Abstract
Objectives:
Reversing HIV-1 latency has been suggested as a strategy to eradicate HIV-1. We investigated the effect of romidepsin on the HIV transcription profile in participants from the REDUC part B clinical trial.
Design:
Seventeen participants on suppressive antiretroviral therapy were vaccinated with six doses of the therapeutic vaccine Vacc-4x followed by treatment with three doses of romidepsin. Samples from nine study participants were available for HIV transcription profile analysis.
Methods:
Read-through, total (TAR), elongated (longLTR), polyadenylated (polyA) and multiply-spliced (TatRev) HIV transcripts and total HIV DNA were quantified at baseline (visit 1) and 4 hours after the second (visit 10b) and third (visit 11b) romidepsin infusions.
Results:
Read-through, total, elongated, and polyadenylated HIV transcripts increased after romidepsin infusion (p=0.020, p=0.0078, p=0.0039, p=0.027, respectively), but no changes were observed in multiply-spliced HIV RNA or HIV DNA. No change was observed in the ratio of read-through/total HIV transcripts. The ratio of elongated/total HIV RNA increased after romidepsin (p=0.016), while the ratio of polyadenylated/elongated HIV decreased. Both elongated HIV transcripts and total HIV DNA correlated negatively with the time to viral rebound after interruption of ART.
Conclusions:
In these patients, romidepsin increased early events in HIV transcription (initiation and especially elongation), but had less effect on later stages (completion, multiple splicing) that may be required for comprehensive latency reversal and cell killing. Without cell death, increased HIV transcription before or after latency reversal may hasten viral rebound after therapy interruption.
Keywords: HIV-1, latency, humans, romidepsin, transcription, RNA, RNA splicing
Introduction:
Antiretroviral therapy (ART) cannot eliminate the HIV genomes integrated in latently-infected cells, which are a major barrier to cure HIV and are responsible for viral rebound after ART discontinuation[1–4]. One strategy to eradicate HIV consists of reactivating viral transcription with latency reversing agents (LRAs), such as histone deacetylase inhibitors (HDACi). Several clinical trials have shown HIV reactivation after HDACi administration[5–14]. A recent clinical trial, REDUC part B, analyzed the administration of a peptide-based therapeutic HIV vaccine (Vacc-4x), plus recombinant human granulocyte macrophage colony-stimulating factor (rhuGM-CSF) as local adjuvant, in combination with the HDACi romidepsin[10]. This approach showed an increase in unspliced cell-associated HIV RNA and residual plasma viremia after romidepsin infusions, along with a reduction in total HIV DNA[10]. However, the mechanism by which romidepsin reverses HIV latency in vivo remains unclear.
In this study, we characterized the HIV transcription profile before and after romidepsin therapy in available samples from the REDUC part B study using a recently-described panel of HIV RNA assays[15]. This novel panel quantifies read-through, total, elongated, polyadenylated, and multiply-spliced transcripts, allowing one to measure different blocks to HIV transcription and the degree to which they are reversed after LRA therapy[15].
Methods:
Study design
This study is a follow-up to the REDUC part B clinical trial. In this trial, 17 HIV-1 infected ART-suppressed individuals received a series of six intradermal immunizations over 12 weeks with Vacc-4x (Bionor Pharma) and rhuGM-CSF (Genzyme) as local adjuvant, followed by intravenous infusions of 5 mg/m2 romidepsin (Celgene) once weekly for three weeks[10]. Subsequently, 16 participants underwent an analytic treatment interruption (ATI)[10]. Trial design, participant characteristics, and levels of HIV DNA and unspliced HIV RNA have been published previously[10].
Ethics statement
The trial (registered at http://clinicaltrials.gov[NTC02092116]) was approved by the Danish Health and Medical Authorities, the Danish Data Protection Agency, and the National Committee on Health Research Ethics (#M-2013–364–13)[10]. Each participant provided written informed consent[10].
