A novel array of human lung glycans reveals influenza A virus binding to phosphorylated as well as sialylated N-glycans.
Abstract
Influenza A viruses can bind sialic acid–terminating glycan receptors, and species specificity is often correlated with sialic acid linkage with avian strains recognizing α2,3-linked sialylated glycans and mammalian strains preferring α2,6-linked sialylated glycans. These paradigms derive primarily from studies involving erythrocyte agglutination, binding to synthetic receptor analogs or binding to undefined surface markers on cells or tissues. Here, we present the first examination of the N-glycome of the human lung for identifying natural receptors for a range of avian and mammalian influenza viruses. We found that the human lung contains many α2,3- and α2,6-linked sialylated glycan determinants bound by virus, but all viruses also bound to phosphorylated, nonsialylated glycans.
INTRODUCTION
Influenza A viruses (IAVs) are a substantial annual burden on human health and the economy, and novel pandemic strains emerge from wild waterfowl hosts at unpredictable intervals. Sialic acid (Sia)–terminating cell surface glycans have been identified as receptors for IAV hemagglutinin (HA), and Sia linkage specificity is thought to provide a critical barrier for cross-species transmission, with avian viruses binding α2,3-linked sialylated glycans and human viruses preferring α2,6-linked sialylated glycans (1–7). Many studies have examined the binding characteristics and the structural interactions of avian and human IAV HAs with glycan receptors and the effects caused by mutations in the HA binding site [reviewed in (8, 9, 10)]. However, a preponderance of the historical data on IAV receptor binding is based on agglutination of erythrocytes from different species (or α2,3- or α2,6-linked resialylated erythrocytes), binding to synthetic sugars or binding to poorly defined surface ligands on cells or tissues. There appears to be a difference in the abundance of sialylated glycan receptors at sites of infection in the natural hosts, i.e., α2,3-linked sialylated glycans are more prevalent in the intestinal tract of birds and α2,6-linked sialylated glycans are enriched on mucosal surfaces of the human upper respiratory tract, largely based on the presumption of specificity of Sia binding by the plant lectins Sambucus nigra agglutinin (SNA; α2,6-linked Sia) and Maackia amurensis lectin (MAL-I; α2,3-linked Sia) (11–13).
Here, we report our characterization of the N-glycome from human lung tissue and our generation of the first human lung–shotgun N-glycan microarray (HL-SGM) to define the natural receptors recognized by a range of IAV. We identified a broad spectrum of both α2,3- and α2,6-linked sialylated N-glycans of various branch patterns and chain length, with and without core fucose modifications, demonstrating a multitude of potential sialylated glycan receptors for IAV. A range of avian, swine, and human IAV of various subtypes exhibited differing spectrums of binding profiles, from very broad binding to a variety of glycans to highly selective ligand specificity. We found that all IAV strains examined also bind well to phosphorylated, nonsialylated N-glycans. While Sia recognition by HA is an established feature of IAV, our findings suggest a possible role for alternative ligands, such as the phosphorylated glycans identified here, as cofactors or facilitators of virus entry, possibly opening new/additional routes to influenza treatment.
RESULTS
Human lung–shotgun N-glycan microarray
To identify the endogenous glycans recognized by IAV, we generated an HL-SGM using N-glycans isolated directly from human lung tissue (Fig. 1A). N-glycans were fluorescently labeled, separated by two-dimensional (2D) high-performance liquid chromatography (HPLC), and 120 fractions were collected and concentrated to equal volume to reflect the natural abundance in the lung and then were covalently printed on N-hydroxysuccinimide (NHS)–activated slides. We first interrogated the HL-SGM with a variety of lectins to confirm the functional immobilization and recognition of glycans and to broadly survey some major structural features of lung N-glycans (Fig. 1B). SNA and MAL-I, which recognize α2,6- and α2,3-linked sialylated glycans, respectively, bound to divergent glycan fractions with higher chart IDs (50 to 138), indicating the presence of both α2,3- and α2,6-linked Sia in the human lung. The lectins concanavalin A, wheat germ agglutinin, and Ricinus communis agglutinin, which bind to glycan determinants with terminal mannose, N-acetylglucosamine/Sia, and galactose/Sia residues, respectively, bound a variety of glycans. The fucose-binding lectin Aleuria aurantia lectin bound many glycans, demonstrating that fucosylation is common in lung N-glycans. The terminal galactose-binding lectin Erythrina cristagalli lectin bound relatively poorly to the HL-SGM, but its binding was enhanced following exposure of galactosyl residues upon removal of Sia by neuraminidase (NA) treatment (fig. S1). Lycopersicon esculentum (tomato) lectin, which recognizes LacNAc repeats within poly-N-acetyllactosamine sequences (-3Galβ1-4GlcNAcβ1-)n (14), bound weakly to a number of fractions, suggesting that LacNAc repeats are not abundant. To further analyze the sialylated glycans and the specificities of SNA and MAL-I, we treated HL-SGM with Arthrobacter ureafaciens NA, which cleaves α2,3-, α2,6-, and α2,8-linked Sia. This treatment completely eliminated binding of SNA, indicating complete removal of α2,6-linked Sia, but MAL-I binding was only partially reduced (fig. S1). This is consistent with our recent findings, demonstrating that MAL-I can also detect certain nonsialylated, galactose-terminating, branched complex-type N-glycans (15) and, therefore, binding by MAL-I does not directly correspond to the presence of sialylated glycans (16).
Fig. 1. An HL-SGM was generated and validated by lectin binding.
