 6). Most of these results are consistent with the present study. Therefore, combined with previous transcriptional regulatory relationships and KEGG pathway enrichment results, we speculated that elevated APC, BRIP1, and PTTG1 leads to high FGF22 expression, thereby influencing multiple signal transduction pathways and carcinogenic pathways, ultimately exacerbating the progress of lung cancer (Figure 5E).
Figure 5.
The effect of fibroblast growth factor 22 (FGF22) on prognosis and its potential regulatory mechanism. A, Effect of significantly deregulated genes in all stages on patients’ overall survival. B, Effect of FGF22 on patients’ overall survival at stage I. C, Effect of FGF22 on patients’ overall survival at stage II. D, Effect of FGF22 on patients’ overall survival at stage III and stage IV. E, Potential regulatory mechanism of APC, BRIP1, PTTG1, and FGF22.
Discussion
The present findings demonstrated that as lung cancer progresses, the number of deregulated TFs and anti-lung cancer target genes increases. The complexity of the transcription regulatory network was increased with the disease progress. By combining the target genes with the maximum number of coexpressed TFs and prognostic-related genes, FGF22 was identified to serve as a potential therapeutic target and prognostic factor for patients with LUAD.
Several anti-lung cancer target genes were deregulated in all stages. These genes play crucial roles in the progression of lung cancer. The rat sarcoma (RAS)–rapidly accelerated fibrosarcoma (RAF)–mitogen-activated protein/extracellular signal-regulated kinase (MEK)–extracellular signal-regulated kinase (ERK) mitogen-activated protein kinase pathway is one of the most important canonical cancer signaling pathways, mediating cellular responses to growth signals essential for cell proliferation and survival.15 BRAF is a member of the serine/threonine kinase RAF family and is regulated by binding to RAS and directly activating MEK1/2, which can further phosphorylate ERK1/2. In the present study, BRAF showed the greatest maximum reduction among all deregulated target genes. Studies have shown that mutations in BRAF affect patient prognosis and have identified multiple risk sites.16 PSMA7 encodes α subunit 7 of the proteasome. A previous study reported that PSMA7 inhibits the proliferation, tumorigenicity, and invasion of A549 human LUAD cells in vitro, suggesting that PSMA7 is a potential tumor biomarker.17 In this study, PSMA7 was upregulated in all stages, which indicated that PSMA7 may be a protective factor for tumors. In the developing nervous system, AKT is a critical mediator of growth factor-induced neuronal survival. AKT1 and the related AKT2 are activated by platelet-derived growth factor. In the present study, AKT1 was downregulated in all stages. A recent study has shown that inhibition of AKT1 enhances migration and invasion in KRAS- or EGFR-mutant A549 cells, suggesting that development of myristoylated alanine-rich C-kinase substrate targeted therapy may improve the therapeutic benefit of AKT inhibitors in patients with NSCLC.18 Fibroblast growth factor 4 is a ligand of FGFR2. The FGF4 was upregulated at stage II, stage III, and stage IV in the present study. A previous study using 2 LUAD cell lines (A549 and H1299) has demonstrated that FGF4 alters cell morphology; promotes EMT-associated protein expression; and enhances cell proliferation, migration/invasion, and colony initiation.19 The above studies provide sufficient evidence that these target genes have important influence on the pathological process, treatment, and prognosis of lung cancer.