Samples
Cryopreserved peripheral blood mononuclear cells (PBMCs) were available from nine participants (Supplementary Table 1) at baseline (visit 1, day −21) and 4 hours after the second (visit 10b, day 112) and the third (visit 11b, day 119) romidepsin infusions. Eight of the nine participants underwent an ATI.
HIV levels
PBMCs were pelleted and nucleic acids were extracted using TRI Reagent[15]. Cell-associated HIV transcripts (Read-through, TAR, longLTR, polyA and Tat-Rev) and total HIV DNA (longLTR) were quantified in duplicate using droplet digital PCR, as previously described[15].
Statistics
Wells without positive droplets were quantified as the limit of quantification (mean copies for 1 positive droplet in two wells) divided by 2. Longitudinal changes were evaluated using the Wilcoxon signed-rank test. Spearman correlations were used to evaluate the association between each HIV RNA or DNA and: 1) time to rebound after ATI (days to VL [viral load] >50copies/ml and VL>1,000copies/ml); and 2) time to suppression after ART reinitiation (days to VL<50copies/ml). GraphPad Prism version 7.0 was used for all statistical analyses.
Results:
Romidepsin increases read-through, total, elongated, and polyadenylated but not multiply-spliced transcripts
We quantified 5 RNA regions that define different HIV transcripts: read-through, TAR(total), longLTR(elongated), polyA(polyadenylated), and Tat-Rev(multiply-spliced)[15] (Figure 1A). We observed a significant increase in read-through (1.7-fold, p=0.02), total (1.9-fold, p<0.01), elongated (2.4-fold, p<0.01) and polyadenylated (1.9-fold, p=0.03) HIV RNA/106 PBMCs after the second romidepsin infusion (visit 10b), and a 1.9-fold increase in elongated transcripts after the third romidepsin infusion (visit 11b) (p<0.01) (Figure 1B). Equivalent results were observed when HIV RNA levels were normalized to cellular transcription (μg RNA) or proviral DNA. No significant changes were observed in multiply-spliced HIV RNA or HIV DNA (Figure 1C). In one study participant, romidepsin led to a second population of TAR RNA+ droplets with a higher amplitude that was barely detected at baseline (Supplementary Figure1).
Figure 1. HIV transcriptional profile, HIV DNA, and blocks to HIV transcription before and after romidepsin therapy.