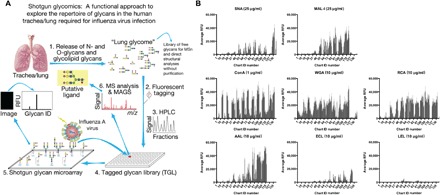
(A) To identify endogenous receptors recognized by IAV, we generated a shotgun glycan microarray comprising the N-glycans from the human lung (7, 42, 43). Lungs (provided by LifeLink) were processed following the oxidative release of natural glycans (ORNG) method (17) and labeled with the bifunctional linker, 2-amino-N-(2-aminoethyl)-benzamide (AEAB) (40). The labeled N-glycans were printed on glass slides to generate the HL-SGM. RFU, relative fluorescence units; m/z, mass/charge ratio; MS, mass spectrometry; MSn, tandem mass spectrometry; MAGS, Metadata Assisted Glycan Sequencing. (B) Biotinylated lectins were bound to the HL-SGM at the noted concentrations and were detected with cyanine 5–conjugated streptavidin. ConA, concanavalin A; WGA, wheat germ agglutinin; RCA, R. communis agglutinin; AAL, A. aurantia lectin; ECL, E. cristagalli lectin; LEL, L. esculentum lectin.
Binding of influenza viruses to HL-SGM
We analyzed binding to the HL-SGM of a panel of 11 different IAVs, including avian, swine, and human strains of differing subtype, geographic location, and date of isolation (table S1). Each IAV exhibited differential binding to the HL-SGM (Fig. 2A). The human H1N1 vaccine and seasonal strains, A/Brisbane/59/2007 and A/Pennsylvania/08/2008 (Penn), displayed very broad binding profiles to many glycans, while the H1N1 pandemic isolates A/California/04/2009, A/Texas/15/2009, and A/Mexico/InDRE4487/2009, as well as the H3N2 seasonal strain A/New York/55/2004, bound in a more restricted fashion, preferring glycans with lower number chart IDs (1 to 48), which generally correspond to less sialylated glycans. The swine isolates, A/sw/Minnesota/02719/2009 and A/sw/Illinois/02860/2009, exhibit broad binding, while the binding profile for A/sw/Minnesota/02749/2009 was also restricted to the lower numbered chart IDs (1 to 48). The avian isolates display wide-ranging binding profiles.
Fig. 2. A range of IAVs all displays binding on the HL-SGM to chart IDs not bound by sialylated glycan binding lectins, SNA, and MAL-I.
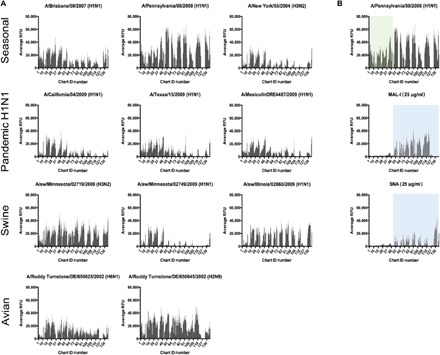
(A) Fluorescently labeled viruses, representative of different subtypes and host species, were bound to the array and display divergent binding profiles. (B) Penn binding to HL-SGM and comparison to SNA and MAL-I. IAV virus Penn was labeled with Alexa Fluor 488 and bound to the HL-SGM. The panels for MAL-I and SNA binding are included to demonstrate the nonoverlap between the Sia-binding lectins (light blue boxes) and the virus binding in the lower fraction numbers (green box).
We selected Penn for more detailed studies, as this virus exhibits a robust binding signal and a broad receptor recognition profile. For each fraction collected by HPLC, a comparison of virus binding activity to fluorescence signal [due to 2-amino-N-(2-aminoethyl)-benzamide (AEAB) label on all glycans] showed no correlation between binding specificity and glycan abundance (fig. S2). The Penn strain bound to glycans of lower fraction numbers, e.g., chart IDs (1 to 48), which are relatively nonsialylated (Fig. 2B), and to those in higher chart IDs (53 to 138), which are sialylated and bound by both MAL-I and SNA. To examine the Sia requirement for virus binding in more detail, we treated the HL-SGM slides with A. ureafaciens NA (Fig. 3A). As a control, we treated a defined sialylated N-glycan microarray side by side to establish reaction conditions on a known array that ensures the complete removal of Sia (fig. S3A). We considered the treatment complete once the binding of SNA to the N-glycan microarray was reduced to background levels. Penn retained binding to the HL-SGM after removal of Sia, and binding was mainly to glycans with lower chart IDs (1 to 48). These glycans did not bind SNA and MAL-I, indicating the presence of additional glycan binding determinants for IAV independent of Sia. We also examined avian and swine influenza strains on a desialylated slide, and both retained binding to the lower numbered fractions as observed with Penn, indicating that binding to nonsialylated glycans is not limited to the Penn strain (fig. S3B).
Fig. 3. The Penn strain binds to sialylated glycans and to glycan fractions containing phosphorylated structures.
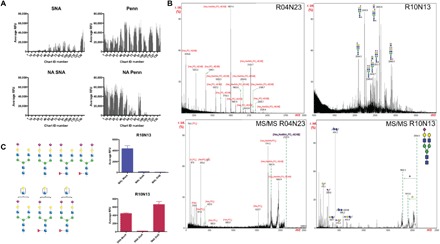
(A) NA treatment of HL-SGM and interrogation with SNA (25 μg/ml; detected with cyanine 5–conjugated streptavidin) and Penn reveal that IAV binds to array after removal of Sia as determined by loss of SNA binding. (B) Matrix-assisted laser desorption/ionization–time-of-flight MS (MALDI-TOF-MS) analysis of fractions, R10N13 (chart ID 118; high binding) indicating sialylated N-glycans and R04N23 (chart ID 47; high binding after NA treatment) indicating phosphorylated glycans, and tandem MS (MS/MS) of selected peaks. (C) Neuraminidase treatment of highest binding fraction R10N13 (predicted structures shown to the left; blue square, N-acetylglucosamine; red triangle, fucose; green circle, mannose; yellow circle, galactose; purple diamond, Sia) and binding of SNA (red bars) and MAL-I (blue bars) reveal the presence of both linkage types within the fraction.
