Abstract
The clinical field of wound healing is challenged by numerous hurdles. Not only are wound-healing disorders complex and multifactorial, but the corresponding patient population is diverse, often elderly and burdened by multiple comorbidities such as diabetes and cardiovascular disease. The care of such patients requires a dedicated, multidisciplinary team of physicians, surgeons, nurses and scientists. In spite of the critical clinical need, it has been over 15 years since a treatment received approval for efficacy by the FDA in the United States. Among the reasons contributing to this lack of effective new treatment modalities is poor understanding of mechanisms that inhibit healing in patients. Additionally, preclinical models do not fully reflect the disease complexity of the human condition, which brings us to a paradox: if we are to use a “mechanistic” approach that favours animal models, we can dissect specific mechanisms using advanced genetic, molecular and cellular technologies, with the caveat that it may not be directly applicable to patients. Traditionally, scientific review panels, for either grant funding or manuscript publication purposes, favour such “mechanistic” approaches whereby human tissue analyses, deemed “descriptive” science, are characterized as a “fishing expedition” and are considered “fatally flawed.” However, more emerging evidence supports the notion that the use of human samples provides significant new knowledge regarding the molecular and cellular mechanisms that control wound healing and contribute to inhibition of the process in patients. Here, we discuss the advances, benefits and challenges of translational research in wound healing focusing on human subject research.
Keywords: chronic ulcers, human specimens, pre-clinical models, wound healing
1 |. THE BURDEN OF NON-HEALING WOUNDS
Impaired wound healing is associated with pain, disability, loss of productivity, depression and social isolation for more than 6 million patients, costing the U.S. healthcare system approximately $20–25 billion each year.[1] Although they differ in etiology, chronic wounds such as venous leg ulcers (VLU), diabetic foot ulcers (DFU) and pressure ulcers (PU), all share an enormous impact due to high incidence with frequent relapses and associated complications.[2–4] VLU develop as a consequence of venous valvular incompetence causing venous reflux, obstruction and hypertension.[5–8] Chronic DFU are a widespread complication in patients suffering from diabetes mellitus (DM). DM affects 9.9% of the population older than 40 in the U.S. alone, of which 30% suffer from lower extremity lesions.[9] Diabetic patients who develop foot ulcer have a significantly increased mortality of 50% at 5 years, almost analogous to mortality rates seen in colon cancer.[10] PU usually occur over bony prominences due to prolonged pressure, or pressure in combination with shear and/or friction. PU particularly affect geriatric patients with multiple morbidities, particularly those that are bed- or wheelchair-bound.[4] Despite the severity of the clinical problem, there are currently only three federal drug administration (FDA) approved therapies for efficacy for the treatment of chronic VLU and DFU. These include the recombinant human PDGF (Regranex) and the tissue-engineered living human skin substitutes, Apligraf and Dermagraft.[4] However, over 50% of DFUs[11] and over 70% of VLU[12] fail to heal depending on the wound size and duration, and there is no efficacious therapy approved for PU treatment, emphasizing the urgent need to better understand mechanisms of healing inhibition for the development of novel therapeutic modalities. Faced with this “silent” epidemic of chronic wounds,[13] we encounter limitations in mechanism- and pathophysiology-based clinical management, owing to an impediment of therapeutic development deriving from poor understanding of mechanism at a molecular level as well as a shortage of appropriate preclinical models.
2 |. “PROS AND CONS”: ANIMAL VS HUMAN WOUND-HEALING MODELS
Various animal wound-healing models have been successfully utilized to better understand the basic mechanisms of the acute wound-healing process, and to some extent, its pathophysiology.[4,14–19] The most commonly used animal models for wound healing, such as murine, rat, porcine and rabbit, are widely available and share important anatomical and physiological characteristics with the human wound-healing process. It is generally accepted that porcine wound-healing model provides the major advantages over other animal models due to high structural similarity to human skin and healing primarily by epithelialization resembling human wound closure, in contrast to rodent wounds that heal primarily by contraction.[15,16,20] Furthermore, particular models have been subject to modifications, such as the presence of diabetes, ischaemia, ischaemia-reperfusion, wound biofilm or mechanical pressure, in efforts to mimic a wound environment commonly encountered in the clinical setting.[15,20–25] As such, animal models of wound healing remain an invaluable resource for testing safety, efficacy and toxicology of novel therapies in the preclinical phase. However, most therapeutic agents that were shown to be successful in enhancing wound healing in animal models have been unsuccessful in significantly improving healing in patients, resulting in limited advanced therapy options. Despite large overlap between molecular and cellular mechanisms, significant anatomic and physiologic divergence has been found between human and the widely used rodent models, primarily in terms of immune response and mechanisms of re-epithelialization.[14,15,26] The most recent advances in developing humanized mouse models are holding a new promise in reproducing clinical hallmarks.[27–30] Mice engrafted with human CD34+ hematopoietic stem cells can be successfully utilized for studying the process of human inflammation in response to tissue injury,[27] while the human skin graft transplanted onto the back of NOD/SCID mice is useful for studying the human wound healing in both wild type and diabetic background.[28,29] However, chronic ulcers are often complicated by complex microbial communities, bacterial biofilms, multiple comorbidities and polypharmacy, all contributing to local and systemic effects on the wound and its environment. One can see how such factors contribute to the complexity of human wounds, posing a challenge when faced with questions such as: “at what point does the healing halt?” “how do these factors interact to produce an environment that interrupts normal healing?” or “what causes some wounds to respond to standard of care and advanced treatment while others do not?” In addition, the discovery of specific genes unique to the human genome that play a role in the wound-healing process underscores the need to use human model(s) and human samples.[31] That said, the value of a mechanistic approach using animal models to test specific mechanisms in a highly controlled setting is not to be undermined.[14] However, animal models cannot take into account the multitude of factors frequently encountered in a clinical setting, resulting in an oversimplification of actual chronic wound conditions in humans. The more we try to control for outside factors to achieve the “ideal” experimental setting, the farther we depart from the “applicable preclinical model” that could yield more successful translation of therapeutics into the clinic. As an example, several in vivo studies utilizing animal models have been successful in facilitating wound healing by exogenously applying recombinant transforming growth factor-β (TGF-β).[32–34] Although early clinical trials for the application of exogenous TGF-β2 to venous leg ulcers appeared promising,[35] TGF-β therapy was ultimately unsuccessful in clinical trials and failed to get approval from the FDA despite its success to accelerate wound healing in pre-clinical animal models. The dearth of translation from animal studies to patients was elucidated a decade later through studies of human tissue biopsies dissecting the components of TGF-β signalling. Alas, there is a functional loss of TGF-β receptors and downstream Smad signalling cascade in the epidermis of chronic wounds, providing an explanation for the limited ability of exogenous TGF-β to accelerate wound healing in patients.[36,37] Unfortunately, this suppression of TGF-β signalling cascade in patients who have chronic wounds was unknown at the time of clinical trial design. This data underscore the importance and value of research utilizing human samples to understand the chronic wound pathophysiology and mechanisms of action prior to proceeding to an advanced phase of a clinical trial.
