Abstract
Background:
Hypercoagulable states (HS) can result from several different inherited and acquired disease conditions that cause abnormalities in the genes, proteins and cellular factors involved in the coagulation cascade. Novel insight into the molecular mechanisms involved in the coagulation pathways can provide a framework to develop improved therapeutics to treat patients with coagulation disorders. Therefore, investigating the genetic abnormalities present in patients with coagulation disorders can offer critical insight into disease pathogenesis. Our study aimed to assess the promoter methylation patterns of the phosphatase and tensin homologue (PTEN) gene as a potential underlying factor involved in HS.
Methods:
To measure the differences between the mRNA expression of PTEN in HS patients and healthy individuals we used qRT-PCR. Following bisulfite conversion, the promoter methylation status was analyzed using methylation specific PCR. The two-tailed student t-test was used to analyze the quantitative data. The data was considered statistically significant with a p value <0.05.
Results:
Our findings reveal PTEN to be down-regulated by 30% in the blood samples of HS patients when compared to healthy controls. The MSP data showed the PTEN promoter region to be un-methylated in both patients and healthy individuals.
Conclusion:
Since no differences in the methylation patterns of the PTEN gene was found between HS patients and controls, this suggests that DNA methylation of the PTEN promoter may not be a significant contributing epigenetic modification involved in the development HS. However, MSP may not be able to detect subtle changes in DNA methylation status. Thus, using an alternative high resolution technique may more accurately indicate differences in the PTEN promoter methylation status in HS patients.
Key Words: Hyperquagulable State, Promoter methylation, PTEN
Introduction
The coagulation system is composed of wide spectrum of cells and proteins that hold integral roles in orchestrating the coagulation cascade in response to blood vessel damage. Normally, the coagulation system maintains a fine balance between the pro- and anti-coagulation factors. The coagulation system can reach an imbalanced state during a perioperative period or times of critical illness. This imbalance may be secondary to certain inherited or acquired factors predisposing an individual to develop thrombosis, otherwise known as a hypercoagulable state (HS) (1).
Several studies have shown a connection between tumors and blood coagulation, such as the occurrence of blood vessel remodeling and systemic coagulopathy in various human cancers (2, 3). It has been previously hypothesized that certain aberrations in critical components involved in blood coagulation, such as the over-expression of tissue factor (TF) or thrombin receptor, PAR1 are involved in certain cancers. These factors have been shown to be regulated in part by the activation of various oncogenic factors such as K-ras and epidermal growth factor receptor (EGFR), or by the inactivation of the tumor suppressor, p53 (4, 5). Alterations to these oncogenic factors significantly influences TF in both soluble and cell-associated forms, which can lead to the development of HS (6). Emerging evidence has supported a role for additional genes including, PML-RAR alpha, PTEN, and MET in cancer coagulopathies (7).
The PTEN (phosphatase and tensin homologue) gene encodes for an enzyme that is found ubiquitously in the tissues of the body. The enzyme encoded by PTEN acts as a tumor suppressor through regulating cell division preventing rapid and uncontrolled cell growth and proliferation. The PTEN enzyme holds a role in mediating apoptotic signaling pathways, regulates cell migration, adhesion, and enhances angiogenesis (8). These functions may constitute critical events in cancer coagulopathy, angiogenesis, and tumor metastasis. Further investigation into the molecular events underlying HS will allow for improved therapeutic design and treatment for HS patients (9). Evaluating the molecular mechanisms by which PTEN is regulated in HS patients may therefore provide valuable information into the pathogenesis of HS.
Research has revealed a critical role for epigenetic modifications such as DNA methylation, acetylation and phosphorylation, in the development of HS (10, 11). Epigenetic factors cause heritable modifications to gene expression that can lead to the suppression or activation of particular genes without directly altering the DNA sequence (12, 13). Several lines of evidence have highlighted the significant influence methylation-based modifications have on the function of gene promoter regions encoding for various clotting factors (12, 14).