Mass spectrometry of high binding fractions of HL-SGM
We analyzed the HL-SGM fractions broadly recognized by a range of viruses. Matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF-MS) analyses of glycans revealed both sialylated glycans and those that were phosphorylated (Fig. 3B and fig. S4). The presence of sialylated glycans was exemplified by the MS analysis of the fraction R10N13 (chart ID 118). We detected major molecular ions [e.g., mass/charge ratio (m/z) 2094, 2240, 2605, and 2896] with compositions that represent mono- or disialylated complex-type N-glycans. The glycans in this fraction, which exhibited the highest relative fluorescence units (RFUs) for Penn binding, are predicted biantennary N-glycans, some with a core fucose, some presenting an additional N-acetyllactosamine unit (Galβ1-4GlcNAcβ1-). All the glycans are sialylated, although only some are sialylated on both branches (Fig. 3C). To determine the terminal Sia linkage, we treated the HL-SGM with A. ureafaciens NA and with Streptococcus pneumoniae NA, the latter of which cleaves primarily α2,3-linked Sia. We examined the SNA and MAL-I binding to fraction R10N13 (chart ID 118) before and after the NA treatments and found that MAL-I binding is decreased after both NA treatments, indicating that α2,3-linked Sia was present, and that SNA binding was decreased after A. ureafaciens NA treatment, but not after S. pneumoniae NA treatment, indicating that α2,6-linked Sia was also present (Fig. 3C). The MALDI-TOF-MS profile of the fraction R04N23 (chart ID 47), a fraction for which binding activity is not reduced by neuraminidase treatment, revealed the presence of a series of phosphorylated glycans with the compositions of Hex5–12HexNAc0–3PO3. On the basis of the knowledge of biosynthetic pathways of N-glycans, structures detected in this fraction are likely high mannose–type N-glycans with a phosphate moiety. Loss of the reducing end HexNAc units of a few molecular ions (e.g., m/z 1070, 1395, 1719, and 2043) was due to processing by the oxidative release of natural glycans (ORNG) method, as previously described (17). We also identified molecular ions corresponding to N-glycans as Hex9, e.g., that could be Man9-containing glycans, or with further extension of hexoses that could contain additional glucose residues (m/z 1881, 2043, and 2205). Three molecular ions (m/z 2004, 2166, and 2328) were observed to display an additional HexNAc unit, which was proposed to be in phosphodiester linkage to the high-mannose structures via a phosphate group, as previously described (18). The fragment ion at m/z 79 could not be generated from a sulfate group (m/z 97, HSO4−) by losing a water molecule. Therefore, the detection of the fragment ions at m/z 79 (PO3−) and 97 (H2PO4−) indicated that the molecular ion at m/z 2125 contained a phosphate moiety. In confirmation of this interpretation, treatment of the fractions R05N15 (chart ID 58) and R02N23 (chart ID 21) with NA and/or phosphatase caused peak shifts in the HPLC profiles that correspond to the loss of Sia or the loss of phosphorylation (fig. S5).
Significance of phosphorylated glycans for IAV binding
To define whether phosphorylation of glycans is required for binding by Penn, we treated the HL-SGM with bovine alkaline phosphatase, A. ureafaciens NA, or a combination of both (Fig. 4). We confirmed that alkaline phosphatase was efficient, using a synthetic glycan array containing a few phosphorylated glycans, in which alkaline phosphatase caused loss of binding of the single-chain variable domain antibody fragment (scFv) M6P-1, specific for selected mannose-6-phosphate (Man6P)–containing glycans (fig. S6) (19). Notably, the dephosphorylation of the HL-SGM by alkaline phosphatase treatment following NA treatment resulted in a total loss of virus binding (Fig. 4). Treating the arrays separately with NA or phosphatase individually leads to the retention of some binding, although the profiles are separate and distinct, with IAV binding on the NA-treated slide to glycans of chart IDs 1 to 48, whereas IAV binding on the phosphatase-treated slide occurred to glycans in chart IDs 50 to 120. We also treated the HL-SGM with NA and then completed the virus binding experiments in the presence of Man6P scFv (Fv M6P-1). The Fv M6P-1 competitively inhibited virus binding to the NA-treated slide (Fig. 4). The complete reduction of virus binding to background levels following phosphatase treatment, combined with the competitive inhibition exhibited by Fv M6P-1, confirms that glycan phosphorylation is a key contributor to the Sia-independent binding by Penn.
Fig. 4. A combination of NA and phosphatase treatment of the HL-SGM results in no binding of Penn, while single enzymatic treatments allow for binding to certain glycan fractions.
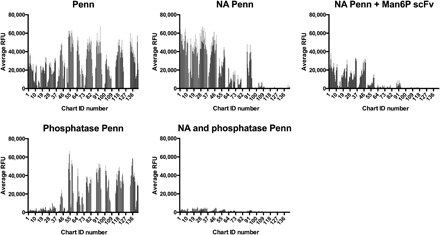
Binding inhibition with Fv M6P-1 (100 μg/ml) is also shown.
To further confirm the importance of glycan phosphorylation to IAV binding to the HL-SGM, we performed hapten inhibition studies (Fig. 5). Incubation of Penn with 20 mM Sia inhibited the binding to those sialylated glycans recognized by MAL-I and/or SNA. The binding profile of Sia inhibition mimics that of an NA-treated slide. Binding competition with 6-sialyllactose produces the same effect (fig. S7A). These results suggest that IAV binds sialylated glycans through the canonical Sia receptor–binding site (RBS) on HA. In competition experiments with 20 mM Sia on an NA-treated slide, IAV retains binding, albeit at somewhat lower RFUs, to the glycans not recognized by either MAL-I or SNA, suggesting that the binding is occurring in a Sia-independent manner and not via the RBS. When we included Man6P on a slide with the sialylated glycans intact, the hapten had no effect on binding to sialylated glycans, indicating that it is not interfering with the canonical RBS on HA. However, following desialylation of glycans on the HL-SGM, the inclusion of Man6P caused a reduction in binding to most of the glycans. This effect is most pronounced for Man6P; other sugars, phosphorylated or sulfated in the 1 or 6 position, including mannose-6-sulfate (Man6S) and fructose-6-phosphate (Fruc6P), had more limited inhibition of IAV binding. Inclusion of glucose-1-phosphate (Glc1P) and Glc6P reduced IAV binding to certain fractions such as chart IDs 26 to 33. Overall, these results indicate that Sia is an inhibitor of IAV binding to sialylated glycans and that Man6P is an inhibitor of binding to phosphorylated glycans.