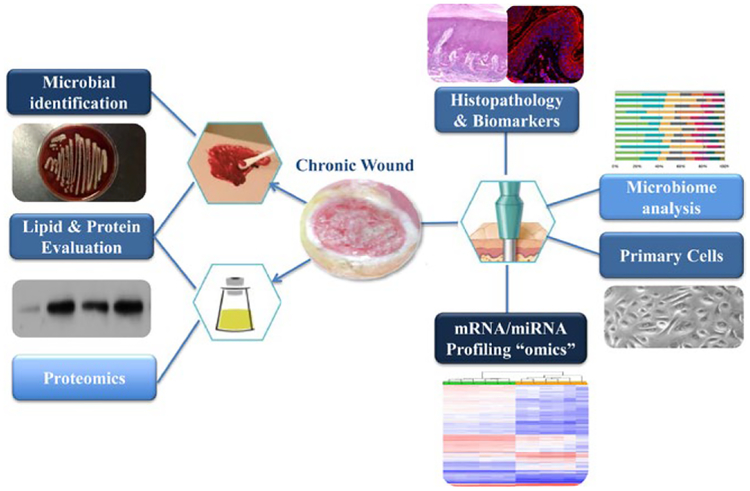
Human samples, coupled with available modern technologies[38–48] and system data integration including medical electronic records,[49,50] can lead to a better appreciation for the complexity of underlying factors affecting wound healing. These include variables such as associated infection, treatment compliance, other medical conditions, comorbidities and medications, which all effect the patho-physiology of wound healing. An additional advantage is the accessibility of chronic wound samples and feasibility of collection using minimally invasive methods. Wound fluid, wound swabs and tissue specimens from chronic ulcers can be obtained during outpatient visits as an integral part of standard of care, while providing biomaterial for a wide spectrum of analyses including primary cell isolation and histopathological evaluation in addition to analyses of gene expression, proteomics, lipidomics, metabolomics and wound microbiome (Figure 1). Wound tissue samples prove to be especially useful because they provide unique opportunities to perform not only histological analyses, but also yields useful biomaterial for primary cell isolation, RNA, protein, lipid and even microbiome analyses. Wound fluid and swabs are relatively easy to obtain, and, although they do not provide histology assessments, they provide invaluable information related to enzyme activity, proteomics, metabolomics and microbiome environment of the chronic wound (Figure 1). Research studies using human tissue samples have already proved successful in dissecting the mechanisms of action for approved therapies even in small patient populations (Table 1).[44] Such studies may not only help unravel an order of importance and interplay between differing elements affecting a patient’s wound, but also discover precise biomarkers to classify them based on their “healing” or “non-healing” characteristics to personalize treatment modalities. Most importantly, we can utilize this knowledge to further optimize preclinical models that will better account for clinical hallmarks, confirm the findings from human tissue samples and test for potential effects of novel therapeutic modalities in a complex but more “improved” setting. Recent advances in organotypic skin models utilizing patient’s cells[51,52] and micro-engineered models of functional human skin, known as organs-on-chip,[53] could provide the basis for preclinical assays with greater translational power. A chronic wound organotypic model assembled with primary DFU-derived fibroblasts and healthy keratinocytes was established, and it maintained the main pathological phenotype of DFUs.[51] More advanced organ-on-a-chip technologies mimicking multilevel organ functions are also being developed and may circumvent the need for animal wound models altogether.[54] Cutaneous organ-on-a-chip models focus on accommodating a bioengineered skin equivalent that is continuously perfused through a microvascular channel. While there are several barriers to overcome before a full, multilevel, organ-on-a-chip skin equivalent can be fully utilized for wound-healing studies, the power of this technology has already been made evident.[53] What remains is to recreate skin’s architecture; complete with sensory organs, appendages, and a full vascular network utilizing patient-derived cells; which would open the doors to more powerful mechanistic studies and treatment screening methods for wound-healing disorders.
FIGURE 1.
Samples from chronic wounds provide a unique resource of biomaterial for mechanistic studies in wound-healing research. Chronic wound tissue can be utilized for histopathology, microbiology assessment, generation of primary cells as well as RNA, protein and lipid isolation for downstream “omics” analyses. Chronic wound fluid and swabs can also be utilized for lipid and protein evaluation as well as microbial DNA isolation followed by microbiome analyses
TABLE 1.
Representative findings resulting from studies using chronic wound tissue as biomaterial
| Discovery | Type of analysis | Ref |
|---|---|---|
| VLU | ||
| Deregulation of epidermal stem cell niche and deprivation of ESCs contribute to pathogenesis of non-healing venous ulcers (n=10) | Histology/IHC, gene arrays, | [38] |
| Mechanism of action of bioengineered bilayered living cellular constructs in VLUs by remodelling inflammatory response. Identified nuclear β-cateni as biomarker of non-healing phenotype (n=30) | Gene arrays, IHC | [44] |
| miR-16, -20a, -21, -106a -130a and -203 were overexpressed in chronic wounds (n=10) | Histology/IHC, omics | [89] |
| Altered expression of genes in VLU code for mediators of inflammation and apoptotic pathways (n=10) | Histology, gene arrays | [130] |
| Epidermis of chronic VLU is hyperproliferative and non-migratory due to nuclear presence of beta-catenin and c-myc (n=10) | IHC, gene expression | [99] |
| Antimicrobial peptide LL37 is diminished in chronic wounds and impairs re-epithelialization (n=9) | IHC, gene expression, microbiology | [127] |
| The suppression of TGF-β receptors in non-healing chronic venous ulcers contributes to wound chronicity (n=12) | Histology, gene arrays | [131] |
| TIMP-1 is overexpressed near the basement membrane in acute wounds, but not in chronic wounds (n=8) | Gene expression, IHC | [132] |
| DFU | ||
| miR15b is induced by S. aureus infection in DFU and contributes to suppressed DNA repair and diminished inflammation (n=12) | Gene expression arrays, IHC | [43] |
| miRNA-132 is suppressed in DFU, and its local replenishment may have therapeutic potential (n=29) | IHC, gene expression | [92] |
| The number of circulating progenitors is correlated with healing outcome of chronic DFUs (n=100) | Flow cytometry, genomics | [86] |
| Higher levels of pro-inflammatory M2 type macrophages favour wound healing in DFU (n=10) | Gene expression | [133] |
| Healing DFUs have higher number of Langerhans cells compared with non-healing DFU (n=12) | Histology, IHC | [126] |
| Quality assessment for use of DFU tissue specimens in research found that only two-thirds of collected specimens was usable for translational studies (n=25) | Histology, IHC RNA isolation | [117] |
| DFUs have increased levels of miR-198, an important regulatory switch controlling wound closure (n=8) | IHC, gene expression | [134] |
| Stem cell mobilization is increased in DFU patients undergoing hyperbaric oxygen therapy (n=12) | IHC, flow cytometry, | [135] |
| Persistently elevated MMP concentrations and imbalance of their inhibitors provide evidence for a hostile molecular environment in DFU (n=20)[82] and VLU (n=5)[70] | Proteomics, enzyme activity | [70,82] |
| Other types of chronic wound | ||
| Extensive neutrophil infiltration in chronic pressure ulcer granulation tissue contributes to chronicity (n=11) | Histology, enzyme activity | [136] |
| Laser capture microdissection was established for endothelial cell followed by microarray analysis and validation (n=3) | Histology, gene expression | [39] |
| High prevalence of Staphylococcus, Pseudomonas and anaerobic bacteria found in chronic VLU and PU (n=2,963) | 16S rDNA sequencing | [57] |
IHC, immunohistochemistry; n, number of human subjects.