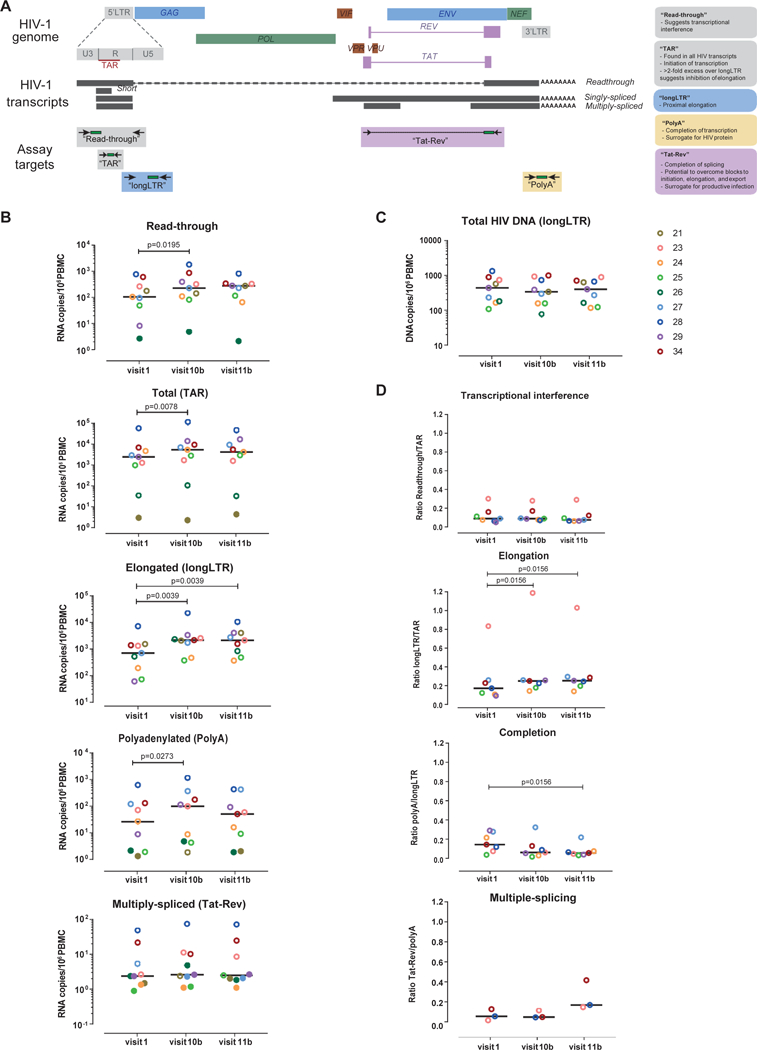
(A) Diagram of the assays used to characterize the HIV transcriptional profile; (B) Dynamics of each HIV transcript per million PBMCs; (C) Dynamics of total HIV DNA; and (D) Changes in transcriptional interference, elongation, completion, and multiple-splicing. Each color represents a different individual from the REDUC part B study; bars represent medians. Determinations below the limit of quantification (LOQ) are represented as solid dots.
Romidepsin increases elongation but not completion or multiple-splicing
We also quantified measures of transcriptional interference (ratio of read-through/total transcripts) and progression through blocks to HIV transcriptional elongation (elongated/total transcripts), completion (polyadenylated/elongated), and multiple-splicing (multiply-spliced/polyadenylated). No change was observed in read-through/total HIV RNA. We observed a significant increase in elongation (elongated/total) after both the second and the third romidepsin infusions (p=0.02). However, we detected no increase in completion (polyadenylated/elongated), which actually decreased after the third infusion (visit 11b) (p=0.02), and no change in multiple-splicing (Figure 1D).
HIV DNA and elongated transcripts predict time to viral rebound
We also analyzed whether there was an association between total HIV DNA or each HIV RNA and time to viral rebound after ATI or suppression after ART reinitiation. A strong negative correlation was observed between HIV DNA and the time to rebound (VL>50copies/ml) at visit 1, 10b, and 11b (Rho=−0.81, −0.88, and −0.91; p=0.02, p<0.01, and p<0.01, respectively). HIV DNA also tended to correlate negatively with time to rebound >1,000copies/ml and positively with time to suppression (Figure 2A). Levels of all HIV RNAs tended to correlate negatively with the time to rebound. This association was strongest for the comparison between elongated transcripts and time to VL>1,000copies/ml after romidepsin administration (Rho=−0.78, p=0.03 at visit 10b; Rho=−0.77, p=0.03 at visit 11b) (Figure 2B). HIV DNA also tended to correlate with levels of each HIV RNA (Figure 2C).
Figure 2. Association between HIV DNA and HIV transcription profile before and after romidepsin infusion, and time to rebound after ATI and subsequent suppression.