By combining transcriptional regulation with survival analysis, we showed that APC, BRIP1, and PTTG1 may lead to upregulation of FGF22, subsequently triggering a series of changes in the carcinogenic pathway. Feng et al showed that the promoter methylation of APC may be a prognostic marker in patients with NSCLC.20 However, another study found no significance of APC hypermethylation in lung cancer.21 A previous study found a rare frameshift mutation in the BRIP1 gene increases the ovarian cancer risk.22 Recent studies showed BRIP1 loss-of-function mutation and protein-truncating mutation all significantly increase the risk of ovarian cancer.23,24 However, the relationship between BRIP1 and lung cancer remains unknown. Furthermore, the increased expression of PTTG1 regulates tumor growth and progression was verified in several studies.25,26 Fibroblast growth factor 22 belongs to the FGF family. The FGF family members possess broad mitogenic and cell survival activities and are involved in a variety of biological processes, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth, and invasion.27 The receptor of FGF22 is FGFR2b, which is encoded by the FGFR2 gene. Gene amplification or missense mutation of FGFR2 occurs in lung cancer, gastric cancer, breast cancer, and other cancers.28 There are currently several drugs targeting nonspecified subtype FGFs for the treatment of cancer that are already on the market or are entering the clinical stage. A previous study has reported that FGF22 is associated with several cancers. A mouse model has suggested that FGF22 has a potential pro-oncogenic role in skin.29 Another study has shown that FGF22 and BRCA1 affect the thyroid hormone pathway in ovarian cancer.30 Our results suggested that FGF22 and related TFs may affect signaling transduction pathways and carcinogenic pathways. However, to our knowledge, no study has reported the effect of FGF22 on lung cancer.
Conclusion
In conclusion, this study identified several anti-lung cancer target genes that were deregulated in all stages. Furthermore, through comprehensive analysis of differential expression genes, transcriptional regulation networks, and clinical data, we revealed a possible mechanism that overexpressed APC, BRIP1, and PTTG1 may lead to upregulation of FGF22, subsequently influencing multiple signaling transduction pathways and other tumorigenic processes. These results need further verification.
Supplemental Material
Supplemental Material, Supplementary_Material for Integrated Analysis of Transcriptome and Prognosis Data Identifies FGF22 as a Prognostic Marker of Lung Adenocarcinoma by Hong-Yan Liu, Hui Zhao, and Wen-Xing Li in Technology in Cancer Research & Treatment
Abbreviations
- AJCC
American Joint Committee on Cancer
- ERK
extracellular signal-regulated kinase
- FC
fold-change
- FDR
false discovery rate
- FGF
fibroblast growth factor
- GDC
Genomic Data Commons
- HR
hazard ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MEK
mitogen-activated protein/extracellular signal-regulated kinase
- NSCLC
non-small cell lung cancer
- RAF
rapidly accelerated fibrosarcoma
- RAS
rat sarcoma
- TF
transcription factor
- VEGF
vascular endothelial growth factor.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Foundation of China (Grant No. 81670060).
ORCID iD: Wen-Xing Li, PhD  https://orcid.org/0000-0001-9984-8439
https://orcid.org/0000-0001-9984-8439
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Pearce A, Haas M, Viney R, et al. Incidence and severity of self-reported chemotherapy side effects in routine care: a prospective cohort study. PLoS One. 2017;12(10):e0184360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gazdar AF. Should we continue to use the term non-small-cell lung cancer? Ann Oncol. 2010;21(suppl 7):vii225–vii229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zonderman AB, Ejiogu N, Norbeck J, Evans MK. The influence of health disparities on targeting cancer prevention efforts. Am J Prev Med. 2014;46(3 suppl 1):S87–S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rami-Porta R, Asamura H, Travis WD, Rusch VW. Lung cancer—major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(2):138–155. [DOI] [PubMed] [Google Scholar]