Major type of analyses utilized in the study is outlined. We apologize to authors who have relevant literature not included in this table due to space limitations.
Although there is a fundamental and important body of knowledge regarding cutaneous wound healing that continues to derive from animal studies, human studies also represent a valuable resource that should not be underestimated. These types of studies offer the unique opportunity to decipher mechanisms of healing impairment that can further facilitate future development of novel therapeutic modalities.
3 |. WHAT DID WE LEARN SO FAR FROM STUDIES UTILIZING HUMAN WOUND SAMPLES?
The use of human samples in conjunction with analyses involving standard cell, molecular and more advanced “omics” technologies led to the better understanding of molecular and cellular processes deregulated in chronic wounds (Tables 1 and 2). Although the etiology of VLU, DFU and PU is multifactorial, these wounds share some common recognizable defects.[4] These common features of non-healing wound pathophysiology include hyperproliferative non-migratory epidermis, unresolved inflammation, impaired fibroblasts function, extracellular matrix deposition, increased levels of pro-teases, decreased angiogenesis and complex microbial communities associated with biofilms.[38,43,46,52,55–61]
TABLE 2.
Representative findings resulting from studies focusing on chronic wound pathophysiology using biomaterial other than tissue specimens (blood, wound fluid, swabs)
| Discovery | Type of analysis | Ref. |
|---|---|---|
| Genetic variation in NOS1AP gene may diminish circulating stem cell response and is associated with impaired healing of DFU (n = 47) | Genomics | [87] |
| Feasibility of modern spectroscopy utilized to associate metabolic products and bacterial infection in chronic wound exudate (n = 20) | Metabolomics microbiology, | [105] |
| Bacteria isolated from chronic VLUs can secrete proteases that degrade extracellular matrix important for wound healing (n = 6) | Microbiology, enzyme activity | [137] |
| Microbial instability and diversity were associated with healing DFU (n = 100) | Microbiome analyses | [56] |
| Specific fungal species and polymicrobial bacteria-fungus biofilms correlate with non-healing outcomes in DFUs (n = 100) | Mycobiome sequencing | [97] |
| Unique thrombin-derived peptides identified in non-healing wounds infected with S. aureus and Pseudomonas aeruginosa | Proteomics | [93] |
| The biological microenvironment of chronic wounds exhibits diminished angiogenesis, increased inflammation and higher cell death compared with acute healing process (n = 10) | Proteomics | [102] |
| Elevated levels of matrix metalloproteinase (MMP) activity are associated with healing impairment in chronic ulcers (n-28)[78] (n = 56)[85] (n = 25)[104] (n = 290)[108] | Proteomics, enzyme activity | [78,85,104,108] |
| Chronic wound fluid has high levels of matrix metalloproteinases and is capable of fibronectin degradation (n = 10) | Proteomics | [138] |
| Environment of non-healing ulcers has high levels of activated MMPs which contribute to poor wound healing. (n = 6)[74] (n = 56)[80] | Enzyme activity, proteomics | [74,80] |
n, number of human subjects.
Major type of analyses utilized in the study is outlined.
In vitro studies have shown that primary cells isolated from chronic wounds maintain tissue characteristics and show altered proliferative and migratory capacity accompanied with an inability to respond to wound-healing stimuli such as growth factors and cytokines.[51,52,55,62,63] While the actions of multiple cell types are tightly regulated by growth factors, cytokines and signalling molecules during the acute wound-healing process,[64–66] this regulation, and interplay between different signalling factors, is dysregulated in chronic wounds.[4,36,67,68] Furthermore, chronic wounds exhibit high levels of metalloproteinases (MMP) and a perturbed ratio between MMP and tissue inhibitors of metalloproteinases (TIMPs), which contribute to a hostile wound environment, lack of growth factor and cytokine signalling and subsequently delayed healing (Table 2).[69–85] In-depth characterization of VLU wound edge tissue has revealed a loss of the epidermal stem cells niche, deregulation of epidermal differentiation and activation of β-catenin pathway coupled with attenuation of TGF-β and epidermal growth factor (EGF) signalling pathways.[36,38,40,41] Diminished concentration of circulating stem progenitor cells was also found to be associated with impaired foot ulcer healing in patients with diabetes.[86,87]
3.1 |. “Omics” approaches and associated findings
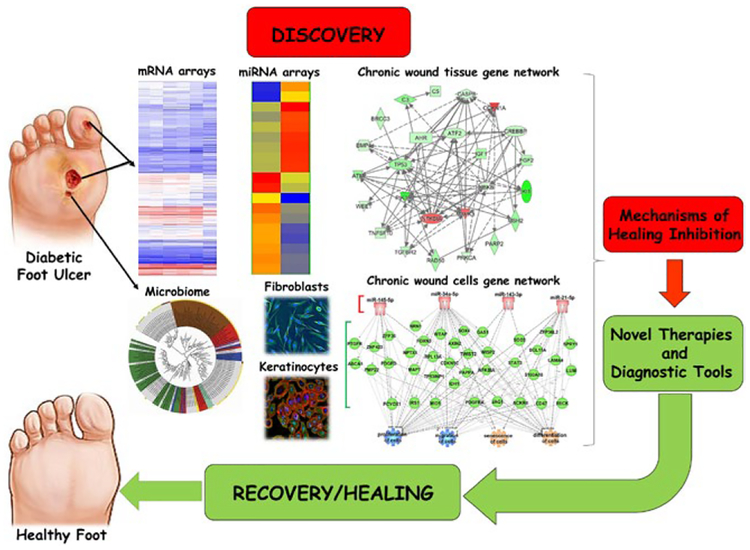
As a complex, multistep process involving multiple cell types and interacting regulatory pathways, wound healing is well suited for an “omics” approach such as gene expression profiling, RNA sequencing, proteomics, metabolomics or microbiome/metagenomics technologies (Figure 2).[38,40–47,88] These technologies provide the ability to simultaneously analyse multiple targets and thereby identify pathways responsible for impaired wound healing. They allow analysis beyond individual components enabling studies investigating global physiology and pathology of wound healing along with interactions of the cellular components participating in various repair processes (Figure 2). Although “omics” approaches were traditionally classified as “descriptive” science and “fishing expeditions”, the “hypothesis generating” (as opposed to more traditional “hypothesis driven”) experimental designs is gaining more acceptance. Here, we outline studies that have successfully utilized a combined approach of using “omics” and patient samples to pinpoint specific processes and molecules associated with healing impairment. Gene expression profiling has been employed to identify specific sets of genes and corresponding microRNAs (miRs) that are deregulated in VLU and DFU tissue samples.[40,41,43,44,52,89] Furthermore, comparative genomic analyses were employed to elucidate transcriptional features that distinguish chronic VLU from acute healing wounds (AW).