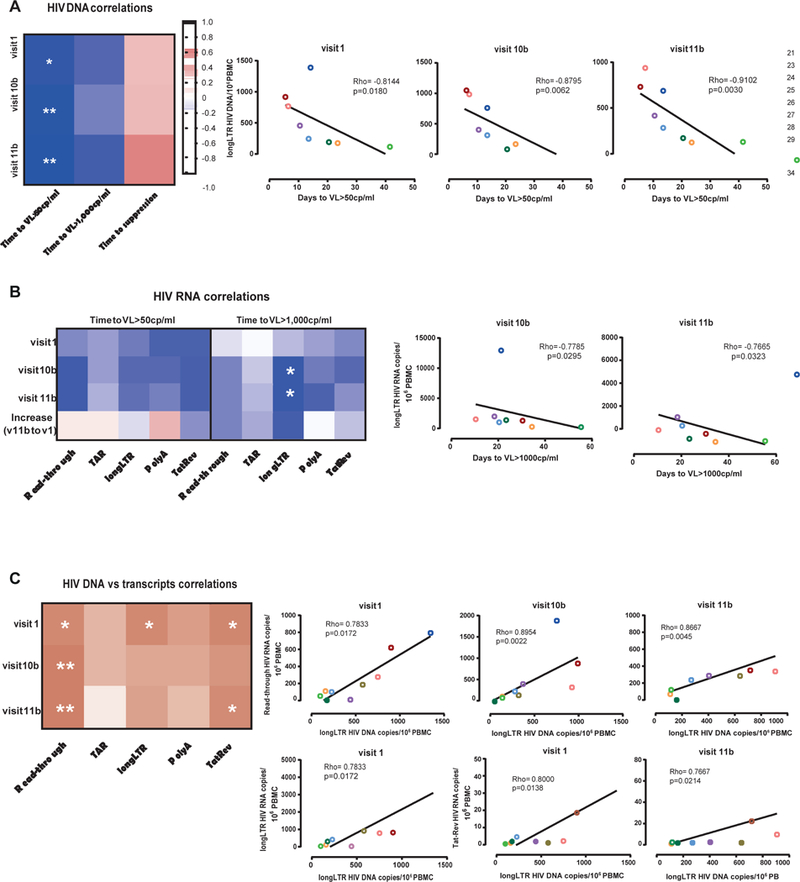
(A) Spearman correlations between the total HIV DNA per million PBMCs and time to rebound (measured as days to VL>50 copies/ml and VL>1,000 copies/ml of plasma), and time to suppression (quantified as days to VL<50 copies/ml of plasma after ART reinitiation); (B) Spearman correlations between the different HIV transcripts per million PBMCs and time to viral rebound (measured as days to VL>50 copies/ml and VL>1,000 copies/ml of plasma); and (C) Spearman correlations between total HIV DNA per million PBMCs and the different HIV transcripts per million PBMCs. Each color represents a different individual from the REDUC part B study.
Discussion:
We characterized the levels of HIV RNAs and transcriptional blocks before and after the “shock and kill” strategy consisting of the HIV therapeutic vaccine Vacc-4x, plus rhuGM-CSF, and the HDACi romidepsin[10]. This is the first study that evaluates in vivo the specific effect of an LRA on the transcriptional blocks implicated in HIV latency.
Limitations of the study should be acknowledged. First, the parent study had sequential study interventions (vaccination and then romidepsin) and we did not have access to samples between vaccination and romidepsin. However, unspliced HIV RNA did not change after vaccination and increased after romidepsin in the parent study, suggesting the effects on HIV transcription are due to romidepsin. Second, samples were only available from 9 of 17 trial participants. Third, the use of PBMCs (instead of CD4+ T) and presence of non-B subtypes may have affected HIV levels and detection frequencies. While we detected a statistically significant increase in the expression of read-through, total, elongated, and polyadenylated HIV transcripts after romidepsin administration, it is difficult to exclude a small effect on multiply-spliced transcripts due to the number of undetectable determinations.
We detected no change in total HIV DNA after romidepsin infusion, which accords with other trials of HDACi[5–9] but differs from the results of the parent REDUC-B study[10]. The latter difference may reflect patient-specific differences in our subset of 9 individuals (of whom only one had an increase in plasma viral load) or the fact that we quantified HIV DNA at an earlier time point using a different assay.
After romidepsin infusion, we observed an increase in read-though transcripts. The increase in read-through transcripts accords with one[16] but not another in vitro study[15], suggesting that romidepsin activated transcription of nearby cellular genes whose transcripts continue into the provirus. However, no change was observed in the fraction of total HIV transcripts that are read-through (read-through/TAR).
The fold increase in total HIV (TAR) transcripts was slightly greater than that of read-through transcripts, and the absolute change in TAR was much greater, suggesting that romidepsin also increased transcriptional initiation from the HIV promoter. In one study participant, romidepsin induced a second population of TAR RNA+ droplets with a higher amplitude. Droplet digital PCR can detect sequence variability using a single primer/probe set[17]. The most likely explanation is that romidepsin reactivated a provirus that was not previously transcribed, possibly one with fewer sequence mismatches and an epigenetic block to HIV transcriptional initiation.