Fig. 5. Different phosphorylated sugars produce no inhibition to Penn on the HL-SGM in hapten competition assays, indicating that charge alone is not mediating virus binding to nonsialylated glycans.
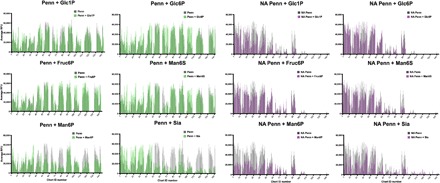
The IAV-only RFUs are represented in gray, while the IAV and hapten RFUs are in green. In addition, the HL-SGM was treated with neuraminidase A (denoted as NA) to remove Sia before binding experiments, and the IAV-only RFUs are in gray, while the IAV and hapten RFUs are in purple.
Proteomics of virus particle
Consistent with the above results, Man6P does not inhibit binding to sialylated N-glycans on the defined N-glycan array, again suggesting that the inhibition is not due to competition with the RBS (fig. S7B). This result suggests that IAV may have an alternative binding site for phosphorylated glycans. We also considered the possibility that IAV might contain host cell–derived Man6P receptors (MPRs). We tested binding of IAV to a glycan microarray containing a few synthetic glycans with Man6P (fig. S7C) and found no virus binding. In previous studies, we reported that the recombinant cation-independent MPR (CI-MPR) and cation-dependent MPR (CD-MPR) (20), as well as Fv M6P-1 (19), all bound well to the Man6P-containing glycans. These results demonstrate that the IAV is not using either the CI-MPR or the CD-MPR to bind to glycans with Man6P modifications. Nevertheless, we conducted proteomic analysis of virus preparations, propagated in Madin-Darby canine kidney (MDCK) cells, for the presence of the host CI-MPR and/or the CD-MPR and detected the presence of peptides corresponding to the canine CI-MPR (see Supplementary Methods and fig. S8) in addition to other host proteins, although relative abundance compared to viral proteins is unknown. The liquid chromatography–tandem MS data have been uploaded to the MassIVE repository as part of the University of California San Diego Center for Computational Mass Spectrometry (UCSD/CCMS) MS datasets. The raw files are housed in the repository and linked with our publication title. Thus, while our analysis reveals that the canine MPR is present on the virus particle, the inability of the virus to bind to the Man6P-containing glycan array demonstrates that the recognition of phosphorylated glycans is not mediated by the MPR and/or requires additional complex glycan determinants.
DISCUSSION
Our findings provide novel insights into the interaction of influenza viruses with glycans derived from the human lung. While many previous studies have demonstrated that IAV can bind Sia-containing glycans, the types of glycans in the human lung recognized by IAV are not well studied. Through our generation of the HL-SGM representing the human lung N-glycans, we explored more broadly the types of natural and endogenous glycans bound by IAV. Our studies led to the unexpected discovery that, in addition to binding select sialylated glycans, many IAV strains can also bind phosphorylated glycans.
The discovery of phosphorylated binding determinants for IAV HA suggests new possibilities for the development of therapeutics for IAV infection.
The development of the human lung N-glycome and generation of an HL-SGM was prompted by the fact that many previous studies on IAV binding to glycans were performed on defined glycan microarrays of limited relevance to endogenous glycans in the human lung or were performed with erythrocytes, which may not be ideal surrogates for defining IAV glycan specificity. While erythrocytes may have many sialylated glycans, there are few studies on the structures of N-glycans in animal erythrocytes and no studies on specific erythrocyte-derived glycans bound by IAV. Defined glycan microarrays, while obviously useful for exploring glycan recognition, have not been generated to replicate the natural N-glycome of human lung.
A few reports have characterized a selection of available receptors for IAV in natural tissues (7, 21, 22); however, our HL-SGM provides the first natural glycan microarray for the human lung and allowed us to identify a broad range of endogenous natural glycan receptors for IAV. We identified a wide variety of sialylated N-glycan determinants with α2,3 and/or α2,6 linkages including bi-, tri-, and tetraantennary glycans of varying sizes. If both Sia linkage types can be found on cell surfaces at sites of infection, then receptor availability may not be the only binding determinant that drives cross-species transmission. Mutations in HA that lead to dual recognition of both α2,3- and α2,6-linked Sia have been linked to avian-to-mammalian transmission of H5 and H7 subtype strains in experimental settings (23–28); however, the loss of α2,3-binding specificity may actually be more important for sustained transmission than a gain of α2,6-binding specificity (29), perhaps as a mechanism for avoiding interactions with mucins that are rich in α2,3-linked Sia (30). Our HL-SGM binding studies here and previous work on a pig lung shotgun glycan microarray indicate that the same preferred receptor, the sialylated biantennary N-glycan, is present in both human and pig lungs (7), supporting the view that receptor availability constitutes only a minor hurdle for cross-species transmission between pigs and humans. With regard to SNA and MAL-I that have been used historically to identify receptor distribution in different host species (1–4, 6, 7) and tissues (11–13), our results revealed that nonsialylated glycans from the lung are recognized by MAL-I. This finding raises important reservations concerning the interpretation of glycan expression relative to IAV binding from lectin binding data alone.