[44,90] Comprehensive genomic data analyses revealed that the prolonged inflammation present in non-healing VLU is at a suboptimal level and insufficient to facilitate progression of healing in comparison with the normal acute wound inflammatory response.[44] Biological processes of immune cell differentiation, activation, migration and signalling were strikingly absent in chronic non-healing VLU.[44] Calcium mobilization, required for epithelial restitution in in vivo models of wound repair, was another process found enriched in acute wounds but not in chronic VLU.[44] Calcium mobilization also functions in immunity, signalling downstream of wound-healing cytokines to stimulate migration and repair.[44,91] Importantly, testing the response of VLU to FDA-approved bilayered skin construct (BLCC) identified that BLCC shifts the genomic profile from a non-healing VLU towards an acute wound-like profile[44] (Table 1). These data support findings that non-healing wounds are unable to enter and complete the acute wound response to accomplish healing, and that the suboptimal inflammatory response, coupled with impaired cell migration and proliferation, is all contributing features to non-healing.[44] A genomic approach was also utilized to identify overexpressed miRs with a critical role in the pathogenesis of non-healing VLU. Aberrant regulation of miR -21, - 16, - 20a, - 106a - 130a and - 20 specific for VLU contributed to inhibition of healing via multiple mechanisms by targeting growth factor signalling including leptin receptor, TGFβ and EGF (Table 1).[40,89,91] The most recent study focused on identifying genes, associated pathways and miRs in DFU as a result of Staphylococcus aureus colonization. S. aureus induced miR-15b-5p, subsequently repressing DNA repair and diminishing inflammatory response, revealing a novel mechanism of healing inhibition in chronic DFU colonized with this pathogen.[43] DFU are also characterized by a significant reduction of miR-132, which contributes to the deregulation of inflammation-related pathways such as NF-κB, NOD-like receptor, Toll-like receptor and TNF-α signalling.[92] Multiple proteomic studies have utilized chronic wound fluid as an accessible source of biomaterial, revealing not only deregulation of secreted host proteases, but also specific peptides from human thrombin derived as result of wound infection.[46,93] In addition to analysing host response to infection, advances in next-generation sequencing methods have allowed for the identification and quantification of microbial communities in chronic and acute wound setting (Table 2).[48] DNA sequencing methods utilizing the 16S ribosomal RNA gene to evaluate bacterial phylogeny and taxonomy have been successfully utilized to identify the complexity of chronic wound microbial “footprints”,[57,94–96] with a few recent studies addressing the correlation of microbiome with clinical outcomes in wound healing.[56,58,97] Taken together, utilization of human specimens for basic research has shown major advances in understanding mechanisms that control healing and its impairment.
FIGURE 2.
“Omics” approaches utilizing chronic wound samples can lead to discovery of new diagnostic and therapeutic targets. Studies focusing on human chronic wound samples and patient-derived primary cells with utilization of “omics” tools provide excellent resource for discovery and development of diagnostic tools, more personalized approach to treatments and identification of new therapeutic targets
4 |. UTILIZATION OF HUMAN SAMPLES: DIAGNOSTIC TOOLS AND BIOMARKER DISCOVERY
In addition to using human samples to understand pathophysiology, such an approach may be very useful in personalization of clinical care. Deciphering healing and non-healing mechanisms using human samples should provide not only potential therapeutic targets but also valuable potential diagnostic tools. But intriguingly, the development of diagnostic tools is lagging. Biomarkers are molecules that can be objectively quantified and correlated with predictive, diagnostic and indicative outcomes, providing a means for personalized assessment and clinical guidance.[46] The deregulation of molecular pathways involved in wound healing can be exploited in the clinical setting for diagnostic, prognostic and therapeutic purposes. The powerful use of molecular markers to stratify patients’ outcomes, risks or responses to therapy has been widely used to guide clinical management of many other diseases including cancer, cardiovascular disease and diabetes.[98] Besides the extensive research efforts, the application of chronic wound biomarkers has still not found its path into daily clinical practice in a major way.
4.1 |. Wound tissue and wound fluid biomarkers
Identification of potential biomarkers goes hand-in-hand with a better understanding of the mechanisms that inhibit healing. Once molecular markers associated with deregulated mechanistic pathways important for the wound-healing process are identified and correlated with the specific outcomes, such as healing, response to standard of care, response to biologics and risk for deep tissue infections associated with chronic wounds, they may yield very useful diagnostic tools. Because chronic wound patients are often plagued with many comorbidities and polypharmacy, biomarkers may be a powerful tool in the clinical assessment of wound pathophysiology as well as treatment design.[46] Ironically, the same clinical complexity is making the identification of biomarkers very challenging. Ideally, biomarkers are present in specimens that can be obtained using simple, minimally invasive procedures in the clinic. Human tissue specimens collected in the form of either debrided ulcer tissue, biopsies taken from patient wounds, wound fluid or wound swabs have been invaluable to the assessment of potential biomarkers for wound healing (Figure 1). As detailed in previous section, patient specimens provide a means for in-depth study of mechanistic pathways involved with wound healing and current therapies, providing a better understanding for the molecular derangements associated with wound-healing impairment.