Methylation of PTEN has been observed in a variety of different human cancers displaying highly abnormal DNA methylation patterns (15-17). However, investigation into the PTEN methylation pattern of patients with HS is quite scarce. Thus, we aimed to examine the role of PTEN in HS pathogenesis and investigate the DNA promoter methylation patterns in HS patients. Our results provide insight into the molecular events that occur in patients with HS and provide a framework for better therapeutic design for these individuals.
Materials and Methods
Clinical samples
The present study was a case-control study conducted in Qazvin province, Iran. Of the 30 randomly selected participants, 26 were HS patients and 4 were normal individuals. Each participant provided written consent and blood was collected from each individual. The study was approved by the research ethics committee of Qazvin University of Medical Sciences, Qazvin Iran. Since heart or coagulation treatments and medications such as warfarin, may interfere with the methylation status of DNA, patients participating in the study must not have received this kind of medication. The healthy control group was selected randomly among different age groups with no history of HS, heart problems or coagulopathy.
Sample collection, DNA extraction, and bisulfite conversion
Whole blood samples were collected using vacuum tubes containing K3-EDTA as anticoagulant. The samples were used immediately or stored at 4 °C for DNA extraction within 24 hours, or for RNA extraction within 2 hours. DNA extraction from whole blood samples was performed using a commercially available kit (Gene ALL, Seoul, South Korea). The purity and concentration of isolated DNA samples were analyzed using a Nano drop device measuring the ratio of A260/230 and A260/280, respectively.
The DNA samples were then treated with bisulfite using the fast bisulfite kit (Qiagen, Frankfurt, Germany) according to the manufacturer’s instructions. Bisulfite conversion is a process during which the non-methylated cytosine residues convert to uracil, while the 5-methylcytosine residues remain intact. This provides a platform to distinguish converted from intact bases, allowing the methylation status of the target DNA segment to be examined.
Methylation specific PCR (MSP)
To differentiate between methylated and un-methylation cytosine bases we carried out Methylation specific PCR (MSP), which provides a high resolution result of DNA methylation. The specific primers for determining the methylated and un-methylated status of the promoter segment were designed using MethPrimer design tool (http://www.urogene.org/Methprimer/index1.html) The primers were designed such that the sequence around the transcription start site (TSS) was preferentially amplified. A DNA sample treated with the Sss1 Methylase enzyme was used as positive methylated sample control.
To conduct the MSP reactions, the polymerase chain reaction for PTEN-UM and PTEN-M were carried out in a 50 μL volume containing 1× polymerase chain reaction buffer (15 mmol/L MgCl2), 2.5 mmol/L mixture of dNTPs, 10 pM of each primer, 4 U HotStart Taq DNA polymerase (Qiagen, Frankfurt, Germany), and 25 to 50 ng of bisulfite-modified DNA. Amplification was performed in a thermocycler with the following conditions: 95 °C for 15 minutes, cycled at 94 °C for 30 seconds, 60 °C for 1 minute, and 72 °C for 30 seconds (40 cycles) followed by extension at 75 °C for 5 minutes. The reaction samples were then resolved in a 2% agarose gel to visualize the products. The nucleotide sequences of the primers used in the MSP reactions are listed in Table 1.
Table 1.
Specific primer used to amplify the methylated and un-methylated pattern of PTEN gene promoter region. of treated DNA Primer forward and reverse sequence for MSP technique and PTEN expression. MF: methylated forward; MR: methylated reverse; UF: unmethylated forward; UR: unmethylated reverse.