Our discovery of IAV binding to phosphorylated glycan determinants in addition to selected sialylated glycan determinants raises important possibilities for IAV entry. Hapten inhibition experiments show that virus binding to sialylated glycans is inhibited by Sia and not by Man6P, whereas binding to phosphorylated structures is not inhibited by Sia but is inhibited by Man6P. These results suggest that IAV binding to phosphorylated glycans is not occurring via the canonical receptor binding site on HA and that either HA or NA may contain binding sites for the phosphorylated glycans. Secondary binding sites on HA and NA have already been demonstrated for Sia recognition (31, 32). HA interactions with monovalent Sia-containing substrates are relatively weak (33, 34), and successful attachment and entry are thought to rely on multiple HA-receptor interactions in which phosphorylated glycans could participate. This binding to phosphorylated glycans could also explain the observation that desialylation of cells by NA treatment fails to block infection (35–37). The recognition of alternative binding substrates could provide influenza viruses with increased evolutionary plasticity by facilitating attachment or entry, as selective pressures are driving changes in the Sia-binding site of HA, and this could have repercussions with respect to cross-species transmission or vaccine efforts.
MATERIALS AND METHODS
Human tissues
Donor lungs not fit for transplant were provided by LifeLink Foundation Inc. (Norcross, GA). Only lungs from healthy individuals with no smoking history or lung complications were included in the study.
Viruses and cells
Virus stocks representing swine, human, and avian isolates were selected on the basis of their availability and the relevance of the strain with respect to circulation in a population (table S1). As described previously (6, 7), the virus stocks were grown in MDCK cells or MDCK-Siat 1 cells that were maintained using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum and penicillin-streptomycin. Briefly, 85 to 90% confluent MDCK cells grown in T175 flasks were washed twice with phosphate-buffered saline (PBS), overlaid with 10 ml of serum-free DMEM supplemented with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin (1 μg/ml) and infected with a 1:1000 or 1:10,000 dilution of the original virus stock. Cells were incubated with rocking for 1 hour at ambient temperature, the inoculum was removed, and 30 ml of serum-free DMEM supplemented with TPCK trypsin (1 μg/ml) was added. The flasks were incubated at 37°C for 2 to 3 days until monolayers were 80 to 90% destroyed. Infected cell supernatants were clarified by low-speed centrifugation and then pelleted through a 25% sucrose cushion in NTE buffer (100 mM NaCl, 10 mM tris, and 1 mM EDTA). Purified viruses were pelleted by centrifugation in ultra-clear centrifuge tubes (Beckman Coulter, Indianapolis, IN) and a Beckman Coulter SW32 Ti rotor at 133548.5g for 2 hours and were resuspended in 2 ml of PBS buffer at 4°C overnight. The resuspended pellets were combined and subjected to an additional spin in an SW41 Ti rotor at 55408g for 1 hour. These pellets were resuspended in 2 ml of PBS, aliquoted, and frozen at −80°C. Frozen, purified virus strains were later thawed, and the hemagglutination units (HAUs) and plaque-forming units (PFU/ml) titers were determined using standard techniques. HA concentration was monitored for the array experiments, although the viruses were not normalized to the same HA titer. We have found that a minimum neat titer of 256 HAU/50 μl is sufficient to visualize virus binding to the array and have confirmed that the binding pattern does not change, although the RFUs increase, upon higher HA titers. The stocks of all viruses used in these studies were resequenced after the limited amplification in MDCK cells before labeling for glycan array analysis. We detected no sequence changes and no evidence of observable heterogeneity.
Agglutination of erythrocytes
Chicken erythrocytes were acquired from Lampire Biological Laboratories (Pipersville, PA) or Rockland Immunochemicals Inc. (Limerick, PA) as whole-blood preparations. As described previously (6, 7), whole-blood preparations were washed two times with 1× PBS and diluted to 0.5% for hemagglutination experiments, which were performed using standard techniques. Briefly, 0.5% erythrocyte preparations were added to twofold serial dilutions of 50 μl of virus stock for 1 hour to determine the HA titer. This procedure was performed before and after labeling the virus with Alexa Fluor 488 to monitor the HA titer during the labeling process. Infectious titers of virus stocks were also determined by plaque assay on MDCK cell monolayers.
Fluorescent labeling of IAV
Binding of fluorescently labeled IAV was performed as previously described (6, 7, 38, 39). Briefly, 200 μl of virus was incubated with 25 μg of Alexa Fluor 488 (Invitrogen) in 1 M NaHCO3 (pH 9.0) for 1 hour at room temperature. Labeled viruses were dialyzed against PBS using a 7000–molecular weight–cutoff Slide-A-Lyzer minidialysis unit (Thermo Fisher Scientific) overnight at 4°C. In all cases, labeled viruses were used in experiments the following day.
Isolation of N-glycans
The frozen human lung tissue was sectioned to obtain pieces representative of each lobe to a total of ~500 g. The tissue was then blended to homogenize and was processed to release intact N-glycans via ORNG, which has been shown to be specific for N-glycan release (17). Briefly, the tissue was treated with 6% NaCIO, and the reaction was quenched via the addition of formic acid. Precipitates were removed via centrifugation, and the solution was dialyzed before labeling with the fluorescent linker, AEAB (40). The AEAB-labeled glycans were then separated over 2D HPLC. The separated fractions become the samples in the tagged glycan library representative of the human lung. These samples were used both for direct structural analysis and for microarray analysis. For glycan microarray analysis, the fractions were printed on NHS-activated slides to generate the HL-SGM. The fractions were printed without adjustment to the concentration to reflect the abundance of each glycan component in the lung.
HPLC separation of AEAB glycans
An HPLC CBM-20A system (Shimadzu), coupled with an ultraviolet-light detector (SPD-20A) and a fluorescence detector (RF-10AXL), was used for HPLC analysis and separation of AEAB-conjugated glycans. 2D HPLC separation, using reverse phase (Vydac C18, Hichrom, UK), followed by normal phase (ZORBAX NH2, Agilent, USA) columns (39), provided multiple fractions. The quantified fractions obtained after 2D HPLC were then lyophilized and stored frozen as the human lung–tagged glycan library.