One of the first chronic wound tissue biomarkers was identified by studying specimens from VLU, elucidating a correlation with increased nuclear expression of β-catenin and its downstream target, oncogene c-myc,[99] with a hyperproliferative, non-migratory chronic wound epidermis.[38,40,99] These biomolecules are also downstream of the Wnt signalling pathway, which plays an important role in skin development and epidermal stem cell maintenance.[100,101] Nuclear presence of β-catenin and c-myc in tissue specimens is quantifiable by use of immunohistochemistry, highlighting their clinical feasibility as a tissue biomarker.[14,46,99]
Studies of aspirated wound fluid have also shed light on new biomarker discoveries involved in chronic wound healing, including detectable increases in levels of tissue metalloproteinases (MMPs).[58,75,102–105] Technologies using quantitative assessments of MMPs are currently being tested for use as predictive biomarkers for non-healing ulcers in clinical trials.[71] Furthermore, MMP-9/MMP-1 ratios in wound fluid have been utilized as a predictive marker for PU outcomes of healing.[80] Effect of topical negative pressure therapy was also assessed by MMP9/TIMP-1 ratio in wounds, which was shown to be significantly lower after treatment, indicating that this MMP9/TIMP-1 ratio had potential biomarker application in assessing the effect of a therapeutic device.[106] All these findings have enabled further development of a technology that allows for point-of-care measurement of elevated protease activity using the Levine swab technique, which is currently being evaluated in clinical trials.[107,108]
Several other studies have utilized chronic wound biomarkers by correlating them with the response to therapy. Molecular analyses of VLU after treatment with bioengineered living cell constructs (BLCC) found that overexpressed nuclear β-catenin, a hallmark of non-healing VLU, was one of the several molecular targets of BLCC treatment therapy.[44,99] A clinical trial had found that despite improving healing rate by almost 29% compared with standard of care therapies, over 50% of patients with chronic VLU still failed to respond to BLCC therapy.[109] This may be in part due to the fact that not all patients share the same molecular makeup, or immune response and immune tolerance, that can be corrected by use of BLCC, making mechanistic studies of patient tissues key players in deciphering the molecular targets of BLCC treatment.[44] Such studies found that patients responding to BLCC appeared to exhibit less nuclear β-catenin staining and also exhibited decreased expression of casein kinase 2 alpha 2 (CSNK2A2), one of the Wnt/Beta-catenin family members. Thus, nuclear β-catenin and CSNK2A2 have the potential to be used as clinical biomarkers for distinguishing patients who may respond to BLCC treatment.[44] In an other study, MMP levels were utilized to indicate response to dressings impregnated with oak bark extract. Human tissue specimens collected from these wounds were found to have levels of MMP-2 that paralleled with improved healing ability after the application of this specialized dressing. The authors concluded that MMP-2 assessment might have powerful indications for use in the clinical setting to not only predict healing potential of a chronic wound, but also provide indicative measures of therapeutic efficacy for treatment monitoring.[110]
4.2 |. Systemic biomarkers for chronic wounds
Systemic biomarkers from patients’ blood samples have also been correlated with clinical outcomes. Circulating progenitor cells are involved with normal wound healing and are mobilized in response to skin wounding and other trauma.[111] Using flow cytometry of patient blood samples, one study found that the depletion of circulating CD34+/CD45-dim stem/progenitor cells was correlated with a non- healing phenotype in DFU.[86,87] These findings suggest that assaying circulating progenitor cells during the early stages of wound care may be useful for predicting healing response in chronic DFU. Cellular content of hypoxia-inducible factors (HIF 1, 2 and 3) in progenitor cells is also known to be associated with regulating their vasculogenic functions,[111,112] although ratios of pro-angiogenic and anti-angiogenic HIFs was not found to be associated with healing outcomes during the initial 8 weeks of care. The most recent study by Margolis et al associated genetic variation in nitric oxide synthase 1 adaptor protein (NOS1AP) with reduced levels of circulating progenitors and impaired healing in patients with diabetes (Table 2).[87] Additional molecular studies of human DFU tissue have also discovered the role of regulatory miR-200b and miR-191 in modulating angiogenesis, apoptosis, cell proliferation and cellular migration in wound healing. The circulating levels of these and other miRs were found to be altered in the blood of patients with type II diabetes complicated with chronic wounds compared to those with normal healing ability.[113] Investigation of circulating cytokines in these patients also suggested that the underlying inflammation correlated with abnormal wound-healing pathology was a key player in inducing these aberrantly expressed plasma miR levels.[113]
4.3 |. Utilization of the microbiome for diagnostic purposes
The characterization of wound microbiome has revealed unique signatures associated with clinical outcomes of healing.[58,105] Loesche et al evaluated microbiota colonizing DFU using 16S ribosomal RNA gene amplification to create microbial “footprints” and assess microbial diversity in correlation with DFU healing.[56] The DFU microbiomes were clustered into 4 distinct community types based on the predominant taxa colonizing the wound.[56] As healing progressed, microbial composition was found to be dynamic, and ulcers with greater transition frequencies between microbial communities were found to have more rapid healing rates. It was speculated that the microbial instability seen in wounds that healed reflected the patient’s innate ability to ward off infection, preventing any particular “non-healing” microbial community from stabilizing within the wound.[56] In addition to bacterial component, analyses of fungal communities have shed new light on chronic DFU. Kalan et al used DNA sequencing to analyse the extremely diverse and heterogeneous fungal component of DFU microbial milieu.[97] The investigators found that wounds with high Ascomycota abundance at presentation were associated with slower rates of healing, suggesting that mycobiome analysis at initial visits may have ability to predict patients who will not respond to standard of care.[97] Furthermore, fungal-bacterial communities implicated the formation of interkingdom biofilms as an additional cause of ulcer chronicity. Together findings focusing on chronic wound microbiome/mycobiome underlined the complexity of chronic wound microbial communities and the importance of interspecies interactions in correlation with clinical outcomes. This approach comes with certain challenges, as the techniques used to analyse microbiome composition do not distinguish between live and dead microorganisms present, while the significant costs of next-generation sequencing may impede clinical application.
In spite of the challenges, the discovery of chronic wound bio-markers highlights the value of knowledge that can be gained through translational studies focusing on the molecular mechanisms of healing impairment using patient-derived biomaterial. Before any of the biomarkers can be implemented into standard clinical practice, their confirmation and a standardization of their analysis in longitudinal, multicentred prospective studies are warranted. If verified through such clinical trials in the future, many of these above-mentioned biomarkers could potentially serve as a powerful prognostic and diagnostic tool to improve the clinical management of non-healing wounds. Of critical importance are clinically relevant biomarkers that would allow early identification of patients predisposed to fail standard of care therapy before starting treatment, ultimately targeting advanced therapies to appropriate patients/wounds, resulting in improved, more efficacious and cost-effective outcomes. Although all these examples support the notion that using human samples would yield valuable scientific advances that are clinically relevant, such approach inherently carries multiple challenges reviewed below.
5 |. CHALLENGES OF TR ANSL ATIONAL RESEARCH INVOLVING HUMAN SUBJECTS
Research studies utilizing samples from chronic wounds as a preclinical study model are accompanied by multiple challenges. In addition to multifactorial etiology, affected patients more often than not present to the clinic at different times after wound onset, with multiple comorbidities and varying histories of treatment compliance.[114] It is also often challenging to recruit and obtain consent for tissue biopsies from patients with long-standing non-healing wounds even though it has been shown that performing biopsy of the chronic wound does not interfere with healing.[115] This issue results in a relatively small number of samples utilized for the majority of studies utilizing human wound tissue (Table 1). Providing information on the safe procedures for sample collection, in addition to a more thorough description of methods employed, may lead to a more successful recruitment of patients, appropriate control groups and ultimately improved overall collection of tissue specimens. Finally, the tissue itself has limited applicability for traditional mechanistic approaches, such as a “knock-down,” “knock-in” or other various genetic modifications.