| Primer | 5' to -3' sequence |
|---|---|
| MF | 5'-GTTTGGGGATTTTTTTTTCGC-3' |
| MR | 5'-AACCCTTCCTACGCCGCG-3' |
| UF | 5'-TATTAGTTTGGGGATTTTTTTTTTGT-3' |
| UR | 5'-CCCAACCCTTCCTACACCACA-3' |
Total RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the control and patient samples using a commercially available column-based kit (Gene ALL, Seoul, South Korea) to measure the quality and concentration of RNA. The Nano drop technique was used by examining the ratio of light absorption at a 280/260 nm wavelength. Reverse transcription was performed with 1 μg of total RNA. To synthesize cDNA, the Thermos ScientificTM kit was used according to manufacturer's instructions
The qRT-PCR was performed using the Rotor Gene 6000 PCR machine (Corbett). The reactions were performed using the Amplicon PCR Master Mix. The normalized values of mRNA levels were calculated by comparing β-Actin expression levels as the housekeeping gene using the 2-ΔΔCT method. Test specificity was verified via melting curve analysis and agarose gel electrophoresis. The primer sequences used in qRT-PCR are listed in Table 2.
Table 2.
. The primer sequences for relative quantification of PTEN gene by qRT-PCR.
| Primer | 5' to -3' sequence |
|---|---|
| Human PTEN Forward | 5'-CCG AAA GGT TTT GCT ACC ATT CT-3' |
| Human PTEN Reverse | 5'-AAA ATTA TTT CCT TTC TGA GCA TTC C-3' |
| Human β-actin Forward | 5'-CCT GTA CGC CAA CAC AGT GC-3' |
| Human β-actin Reverse | 5'-ATA CTC CTG CTT GCT GATC-3' |
Statistical analysis
For all the quantitative data, the mean value ± one standard deviation is shown. Statistical analysis for each set of experimental means was performed using Graph Pad Prism. A two-tailed Student’s t-test was used and P-values less than or equal to 0.05 were considered to be of statistical significance.
Results
In the sample population of HS patients, 16 subjects were shown to have HS due to the presence of Factor 5 Leiden. Five samples were found to have defects in anti-thrombin, while 3 HS patients had a deficiency in protein C and S which (Table 3).
Table 3.
The most prevalent cause of HS development in the patients under study.
| Cause of HS | Male | Female |
|---|---|---|
| Average adult age | 45 | 52 |
| Average child age | 12 | 11 |
| Factor 5 Leiden defects | 34% | 26% |
| Anti-thrombin defects | 14% | 9% |
| Pro C and Pro S deficiency | 8% | 5% |
| Other reasons | 1% | 3% |
| Hb average | 10.3 | 9.4 |
PTEN gene methylation
The MSP data showed that PTEN methylation remained stable among different phenotypes of HS (various ages, genders and the underline causes). Analysis of band intensities showed that the methylation pattern of PTEN displays an un-methylated status among the HS patients and normal individuals. This was determined by observing very sharp product intensities with un-methylated primers and a weak or invisible band intensities for the methylated primers. Our observations suggest an un-methylated status of CpG Islands in the selected PTEN promoter region (Fig. 1 and Fig. 2).
Fig. 1.
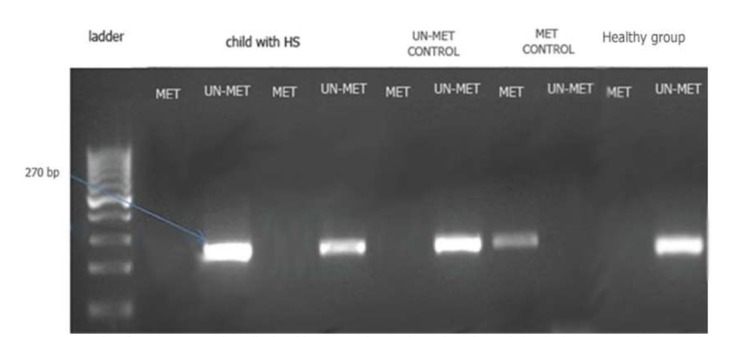
MSP product from PTEN gene in pediatric subjects; Met: the reactions where the methylated primer was used; Un-met: the reactions where the Un-methylated primer was used).
Fig. 2.