MS analysis
All MS analyses of AEAB glycans were carried out using a Bruker Daltonics Ultraflex-II MALDI-TOF/TOF system under reflectron negative mode. 2,5-dihydroxybenzoic acid (10 mg/ml in 50% methanol) was used as matrix.
Microarray preparation and analysis
All samples were diluted 1:5 in sodium phosphate buffer (150 mM; pH 8.5) before printing. Noncontact piezo dispensing was performed using a sciFLEXARRAYER S11 ultralow volume dispensing robot (Scienion AG) onto NHS-functionalized glass slides (SCHOTT Nexterion Slide H, SCHOTT AG). All printed samples were dispensed within 10% variation of 330 pl in replicates of four spots per analyte. Upon completion of dispensing, printed arrays were incubated overnight at 80% relative humidity. All arrays were then blocked via submersion in 100 mM sodium tetraborate buffer (pH 8.5) with 50 mM ethanolamine for 1 hour at room temperature on an orbital shaker, followed by 10× dip washes in 1× PBS with 0.05% polysorbate 20, followed by 10× dip washes in ultrapure water. The arrays were then dried in a slide spinner and stored at −20°C until needed.
Before assay, arrays were rehydrated for 5 min in TSM buffer [20 mM tris-HCl, 150 mM sodium chloride (NaCl), 0.2 mM calcium chloride (CaCl2), and 0.2 mM magnesium chloride (MgCl2)], and the analyses were carried out for lectins and viruses as previously described for 1 hour at 4°C to prevent NA activity (37, 40). Biotinylated lectins (Vector Labs) were used as controls in the binding assay, and the bound lectins were detected by a secondary incubation with cyanine 5–conjugated streptavidin (5 μg/ml; Invitrogen). For multipanel experiments on a single slide, a ProPlate Multi-Well chamber (Grace Bio-Labs) was used to isolate each printed array during the assay. All arrays were scanned using a GenePix 4300A microarray scanner (Molecular Devices) equipped with four lasers covering an excitation range from 488 to 635 nm. Acquired images were quantified using GenePix Pro Microarray Analysis software and then further processed using Microsoft Excel spreadsheets as previously described (38, 41). For cyanine 5 fluorescence, 649 nm (ex) and 670 nm (em) were used. For Alexa Fluor 488 fluorescence, 495 nm (ex) and 519 nm (em) were used. For enzymatic treatment of the microarrays, each array was rehydrated for 5 min in TSM buffer and then incubated with Neuraminidase A (New England BioLabs) diluted 1:6 in 1× GlycoBuffer for 18 hours rocking at room temperature. Following incubation, the array was washed four times each with TSMW (20 mM tris-HCl, 150 mM NaCl, 0.2 mM CaCl2, 0.2 mM MgCl2, and 0.05% Tween-20), TSM, and water. For phosphatase treatment, arrays were treated with bovine alkaline phosphatase (Sigma-Aldrich, Burlington, MA) at a 1:5 dilution in water for 24 hours at 37°C. Following incubation, the array was washed four times each with TSM and water. Hapten sugars Man6P, Fruc6P, Glc1P, Glu6P, and Man6S were obtained from Sigma-Aldrich, and Fv M6P-1 was produced by S.M.-L. (19). 6-sialyllactose was also used for hapten competition and obtained from Carbosynth. Hapten competition experiments were carried out with 20 mM hapten or 100 μg/ml of the Fv M6P-1 premixed with the virus or lectin before incubation of the combined solution on the array. Because of normal variance in absolute RFU signal between replicate experiments, the glycan microarray experiments are considered to be a qualitative assay, with specific emphasis on the binding profile or signature of the sample, rather than a quantitative examination of total RFU. In the case of enzymatic treatment or hapten competition, the reduction in signal is measured pairwise with untreated slides with the same sample preparation to reduce experiment to experiment variance; however, the interpretation of the results is still focused on qualitative changes in the binding profile and not on the addition or subtraction of absolute RFU.
Supplementary Material
Acknowledgments
We would like to thank A. McQuillan (BIDMC), U. Agge, and N. Harmel (both at RCB) for technical assistance and advice. Funding: We acknowledge support by the U.S. Department of Health and Human Services contract HHSN272201400004C (NIAID Centers of Excellence for Influenza Research and Surveillance) and the NIH award P41GM103694 (National Center for Functional Glycomics). J.F.T. was supported by National Institute of Allergy and Infectious Disease (NIAID) of the NIH under award number T32AI106699. Author contributions: L.B.-L., N.J., J.F.T., C.G., and S.D. performed the experiments, with the assistance of S.F.C. and J.H.-M. L.B.-L. and N.J. analyzed the data, with the assistance of J.H.-M. S.M.-L. and T.B. provided the critical reagent. D.A.S. and R.D.C. supervised the experiments. L.B.-L., N.J., D.A.S., and R.D.C. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The antibody fragment Fv M6P-1 can be provided by T.B. and S.M.-L. for noncommercial research purposes pending scientific review and a completed material transfer agreement. Requests for the Fv M6P-1 should be submitted to T. Stachelhaus, Ascenion GmbH (Stachelhaus@ascenion.de). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaav2554/DC1
Supplementary Methods
Table S1. IAV strains used in this work.
Table S2. Raw glycan microarray data (Excel file).
Fig. S1. Success of the HL-SGM print is further validated by lectin binding after A. ureafaciens NA treatment.
Fig. S2. HPLC chromatograms overlaid with the HL-SGM binding profile for Penn indicate that glycan specificity is more important for virus binding than abundance of glycan.
Fig. S3. Conditions for NA treatment of the microarrays are confirmed on a defined N-glycan microarray, and those enzymatic conditions are applied to the HL-SGM for an avian and swine strain, revealing the same Sia-independent binding.
Fig. S4. MALDI-TOF-MS characterization of additional glycan fractions reveals phosphorylated glycans and sialylated glycans.