5.1 |. Sample size
Another important issue is the overall sample size discrepancy. Traditional clinical trials that are testing efficacy typically involve hundreds of patients. Most clinical research sample size collections are markedly smaller, which often challenges their value. Therefore, one has to make a fundamental distinction between clinical trial and clinical research that is not just semantic. As described above, clinical research that focuses on deciphering molecular and cellular mechanisms that inhibit healing use comprehensive multipronged strategies involving modern technologies, and the type of research is often limited by rather moderate research funds and an allotted amount of time to complete it. Therefore, it is unrealistic to expect that the sample size of clinical research studies should be compared to that of clinical trials that are testing efficacy of therapies and interventions.
5.2 |. Obtaining samples and quality control
Debridement is one of the standard of care procedures, which simultaneously enables the collection of specimens that may be used for research to enhance the understanding of wound-healing pathophysiology.[116] However, the variable techniques of debridement procedure and lack of standardized protocol to evaluate the quality of samples collected result in a wide range of collected specimens, which represent another challenge. A single study focusing on debridement tissue collection and subsequent analyses found that quality of specimens depends on depth of surgical debridement of DFU. The resulting specimens collected from debridement can be classified accordingly into three groups: callus only, partial specimens (containing callus with some epidermis) and complete specimens (containing callus, the full thickness epidermis and a portion of the dermis).[117] The presence of a full thickness epidermis and dermis proved to be an essential determinant of a high-quality tissue sample, as only complete specimens were viable for further biomarker assessment, RNA isolation and gene expression analysis.[43,117] However, up to one-third of the total collected biopsies were callus only or partial specimens, and as such did not yield quality biomaterial useful for further analyses. This poses further obstacles to the already limited supply of biomaterial available for mechanistic studies of chronic wound pathophysiology. Immunohistochemistry analysis of collected DFU specimens could not be properly assessed nor quantified, thus resulting in no valuable data. Moreover, RNA isolation and quality analysis demonstrated that up to 91% of incomplete samples had low RNA quality with an RNA Integrity Number (RIN) score of less than 4, making them impractical for further RNA analysis.[43,44,117] Considering that deeper surgical debridement is a challenge in itself, it is not surprising that a large proportion of collected specimens barely extend beyond callus. The wound biopsy with the full thickness epidermis and dermis can assure collection of a high-quality tissue sample required for the sophisticated analyses.[115] It is often difficult to obtain consent for biopsies from patients with healing dysfunction, and clinicians themselves frequently hesitate to debride to a deeper extent, especially when dealing with painful chronic wounds such as VLUs. However, it has been shown that chronic wound biopsies do not inhibit healing, and deeper debridement would not only reap higher quality tissue biopsies for research purposes, but would also ultimately benefit patients, improve diagnostics and also advance the design of large-scale clinical trials.[115] Thus, it is of utmost significance to be aware of these and similar findings and the need to define a standardized protocol, with the ultimate goal to collect and utilize more adequate chronic wound samples for the development of more successful diagnostic and therapeutic modalities.
5.3 |. Controls
As with any research, controls are, arguably, the most important component of experimental design. In addition to challenges associated with tissue standardization and collection, the chronic wound human research field is facing lack of consensus on the appropriate source for controls. Ideally, the patient should be his/her own control, but this may present further difficulties in obtaining an adequate location matched control from non-ulcerated skin, as it may pose a significant risk to the patient. To overcome this challenge, tissue samples obtained from podiatric surgeries have been utilized as controls to identify pathways contributing to delayed healing in DFU.[43,118,119]
These control tissue specimens were collected to match ulcer location and were obtained from age/sex/ethnicity-matched diabetic patients. Importantly, diabetes itself had very subtle changes to the non-ulcerated plantar foot skin, as cutaneous morphology, mRNA and miRNA expression were not found to be majorly affected compared with healthy non-diabetic human skin.[119] However, when advanced to cause secondary complications, such as vascular insufficiency or neuropathy, the changes in molecular pathology are evident.[118] Additional studies have indicated possible differences between the intact diabetic and non-diabetic human skin. It has been shown that the foot skin of individuals suffering from diabetes has a greater density of inflammatory cells and blood vessels in comparison with normal regardless whether it progresses to ulcer development.[120] Further, the number of degranulated mast cells is shown to be greater, while substance P expression appears to be lower in non-ulcerated diabetic forearm and dorsal foot skin. These changes influenced by diabetes may contribute to failure of proper initiation and progression of acute inflammatory response seen in DFUs.[59,121] Comparison of chronic wounds to acute, healing wounds also provides important insight. However, as wound healing is a dynamic process, selection of a specific time-point(s) is very important. Existing human gene expression profiles from acute wounds generated from split-thickness skin graft donor sites of the anterior thigh from healthy patients represent a unique resource for comparative wound-healing studies.[90] An alternative control for translational studies focusing on the wound re-epithelialization process is the human ex vivo wound-healing model.[66,122–126] This human experimental wound model has been extensively utilized to study the re-epithelialization process. Comparative analyses between ex vivo wound model and acute human wounds confirmed comparable expression patterns for multiple genes involved in wound healing,[127] while multiple translational studies focusing on pathophysiology of chronic wound epidermis successfully utilized human ex vivo wounds as a control.[36,43,126,127]
5.4 |. Other variables
In addition to the challenges associated with sample collection and standardization, longitudinal studies encounter difficulties with patient retention and compliance. Because non-healing ulcers represent a major burden in the elderly population[13] and are predominantly treated in an outpatient setting, it becomes difficult to collect biomaterial at different time points as many patients are lost to follow-up.[3,13] Furthermore, it is difficult to control for variability in patient compliance to standard of care,[128] which can significantly influence findings focusing on correlation with wound size reduction or evaluation of novel treatments.[129]
In summary, using human samples to study wound healing, in spite of the limitations and challenges, provides unique resource and valuable information spanning from the fundamental molecular and cellular mechanisms that control wound healing to diagnostic tools and therapy targets. Studies focusing on human samples and utilization of omics tools will streamline diagnostics and facilitate personalized treatment plans. (Figure 2). As such, it has earned its place as a validated scientific approach.
ACKNOWLEDGEMENTS
We are very grateful to our patients, past and current members of our collaborative clinical and research teams as well as our colleagues in the field for their continuous inspiration. We apologize to our colleagues whose contributions were not included due to space limitations. MT-C acknowledges funding from the National Institutes of Health for research support (NR015649; NR013881; 2U24DK076169).
Funding information
National Institute of Diabetes and Digestive and Kidney Diseases, Grant/Award Number: 2U24DK076169; National Institute of Nursing Research, Grant/Award Number: NR013881 and NR015649
Footnotes
CONFLICT OF INTEREST
All authors have declared no conflicting interests.