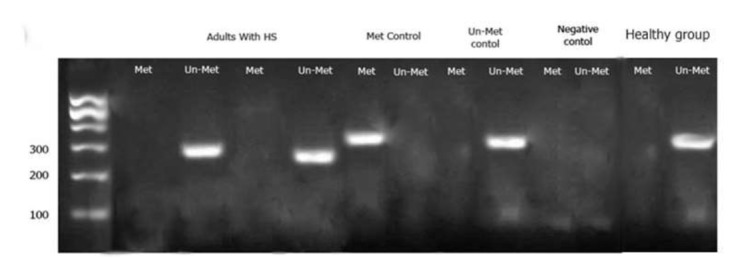
MSP product from PTEN gene in adult; Met: the reactions where the methylated primer was used; Un-met: the reactions where the Un-methylated primer was used).
The expression of PTEN
The PTEN gene expression levels were measured using mRNA relative quantification normalized for β-Actin expression levels. As shown in Figure 3, qRT-PCR analysis revealed that the level of PTEN was down-regulated by 30% compared to normal individuals.
Fig. 3.
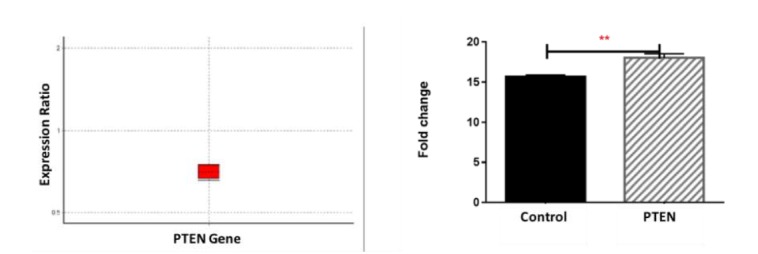
Comparing PTEN expression in patients with HS and healthy control group; as it is observed, the PTEN is down regulated in HS patients (in comparison to control group) by mean factor of 0.702. Analyzing CT in graph pad (right) software reveals the significance of change in PTEN expression by p value<0.05.
Discussion
Several lines of studies have supported the aberrant gene expression pattern and wide range of genetic abnormalities in the patients with hypercoagulability status (HS). In this regard, we attempted to clarify the possible contribution of promoter methylation on the PTEN expression- as a common deregulated gene- in the HS patients, and demonstrated that despite 30% decrease in mRNA expression levels, PTEN is not a target of DNA hypermethylation in the subjects.
DNA methylation primarily occurs at CpG sites, and the majority of cytosine residues are methylated in mammals. However, there are regions of DNA near the promoter of genes that have a higher concentration of CpG sites, known as CpG islands which are not methylated in normal cells while highly methylated in cancer cells. This change in DNA methylation is considered an epigenetic modification commonly found in tumors and marks the early stages of cancer development. Hypermethylation of CpG islands has the potential to turn off various tumor suppressor genes and therefore initiate cancer development (18, 19). Hypermethylation of tumor suppressor genes has been shown to occur in a variety of human cancers (20). Abnormalities in cellular function as a result of tumor-related changes has the potential to trigger coagulopathy. Several studies have aimed to understand the function of tumor suppressor genes and their role in coagulation disorders such as HS (2, 3). In this study, we aimed to determine how the PTEN tumor suppressor gene is dysregulated in HS patients and examine the potential role for DNA methylation. Although our findings revealed a down-regulation of PTEN expression in HS patients, examination of the promoter region of PTEN via MSP showed no changes in the methylation patterns in the HS samples compared with controls.
Our results suggest that the changes in PTEN expression in HS may be due to mechanisms other than DNA methylation at the promoter region. However further investigation is needed to confirm this observation. Using methylation-based methodologies with higher resolution may be able to reveal subtle changes in methylation levels, which are hard to distinguish using the MSP technique. Assessing histone modifications and other epigenetic modifications may provide insight into the epigenetic-based changes controlling PTEN gene regulation in HS.