Fig. S5. Presence of sialylated or phosphorylated glycans within the human lung fractions is confirmed by peak shifts in the HPLC profile after enzymatic treatment due to phosphatase or NA sensitivity.
Fig. S6. Phosphatase conditions for the HL-SGM were optimized on a defined mannose phosphate glycan microarray using binding of Fv M6P-1.
Fig. S7. Hapten competition studies indicate that binding to sialylated glycans can be inhibited by sialyllactose, but not Fv M6P-1, which binds to the mannose phosphate array while Penn does not.
Fig. S8. Proteomics of Penn grown in canine kidney cells identifies the canine MPR protein.
REFERENCES AND NOTES
- 1.Rogers G. N., Paulson J. C., Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127, 361–373 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Rogers G. N., D’Souza B. L., Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173, 317–322 (1989). [DOI] [PubMed] [Google Scholar]
- 3.Connor R. J., Kawaoka Y., Webster R. G., Paulson J. C., Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205, 17–23 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Russell R. J., Stevens D. J., Haire L. F., Gamblin S. J., Skehel J. J., Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj. J. 23, 85–92 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Kumari K., Gulati S., Smith D. F., Gulati U., Cummings R. D., Air G. M., Receptor binding specificity of recent human H3N2 influenza viruses. Virol. J. 4, 42 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley K. C., Jones C. A., Tompkins S. M., Tripp R. A., Russell R. J., Gramer M. R., Heimburg-Molinaro J., Smith D. F., Cummings R. D., Steinhauer D. A., Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (Novel 2009 H1N1). Virology 413, 169–182 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Byrd-Leotis L., Liu R., Bradley K. C., Lasanajak Y., Cummings S. F., Song X., Heimburg-Molinaro J., Galloway S. E., Culhane M. R., Smith D. F., Steinhauer D. A., Cummings R. D., Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 111, E2241–E2250 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Graaf M., Fouchier R. A., Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 33, 823–841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skehel J. J., Wiley D. C., Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Xiong X., McCauley J. W., Steinhauer D. A., Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr. Top. Microbiol. Immunol. 385, 63–91 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Couceiro J. N., Couceiro S. S., Paulson J. C., Baum L. G., Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 29, 155–165 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Barkhordari A., Stoddart R. W., McClure S. F., McClure J., Lectin histochemistry of normal human lung. J. Mol. Histol. 35, 147–156 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Matrosovich M. N., Matrosovich T. Y., Gray T., Roberts N. A., Klenk H.-D., Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 101, 4620–4624 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merkle R. K., Cummings R. D., Relationship of the terminal sequences to the length of poly-N-acetyllactosamine chains in asparagine-linked oligosaccharides from the mouse lymphoma cell line BW5147. Immobilized tomato lectin interacts with high affinity with glycopeptides containing long poly-N-acetyllactosamine chains. J. Biol. Chem. 262, 8179–8189 (1987). [PubMed] [Google Scholar]
- 15.Wang L., Cummings R. D., Smith D. F., Huflejt M., Campbell C. T., Gildersleeve J. C., Gerlach J. Q., Kilcoyne M., Joshi L., Serna S., Reichardt N.-C., Parera Pera N., Pieters R. J., Eng W., Mahal L. K., Cross-platform comparison of glycan microarray formats. Glycobiology 24, 507–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisler C., Jarvis D., Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology 21, 988–993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song X., Ju H., Lasanajak Y., Kudelka M. R., Smith D. F., Cummings R. D., Oxidative release of natural glycans for functional glycomics. Nat. Methods 13, 528–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabas I., Kornfeld S., Biosynthetic intermediates of β-glucuronidase contain high mannose oligosaccharides with blocked phosphate residues. J. Biol. Chem. 255, 6633–6639 (1980). [PubMed] [Google Scholar]
- 19.Blackler R. J., Evans D. W., Smith D. F., Cummings R. D., Brooks C. L., Braulke T., Liu X., Evans S. V., Müller-Loennies S., Single-chain antibody-fragment M6P-1 possesses a mannose 6-phosphate monosaccharide-specific binding pocket that distinguishes N-glycan phosphorylation in a branch-specific manner. Glycobiology 26, 181–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohnsack R. N., Song X., Olson L. J., Kudo M., Gotschall R. R., Canfield W. M., Cummings R. D., Smith D. F., Dahms N. M., Cation-independent mannose 6-phosphate receptor: A composite of distinct phosphomannosyl binding sites. J. Biol. Chem. 284, 35215–35226 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walther T., Karamanska R., Chan R. W., Chan M. C. W., Jia N., Air G., Hopton C., Wong M. P., Dell A., Malik Peiris J. S., Haslam S. M., Nicholls J. M., Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLOS Pathog. 9, e1003223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasik B. R., Barnard K. N., Ossiboff R. J., Khedri Z., Feng K. H., Yu H., Chen X., Perez D. R., Varki A., Parrish C. R., Distribution of O-acetylated sialic acids among target host tissues for influenza virus. mSphere 2, e00379-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai M., Watanabe T., Hatta M., Das S. C., Ozawa M., Shinya K., Zhong G., Hanson A., Katsura H., Watanabe S., Li C., Kawakami E., Yamada S., Kiso M., Suzuki Y., Maher E. A., Neumann G., Kawaoka Y., Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486, 420–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herfst S., Schrauwen E. J., Linster M., Chutinimitkul S., de Wit E., Munster V. J., Sorrell E. M., Bestebroer T. M., Burke D. F., Smith D. J., Rimmelzwaan G. F., Osterhaus A. D., Fouchier R. A., Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell C. A., Fonville J. M., Brown A. E. X., Burke D. F., Smith D. L., James S. L., Herfst S., van Boheemen S., Linster M., Schrauwen E. J., Katzelnick L., Mosterin A., Kuiken T., Maher E., Neumann G., Osterhaus A. D. M. E., Kawaoka Y., Fouchier R. A. M., Smith D. J., The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 336, 1541–1547 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe Y., Ibrahim M. S., Ellakany H. F., Kawashita N., Mizuike R., Hiramatsu H., Sriwilaijaroen N., Takagi T., Suzuki Y., Ikuta K., Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLOS Pathog. 