REFERENCES
- [1].Richmond NA, Lamel SA, Davidson JM, Martins-Green M, Sen CK, Tomic-Canic M, Vivas AC, Braun LR, Kirsner RS, Wound Repair Regen. 2013, 21, 789. [DOI] [PubMed] [Google Scholar]
- [2].Rice JB, Desai U, Cummings AK, Birnbaum HG, Skornicki M, Parsons NB, Diabetes Care 2014, 37, 651. [DOI] [PubMed] [Google Scholar]
- [3].Kirsner RS, Marston WA, Snyder RJ, Lee TD, Cargill DI, Zhang Y, Dickerson JE Jr, Slade HB, Wound Repair Regen. 2013, 21, 682. [DOI] [PubMed] [Google Scholar]
- [4].Eming SA, Martin P, Tomic-Canic M, Sci. Transl. Med 2014, 6, 265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kirsner RS, Marston WA, Snyder RJ, Lee TD, Cargill DI, Slade HB, Lancet 2012, 380, 977. [DOI] [PubMed] [Google Scholar]
- [6].Criado PR, Alavi A, Kirsner RS, Int. J. Low. Extrem. Wounds 2014, 13, 130. [DOI] [PubMed] [Google Scholar]
- [7].Yim E, Vivas A, Maderal A, Kirsner RS, JAMA Dermatol. 2014, 150, 385. [DOI] [PubMed] [Google Scholar]
- [8].Escandon J, Vivas AC, Perez R, Kirsner R, Davis S, Int. Wound J 2012, 9, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gregg EW, Gu Q, Williams D, de Rekeneire N, Cheng YJ, Geiss L, Engelgau M, Diabetes Res. Clin. Pract 2007, 77, 485. [DOI] [PubMed] [Google Scholar]
- [10].Lamont P, Franklyn K, Rayman G, Boulton AJM, Int. J. Low. Extrem. Wounds 2013, 12, 71. [DOI] [PubMed] [Google Scholar]
- [11].Lebrun E, Tomic-Canic M, Kirsner RS, Wound Repair Regen. 2010, 18, 433. [DOI] [PubMed] [Google Scholar]
- [12].Margolis DJ, Berlin JA, Strom BL, Am. J. Med 2000, 109, 15. [DOI] [PubMed] [Google Scholar]
- [13].Gould L, Abadir P, Brem H, Carter M, Conner-Kerr T, Davidson J, DiPietro L, Falanga V, Fife C, Gardner S, Grice E, Harmon J, Hazzard WR, High KP, Houghton P, Jacobson N, Kirsner RS, Kovacs EJ, Margolis D, McFarland Horne F, Reed MJ, Sullivan DH, Thom S, Tomic-Canic M, Walston J, Whitney JA, Williams J, Zieman S, Schmader K, J. Am. Geriatr. Soc 2015, 63, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Elliot S, Wikramanayake TC, Jozic I, Tomic-Canic M, J. Invest. Dermatol 2018, 138, 736. [DOI] [PubMed] [Google Scholar]
- [15].Pastar I, Cao T, Sawaya A, Liang L, Glinos D, Drakulich S, Chen V, Stojadinovic O, C Davis S, Tomic-Canic M, Skin Tissue Models, (Eds: Marques A, Pirraco RP, Cerqueira MT, Reis RL), Amsterdam, Netherlands, Elsevier Inc, 2017. [Google Scholar]
- [16].Sullivan TP, Eaglstein WH, Davis SC, Mertz P, Wound Repair Regen. 2001, 9, 66. [DOI] [PubMed] [Google Scholar]
- [17].Park SA, Covert J, Teixeira L, Motta MJ, DeRemer SL, Abbott NL, Dubielzig R, Schurr M, Isseroff RR, McAnulty JF, Murphy CJ, Wound Repair Regen. 2015, 23, 251. [DOI] [PubMed] [Google Scholar]
- [18].Dunn L, Prosser HC, Tan JT, Vanags LZ, Ng MK, Bursill CA, J. Vis. Exp 2013, 75, e50265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen J, Chen Y, Chen Y, Yang Z, You B, Ruan YC, Peng Y, Cell. Physiol. Biochem 2016, 39, 2262. [DOI] [PubMed] [Google Scholar]
- [20].Gordillo GM, Bernatchez SF, Diegelmann RF, Di Pietro LA, Eriksson E, Hinz B, Kirsner R, Liu P, Parnell LK, Sandusky GE, Sen CK, Tomic-Canic M, Volk SW, Baird A, Adv. Wound Care. (New Rochelle). 2013, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim DJ, Mustoe T, Clark RA, Wound Repair Regen. 2015, 23, 318. [DOI] [PubMed] [Google Scholar]
- [22].Yeung CYC, Holmes DF, Thomason HA, Stephenson C, Derby B, Hardman MJ, Wound Repair Regen. 2016, 24, 1089. [DOI] [PubMed] [Google Scholar]
- [23].Davidson JM, Arch. Dermatol. Res 1998, 290(Suppl), S1. [DOI] [PubMed] [Google Scholar]
- [24].Roy S, Biswas S, Khanna S, Gordillo G, Bergdall V, Green J, Marsh CB, Gould LJ, Sen CK, Physiol. Genomics 2009, 37, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC, PLoS ONE 2013, 8, e56846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, Wakamatsu E, Benoist C, Koller D, Regev A, ImmGen C, Proc. Natl Acad. Sci. USA 2013, 110, 2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Baird A, Deng C, Eliceiri MH, Haghi F, Dang X, Coimbra R, Costantini TW, Torbett BE, Eliceiri BP, Wound Repair Regen. 2016, 24, 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Martinez-Santamaria L, Conti CJ, Llames S, Garcia E, Retamosa L, Holguin A, Illera N, Duarte B, Camblor L, Llaneza JM, Jorcano JL, Larcher F, Meana A, Escamez MJ, Del Rio M, Exp. Dermatol 2013, 22, 195. [DOI] [PubMed] [Google Scholar]
- [29].Escamez MJ, Garcia M, Larcher F, Meana A, Munoz E, Jorcano JL, Del Rio M, J. Invest. Dermatol 2004, 123, 1182. [DOI] [PubMed] [Google Scholar]
- [30].Del Rio M, Larcher F, Serrano F, Meana A, Munoz M, Garcia M, Munoz E, Martin C, Bernad A, Jorcano JL, Hum. Gene Ther 2002, 13, 959. [DOI] [PubMed] [Google Scholar]
- [31].Baird A, Costantini T, Coimbra R, Eliceiri BP, Wound Repair Regen. 2016, 24, 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Puolakkainen PA, Reed MJ, Gombotz WR, Twardzik DR, Abrass IB, Sage HE, Wound Repair Regen. 1995, 3, 330. [DOI] [PubMed] [Google Scholar]
- [33].Puolakkainen PA, Twardzik DR, Ranchalis JE, Pankey SC, Reed MJ, Gombotz WR, J. Surg. Res 1995, 58, 321. [DOI] [PubMed] [Google Scholar]
- [34].Mustoe TA, Pierce GF, Thomason A, Gramates P, Sporn MB, Deuel TF, Science 1987, 237, 1333. [DOI] [PubMed] [Google Scholar]
- [35].Robson MC, Phillip LG, Cooper DM, Lyle WG, Robson LE, Odom L, Hill DP, Hanham AF, Ksander GA, Wound Repair Regen. 1995, 3, 157. [DOI] [PubMed] [Google Scholar]
- [36].Pastar I, Stojadinovic O, Krzyzanowska A, Barrientos S, Stuelten C, Zimmerman K, Blumenberg M, Brem H, Tomic-Canic M, Mol. Med 2010, 16, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim BC, Kim HT, Park SH, Cha JS, Yufit T, Kim SJ, Falanga V, J. Cell. Physiol 2003, 195, 331. [DOI] [PubMed] [Google Scholar]