Treatments capable of reversing the epigenetic changes of patients, such as inhibitors of histone de-acetylation (HDAC inhibitors) and de-methylated factors (DNMT inhibitors), provide an opportunity to reverse harmful gene expression patterns. The relationship between epigenetic modifications and malignant and normal hematopoiesis has gained attention over the recent years. Several lines of evidence support a positive role for epigenetic therapies, such as DNA-demethylating drugs which can act on gene promoter regions encoding for clotting factors and eliminate the harmful epigenetic change in the targeted gene (21, 22).
PTEN is a tumor suppressor gene located on chromosome 10 q23. The loss of this gene has been detected in endometrial, prostate, breast, and brain cancer. This gene plays an important role in cell death and apoptosis. It is also involved in cell migration, differentiation and the downregulation of cell proliferation through inhibiting PIP3 (15). Methylation of the PTEN gene promoter was discovered in metabolic syndrome (MeS), one of the risk factors for type 2 diabetes (T2D). Furthermore, there is evidence indicating that PTEN promoter methylation status may be a protective factor in determining metabolic syndrome. (23). In our study, we aimed examine the association of PTEN methylation in patients with HS. We hypothesized that the methylation pattern of the PTEN promoter in this illness may be involved in disease development. Examination of HS patients and healthy subjects revealed that the PTEN gene promoter was un-methylated, suggesting a role for mechanisms other than DNA methylation for causing PTEN abnormalities in this disease. However, DNA methylation may occur in regions of PTEN other than what was examined in this study, therefore DNA methylation should not be entirely ruled out.
Acknowledgements
More thorough analysis into the DNA methylation patterns of the PTEN promoter region in HS patients requires additional comprehensive examination such that it can be sufficiently determined how epigenetic mechanisms are involved in HS pathogenesis. However, due to the reduced expression of PTEN in HS patients we assume that this gene may have a critical role in the development of the HS disease phenotype.
References
- 1.Palta S, Saroa R, Palta A. Overview of the coagulation system. Indian journal of anaesthesia. 2014;58(5):515. doi: 10.4103/0019-5049.144643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rak J, Joanne LY, Luyendyk J, Mackman N. Oncogenes, trousseau syndrome, and cancer-related changes in the coagulome of mice and humans. Cancer research. 2006;66(22):10643–6. doi: 10.1158/0008-5472.CAN-06-2350. [DOI] [PubMed] [Google Scholar]
- 3.Rickles FR, Edwards RL. Activation of blood coagulation in cancer: Trousseau's syndrome revisited. Blood. 1983;62(1):14–31. [PubMed] [Google Scholar]
- 4.Joanne LY, May L, Lhotak V, Shahrzad S, Shirasawa S, Weitz JI, et al. Oncogenic events regulate tissue factor expression in colorectal cancer cells: implications for tumor progression and angiogenesis. Blood. 2005;105(4):1734–41. doi: 10.1182/blood-2004-05-2042. [DOI] [PubMed] [Google Scholar]
- 5.Versteeg HH, Ruf W, editors. Emerging insights in tissue factor-dependent signaling events. Seminars in thrombosis and hemostasis. 2006 doi: 10.1055/s-2006-933337. Copyright©2006 by Thieme Medical Publishers, Inc., 333 Seventh Avenue, New York, NY 10001, USA. [DOI] [PubMed] [Google Scholar]