The median fold increase in elongated HIV transcripts exceeded that of either total or read-through transcripts, and the ratio of elongated/total transcripts increased after romidepsin. These results, which accord with those observed in vitro[15, 18], suggest that romidepsin specifically increases HIV transcriptional elongation. This increase in elongation accords with the increase in unspliced HIV RNA observed in the parent trial[10]. We also observed an increase in polyadenylated HIV transcripts at the first time point after romidepsin, in contrast to prior in vitro studies[15, 16]. However, the median fold change in polyadenylated transcripts was less than that of elongated transcripts and the ratio of polyadenylated/elongated HIV RNA actually decreased, suggesting a decrease in the fraction of elongated transcripts that complete transcription.
Together, this data suggests that romidepsin increased HIV transcriptional initiation and especially elongation but not completion or multiple-splicing in the PBMCs sampled from these participants. Since polyadenylated HIV RNAs are likely necessary for efficient translation, while Tat and Rev may be necessary for productive infection, these results may explain why 8 of 9 study participants showed no increase in plasma virus. Moreover, the lack of HIV protein could prevent MHC:peptide-mediated recognition and killing of HIV-infected cells even if they transcribe elongated or unspliced transcripts. This finding may explain the lack of decrease in HIV DNA in the 9 participants studied here. However, it should be noted that other participants in the parent study had an increase in viral load and a decrease in HIV DNA, which could reflect effects of romidepsin and/or vaccination that differ by patient or are missed in sampling PBMCs[10, 12, 13].
We observed strong positive correlations between total HIV DNA and HIV RNA, as previously reported[19], but also found correlations with read-through, elongated, and multiply-spliced transcripts. This data suggests that the higher the number of proviruses, the more proviruses that are near transcriptionally-active genes and/or with fewer defects or transcriptional blocks.
HIV DNA and elongated HIV RNA correlated negatively with time to viral rebound, in accord with prior studies showing correlations between time to rebound and HIV DNA[20] or unspliced HIV RNA[21, 22]. However, we found a similar trend for all the different HIV transcripts, suggesting that higher activity at each stage of HIV transcription may hasten time to rebound after ATI. In the 9 patients studied here, the change in elongated HIV transcripts (visit 11b-visit 1) tended to correlate inversely with time to rebound, suggesting a possible small effect of romidepsin. However, it should be noted that this correlation was not statistically significant, and correlations do not indicate causality. Moreover, the ATI was >7 weeks after visit 11b, and no net change in time to viral rebound was observed in the parent trial[10].
These findings have important implications for strategies aimed at HIV cure. Romidepsin increases HIV transcriptional initiation and elongation, and may play a role in strategies to reverse latency, especially if combined with other agents to increase polyadenylated and multiply-spliced transcripts. However, even effective latency reversal may not lead to the death of infected cells[23], and other approaches are needed to augment cell killing. Without cell death, increased HIV transcription may shorten the time to rebound during ATI. Measures of HIV DNA and RNA are strongly predictive of the time to rebound and could be used along with other data to decide which patients may be candidates for ATI.
Supplementary Material
Droplet digital PCR (ddPCR) plot showing the detection of different cell-associated HIV RNA TAR sequences after romidepsin infusion. (A) One dimension ddPCR plot corresponding to channel 1 (FAM) fluorescence amplitude detected at each time point, and (B) Two dimensions ddPCR plots corresponding to channel 1 (FAM) and channel 2 (VIC) fluorescence amplitude detected at each time point.
Participant characteristics.
Acknowledgements:
We acknowledge the participation and commitment of the study participants, which made the study possible. S.M.-L. designed and performed experiments, analyzed data, and (with S.A.Y.) wrote the manuscript. P.K. designed and performed experiments and analyzed data. M.T. and O.S.S. designed the clinical trial, provided samples, and assisted with manuscript preparation. S.A.Y. and J.K.W. designed the study, supervised the work, and edited the manuscript. All authors have read and accepted the final manuscript.