7, e1002068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K., Xu X., Lu H., Zhu W., Gao Z., Xiang N., Shen Y., He Z., Gu Y., Zhang Z., Yang Y., Zhao X., Zhou L., Li X., Zou S., Zhang Y., Li X., Yang L., Guo J., Dong J., Li Q., Dong L., Zhu Y., Bai T., Wang S., Hao P., Yang W., Zhang Y., Han J., Yu H., Li D., Gao G. F., Wu G., Wang Y., Yuan Z., Shu Y., Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Shi Y., Zhang W., Wang F., Qi J., Wu Y., Song H., Gao F., Bi Y., Zhang Y., Fan Z., Qin C., Sun H., Liu J., Haywood J., Liu W., Gong W., Wang D., Shu Y., Wang Y., Yan J., Gao G. F., Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342, 243–247 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Xiong X., Coombs P. J., Martin S. R., Liu J., Xiao H., McCauley J. W., Locher K., Walker P. A., Collins P. J., Kawaoka Y., Skehel J. J., Gamblin S. J., Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497, 392–396 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Ehre C., Worthington E. N., Liesman R. M., Grubb B. R., Barbier D., O’Neal W. K., Sallenave J.-M., Pickles R. J., Boucher R. C., Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc. Natl. Acad. Sci. U.S.A. 109, 16528–16533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauter N. K., Glick G. D., Crowther R. L., Park S. J., Eisen M. B., Skehel J. J., Knowles J. R., Wiley D. C., Crystallographic detection of a second ligand binding site in influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 89, 324–328 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benton D. J., Wharton S. A., Martin S. R., McCauley J. W., Role of neuraminidase in influenza A(H7N9) virus receptor binding. J. Virol. 91, e02293-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauter N. K., Bednarski M. D., Wurzburg B. A., Hanson J. E., Whitesides G. M., Skehel J. J., Wiley D. C., Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: A 500-MHz proton nuclear magnetic resonance study. Biochemistry 28, 8388–8396 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Sauter N. K., Hanson J. E., Glick G. D., Brown J. H., Crowther R. L., Park S. J., Skehel J. J., Wiley D. C., Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: Analysis by proton nuclear magnetic resonance spectroscopy and x-ray crystallography. Biochemistry 31, 9609–9621 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Stray S. J., Cummings R. D., Air G. M., Influenza virus infection of desialylated cells. Glycobiology 10, 649–658 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Oshansky C. M., Pickens J. A., Bradley K. C., Jones L. P., Saavedra-Ebner G. M., Barber J. P., Crabtree J. M., Steinhauer D. A., Tompkins S. M., Tripp R. A., Avian influenza viruses infect primary human bronchial epithelial cells unconstrained by sialic acid α2,3 residues. PLOS ONE 6, e21183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Londrigan S. L., Turville S. G., Tate M. D., Deng Y.-M., Brooks A. G., Reading P. C., N-linked glycosylation facilitates sialic acid-independent attachment and entry of influenza A viruses into cells expressing DC-SIGN or L-SIGN. J. Virol. 85, 2990–3000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heimburg-Molinaro J., Tappert M., Song X., Lasanajak Y., Air G., Smith D. F., Cummings R. D., Probing virus–glycan interactions using glycan microarrays. Methods Mol. Biol. 808, 251–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amonsen M., Smith D. F., Cummings R. D., Air G. M., Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with α2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 81, 8341–8345 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song X., Xia B., Stowell S. R., Lasanajak Y., Smith D. F., Cummings R. D., Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem. Biol. 16, 36–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heimburg-Molinaro J., Song X., Smith D. F., Cummings R. D., Preparation and analysis of glycan microarrays. Curr. Protoc. Protein Sci. 64, 12.10.1–12.10.29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song X., Lasanajak Y., Xia B., Heimburg-Molinaro J., Rhea J. M., Ju H., Zhao C., Molinaro R. J., Cummings R. D., Smith D. F., Shotgun glycomics: A microarray strategy for functional glycomics. Nat. Methods 8, 85–90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith D. F., Cummings R. D., Application of microarrays for deciphering the structure and function of the human glycome. Mol. Cell. Proteomics 12, 902–912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaav2554/DC1
Supplementary Methods
Table S1. IAV strains used in this work.
Table S2. Raw glycan microarray data (Excel file).
Fig. S1. Success of the HL-SGM print is further validated by lectin binding after A. ureafaciens NA treatment.
Fig. S2. HPLC chromatograms overlaid with the HL-SGM binding profile for Penn indicate that glycan specificity is more important for virus binding than abundance of glycan.
Fig. S3. Conditions for NA treatment of the microarrays are confirmed on a defined N-glycan microarray, and those enzymatic conditions are applied to the HL-SGM for an avian and swine strain, revealing the same Sia-independent binding.
Fig. S4. MALDI-TOF-MS characterization of additional glycan fractions reveals phosphorylated glycans and sialylated glycans.
Fig. S5. Presence of sialylated or phosphorylated glycans within the human lung fractions is confirmed by peak shifts in the HPLC profile after enzymatic treatment due to phosphatase or NA sensitivity.
Fig. S6. Phosphatase conditions for the HL-SGM were optimized on a defined mannose phosphate glycan microarray using binding of Fv M6P-1.
Fig. S7. Hapten competition studies indicate that binding to sialylated glycans can be inhibited by sialyllactose, but not Fv M6P-1, which binds to the mannose phosphate array while Penn does not.
Fig. S8. Proteomics of Penn grown in canine kidney cells identifies the canine MPR protein.