- 6.Bluff JE, Brown NJ, Reed MW, Staton CA. Tissue factor, angiogenesis and tumour progression. Breast Cancer Research. 2008;10(2):204. doi: 10.1186/bcr1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falanga A, Russo L, Tartari CJ. Pathogenesis and treatment of thrombohemorrhagic diathesis in acute promyelocytic leukemia. Mediterranean journal of hematology and infectious diseases. 2011;3(1) doi: 10.4084/MJHID.2011.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nature reviews Molecular cell biology. 2012;13(5):283–96. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 9.Rong Y, Post DE, Pieper RO, Durden DL, Van Meir EG, Brat DJ. PTEN and hypoxia regulate tissue factor expression and plasma coagulation by glioblastoma. Cancer research. 2005;65(4):1406–13. doi: 10.1158/0008-5472.CAN-04-3376. [DOI] [PubMed] [Google Scholar]
- 10.Dahlbäck B. Procoagulant and anticoagulant properties of coagulation factor V: factor V Leiden (APC resistance) causes hypercoagulability by dual mechanisms. The Journal of laboratory and clinical medicine. 1999;133(5):415–22. doi: 10.1016/s0022-2143(99)90018-5. [DOI] [PubMed] [Google Scholar]
- 11.Rak J, Klement P, Yu J. Genetic determinants of cancer coagulopathy, angiogenesis and disease progression. Vnitrni lekarstvi. 2006;52:135–8. [PubMed] [Google Scholar]
- 12.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):634–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Azad M, Kaviani S, Noruzinia M, Mortazavi Y, Mobarra N, Alizadeh S, et al. Gene expression status and methylation pattern in promoter of P15INK4b and P16INK4a in cord blood CD34+ stem cells. Iranian journal of basic medical sciences. 2013;16(7):822. [PMC free article] [PubMed] [Google Scholar]
- 14.Tuck-Muller C, Narayan A, Tsien F, Smeets D, Sawyer J, Fiala E, et al. DNA hypomethylation and unusual chromosome instability in cell lines fromICF syndrome patients. Cytogenetic and Genome Research. 2000;89(1-2):121–8. doi: 10.1159/000015590. [DOI] [PubMed] [Google Scholar]
- 15.Pallares J, Bussaglia E, Martínez-Guitarte JL, Dolcet X, Llobet D, Rue M, et al. Immunohistochemical analysis of PTEN in endometrial carcinoma: a tissue microarray study with a comparison of four commercial antibodies in correlation with molecular abnormalities. Modern pathology. 2005;18(5):719–27. doi: 10.1038/modpathol.3800347. [DOI] [PubMed] [Google Scholar]
- 16.Hühns M, Salem T, Schneider B, Krohn M, Linnebacher M, Prall F. PTEN mutation, loss of heterozygosity, promoter methylation and expression in colorectal carcinoma: Two hits on the gene? Oncology reports. 2014;31(5):2236–44. doi: 10.3892/or.2014.3097. [DOI] [PubMed] [Google Scholar]
- 17.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in Genetics. 2000;16(4):168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 18.Berdasco M, Esteller M. Hot topics in epigenetic mechanisms of aging: 2011. Aging cell. 2012;11(2):181–6. doi: 10.1111/j.1474-9726.2012.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang AS, Estecio MR, Garcia-Manero G, Kantarjian HM, Issa J-PJ. Comment on" Chromosomal instability and tumors promoted by DNA hypomethylation" and" Induction of tumors in mice by genomic hypomethylation". Science. 2003;302(5648):1153. doi: 10.1126/science.302.5648.1153a. [DOI] [PubMed] [Google Scholar]
- 20.Datta J, Ghoshal K, Denny WA, Gamage SA, Brooke DG, Phiasivongsa P, et al. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer research. 2009;69(10):4277–85. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Undas A, Brummel-Ziedins KE, Mann KG. Statins and blood coagulation. Arteriosclerosis, thrombosis, and vascular biology. 2005;25(2):287–94. doi: 10.1161/01.ATV.0000151647.14923.ec. [DOI] [PubMed] [Google Scholar]
- 22.Lu G, DeGuzman FR, Hollenbach SJ, Karbarz MJ, Abe K, Lee G, et al. A specific antidote for reversal of anticoagulation by direct and indirect inhibitors of coagulation factor Xa. Nature Medicine. 2013;19(4):446–51. doi: 10.1038/nm.3102. [DOI] [PubMed] [Google Scholar]
- 23.Hashemi M, Rezaei H, Eskandari-Nasab E, Kaykhaei M-A, Taheri M. Association of promoter methylation and 32-bp deletion of the PTEN gene with susceptibility to metabolic syndrome. Molecular medicine reports. 2013;7(1):342–6. doi: 10.3892/mmr.2012.1174. [DOI] [PubMed] [Google Scholar]