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (1R01AI132128 [SY, JW], R56AI116342 [JW], R33AI116218 [JW], R56AI091573 [JW], U19AI096109 [JW]), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) at the National Institutes of Health (1R01DK108349 [SY]), the U.S. Department of Veterans Affairs (IK2 CX000520 [SY], I01 BX000192 [JW]), and the American Foundation for AIDS Research (amfAR) Institute for HIV Cure Research (109301 [SY, JW]).
Footnotes
Conflict of interest:
Bionor Pharma sponsored the clinical study from which these samples were derived and contributed to the design of the clinical trial. The funders had no role in data collection and analysis, decision to publish, or preparation of the manuscript. The remaining authors declare no conflict of interest.
References:
- 1.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94(24):13193–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278(5341):1291–1295. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278(5341):1295–1300. [DOI] [PubMed] [Google Scholar]
- 4.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A 2008; 105(43):16725–16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487(7408):482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 2014; 210(5):728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 2014; 10(10):e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1(1):e13–21. [DOI] [PubMed] [Google Scholar]
- 9.Sogaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS Pathog 2015; 11(9):e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leth S, Schleimann MH, Nissen SK, Hojen JF, Olesen R, Graversen ME, et al. Combined effect of Vacc-4x, recombinant human granulocyte macrophage colony-stimulating factor vaccination, and romidepsin on the HIV-1 reservoir (REDUC): a single-arm, phase 1B/2A trial. Lancet HIV 2016; 3(10):e463–472. [DOI] [PubMed] [Google Scholar]
- 11.Tapia G, Hojen JF, Okvist M, Olesen R, Leth S, Nissen SK, et al. Sequential Vacc-4x and romidepsin during combination antiretroviral therapy (cART): Immune responses to Vacc-4x regions on p24 and changes in HIV reservoirs. J Infect 2017; 75(6):555–571. [DOI] [PubMed] [Google Scholar]
- 12.Winckelmann A, Barton K, Hiener B, Schlub TE, Shao W, Rasmussen TA, et al. Romidepsin-induced HIV-1 viremia during effective antiretroviral therapy contains identical viral sequences with few deleterious mutations. AIDS 2017; 31(6):771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winckelmann A, Barton K, Hiener B, Shao W, Ostergaard L, Rasmussen T, et al. A5 Peripheral blood cells contribute to HIV-1 viremia induced by romidepsin. Virus Evol 2017; 3(Suppl 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winckelmann A, Morcilla V, Shao W, Schleimann MH, Hojen JF, Schlub TE, et al. Genetic characterization of the HIV-1 reservoir after Vacc-4x and romidepsin therapy in HIV-1-infected individuals. AIDS 2018; 32(13):1793–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, et al. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 2018; 10(430). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 2014; 20(4):425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sedlak RH, Liang S, Niyonzima N, De Silva Feelixge HS, Roychoudhury P, Greninger AL, et al. Digital detection of endonuclease mediated gene disruption in the HIV provirus. Sci Rep 2016; 6:20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartholomeeusen K, Fujinaga K, Xiang Y, Peterlin BM. Histone deacetylase inhibitors (HDACis) that release the positive transcription elongation factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. The Journal of biological chemistry 2013; 288(20):14400–14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2013; 9(2):e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams JP, Hurst J, Stohr W, Robinson N, Brown H, Fisher M, et al. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife 2014; 3:e03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 2016; 30(3):343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasternak AO, Berkhout B. What do we measure when we measure cell-associated HIV RNA. Retrovirology 2018; 15(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 2012; 36(3):491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Droplet digital PCR (ddPCR) plot showing the detection of different cell-associated HIV RNA TAR sequences after romidepsin infusion. (A) One dimension ddPCR plot corresponding to channel 1 (FAM) fluorescence amplitude detected at each time point, and (B) Two dimensions ddPCR plots corresponding to channel 1 (FAM) and channel 2 (VIC) fluorescence amplitude detected at each time point.
Participant characteristics.