- 6. Kim HS, Mendiratta S, Kim J, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155(3):552–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346(6206):251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alevizakos M, Kaltsas S, Syrigos KN. The VEGF pathway in lung cancer. Cancer Chemother Pharmacol. 2013;72(6):1169–1181. [DOI] [PubMed] [Google Scholar]
- 10. Bai J, Hu S. Transcriptome network analysis reveals potential candidate genes for squamous lung cancer. Int J Mol Med. 2012;29(1):95–101. [DOI] [PubMed] [Google Scholar]
- 11. Perez-Ramirez C, Canadas-Garre M, Alnatsha A, et al. Interleukins as new prognostic genetic biomarkers in non-small cell lung cancer. Surg Oncol. 2017;26(3):278–285. [DOI] [PubMed] [Google Scholar]
- 12. Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35(24):3209–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han H, Shim H, Shin D, et al. TRRUST: a reference database of human transcriptional regulatory interactions. Sci Rep. 2015;5:11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen-Ngoc T, Bouchaab H, Adjei AA, Peters S. BRAF alterations as therapeutic targets in non-small-cell lung cancer. J Thorac Oncol. 2015;10(10):1396–1403. [DOI] [PubMed] [Google Scholar]
- 16. Anguera G, Majem M. BRAF inhibitors in metastatic non-small cell lung cancer. J Thorac Dis. 2018;10(2):589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan JY, Huang X, Luo YL. PSMA7 inhibits the tumorigenicity of A549 human lung adenocarcinoma cells. Mol Cell Biochem. 2012;366(1-2):131–137. [DOI] [PubMed] [Google Scholar]
- 18. Rao G, Pierobon M, Kim IK, et al. Inhibition of AKT1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with K-RAS or EGFR mutations. Sci Rep. 2017;7(1):7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi L, Song W, Li L, et al. FGF4 induces epithelial-mesenchymal transition by inducing store-operated calcium entry in lung adenocarcinoma. Oncotarget. 2016;7(45):74015–74030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng H, Zhang Z, Qing X, Wang X, Liang C, Liu D. Promoter methylation of APC and RAR-beta genes as prognostic markers in non-small cell lung cancer (NSCLC). Exp Mol Pathol. 2016;100(1):109–113. [DOI] [PubMed] [Google Scholar]
- 21. Ali A, Kumar S, Kakaria VK, et al. Detection of promoter DNA methylation of APC, DAPK, and GSTP1 genes in tissue biopsy and matched serum of advanced-stage lung cancer patients. Cancer Invest. 2017;35(6):423–430. [DOI] [PubMed] [Google Scholar]
- 22. Rafnar T, Gudbjartsson DF, Sulem P, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43(11):1104–1107. [DOI] [PubMed] [Google Scholar]
- 23. Weber-Lassalle N, Hauke J, Ramser J, et al. BRIP1 loss-of-function mutations confer high risk for familial ovarian cancer, but not familial breast cancer. Breast Cancer Res. 2018;20(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramus SJ, Song H, Dicks E, et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. J Natl Cancer Inst. 2015;107(11):pii:djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu X, Cao L, Zhang Y, Yin Y, Hu X, Cui Y. Network analysis of DEGs and verification experiments reveal the notable roles of PTTG1 and MMP9 in lung cancer. Oncol Lett. 2018;15(1):257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li WH, Chang L, Xia YX, et al. Knockdown of PTTG1 inhibits the growth and invasion of lung adenocarcinoma cells through regulation of TGFB1/SMAD3 signaling. Int J Immunopathol Pharmacol. 2015;28(1):45–52. [DOI] [PubMed] [Google Scholar]
- 27. Clayton NS, Wilson AS, Laurent EP, Grose RP, Carter EP. Fibroblast growth factor-mediated crosstalk in cancer etiology and treatment. Dev Dyn. 2017;246(7):493–501. [DOI] [PubMed] [Google Scholar]
- 28. Katoh M. Cancer genomics and genetics of FGFR2 (Review). Int J Oncol. 2008;33(2):233–237. [PubMed] [Google Scholar]
- 29. Jarosz M, Robbez-Masson L, Chioni AM, Cross B, Rosewell I, Grose R. Fibroblast growth factor 22 is not essential for skin development and repair but plays a role in tumorigenesis. PLoS One. 2012;7(6):e39436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu X, Lu J, Liao B, Li X, Qian X, Li K. Driver pattern identification over the gene co-expression of drug response in ovarian cancer by integrating high throughput genomics data. Sci Rep. 2017;7(1):16188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Supplementary_Material for Integrated Analysis of Transcriptome and Prognosis Data Identifies FGF22 as a Prognostic Marker of Lung Adenocarcinoma by Hong-Yan Liu, Hui Zhao, and Wen-Xing Li in Technology in Cancer Research & Treatment