- [38].Stojadinovic O, Pastar I, Nusbaum AG, Vukelic S, Krzyzanowska A, Tomic-Canic M, Wound Repair Regen. 2014, 22, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Roy S, Patel D, Khanna S, Gordillo GM, Biswas S, Friedman A, Sen CK, Proc. Natl Acad. Sci. USA 2007, 104, 14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Brem H, Stojadinovic O, Diegelmann RF, Entero H, Lee B, Pastar I, Golinko M, Rosenberg H, Tomic-Canic M, Mol. Med 2007, 13, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Stojadinovic O, Pastar I, Vukelic S, Mahoney MG, Brennan D, Krzyzanowska A, Golinko M, Brem H, Tomic-Canic M, J. Cell Mol. Med 2008, 12, 2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Becker M, De Bastiani MA, Parisi MM, Guma FT, Markoski MM, Castro MA, Kaplan MH, Barbe-Tuana FM, Klamt F, Sci. Rep 2015, 5, 13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ramirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, Gil J, Davis S, Kirsner R, Tomic-Canic M, J. Invest. Dermatol 2018, 58, 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Stone R, Stojadinovic O, Rosa AM, Ramirez HA, Badiavas E, Blumenberg M, Tomic-Canic M, Sci. Transl. Med 2017, 9, pii: eaaf8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Program NCS, Liechty KW, Segre JA, Proc. Natl Acad. Sci. USA 2010, 107, 14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lindley LE, Stojadinovic O, Pastar I, Tomic-Canic M, Plast. Reconstr. Surg 2016, 138(3 Suppl), 18S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Grice EA, Segre JA, Annu. Rev. Genomics Hum. Genet 2012, 13, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hodkinson BP, Grice EA, Adv. Wound Care (New Rochelle). 2015, 4, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Golinko MS, Margolis DJ, Tal A, Hoffstad O, Boulton AJ, Brem H, Wound Repair Regen. 2009, 17, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kurd SK, Hoffstad OJ, Bilker WB, Margolis DJ, Wound Repair Regen. 2009, 17, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Maione AG, Smith A, Kashpur O, Yanez V, Knight E, Mooney DJ, Veves A, Tomic-Canic M, Garlick JA, Wound Repair Regen. 2016, 24, 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Liang L, Stone RC, Stojadinovic O, Ramirez H, Pastar I, Maione AG, Smith A, Yanez V, Veves A, Kirsner RS, Garlick JA, Tomic-Canic M, Wound Repair Regen. 2016, 24, 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Atac B, Wagner I, Horland R, Lauster R, Marx U, Tonevitsky AG, Azar RP, Lindner G, Lab Chip 2013, 13, 3555. [DOI] [PubMed] [Google Scholar]
- [54].Chan CY, Huang PH, Guo F, Ding X, Kapur V, Mai JD, Yuen PK, Huang TJ, Lab Chip 2013, 13, 4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Brem H, Golinko MS, Stojadinovic O, Kodra A, Diegelmann RF, Vukelic S, Entero H, Coppock DL, Tomic-Canic M, J. Transl. Med 2008, 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Loesche M, Gardner SE, Kalan L, Horwinski J, Zheng Q, Hodkinson BP, Tyldsley AS, Franciscus CL, Hillis SL, Mehta S, Margolis DJ, Grice EA, J. Invest. Dermatol 2017, 137(1), 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, Cox SB, White JS, Wound Repair Regen. 2016, 24, 163. [DOI] [PubMed] [Google Scholar]
- [58].Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA, Diabetes 2013, 62, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tellechea A, Leal EC, Kafanas A, Auster ME, Kuchibhotla S, Ostrovsky Y, Tecilazich F, Baltzis D, Zheng Y, Carvalho E, Zabolotny JM, Weng Z, Petra A, Patel A, Panagiotidou S, Pradhan-Nabzdyk L, Theoharides TC, Veves A, Diabetes 2016, 65, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Park LK, Maione AG, Smith A, Gerami-Naini B, Iyer LK, Mooney DJ, Veves A, Garlick JA, Epigenetics 2014, 9, 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tecilazich F, Dinh T, Pradhan-Nabzdyk L, Leal E, Tellechea A, Kafanas A, Gnardellis C, Magargee ML, Dejam A, Toxavidis V, Tigges JC, Carvalho E, Lyons TE, Veves A, PLoS ONE 2013, 8, e83314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cha J, Kwak T, Butmarc J, Kim TA, Yufit T, Carson P, Kim SJ, Falanga V, J. Dermatol. Sci 2008, 50, 15. [DOI] [PubMed] [Google Scholar]
- [63].Maione AG, Brudno Y, Stojadinovic O, Park LK, Smith A, Tellechea A, Leal EC, Kearney CJ, Veves A, Tomic-Canic M, Mooney DJ, Garlick JA, Tissue Eng Part C Methods 2015, 21, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M, Wound Repair Regen. 2008, 16, 585. [DOI] [PubMed] [Google Scholar]
- [65].Barrientos S, Brem H, Stojadinovic O, Tomic-Canic M, Wound Repair Regen. 2014, 22, 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pastar I, Stojadinovic O, Yin NC, Ramirez H, Nusbaum AG, Sawaya A, Patel SB, Khalid L, Isseroff RR, Tomic-Canic M, Adv. Wound Care (New Rochelle). 2014, 3, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Eming SA, Tomic-Canic M, Exp. Dermatol 2017, 26, 97. [DOI] [PubMed] [Google Scholar]
- [68].Hamdan S, Pastar I, Drakulich S, Dikici E, Tomic-Canic M, Deo S, Daunert S, ACS Cent. Sci 2017, 3, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jun JI, Lau LF, Nat. Cell Biol 2010, 12, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Harsha A, Stojadinovic O, Brem H, Sehara-Fujisawa A, Wewer U, Loomis CA, Blobel CP, Tomic-Canic M, J. Mol. Med. (Berl) 2008, 86, 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gibson DJ, Schultz GS, Adv. Wound Care (New Rochelle). 2013, 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lazaro JL, Izzo V, Meaume S, Davies AH, Lobmann R, Uccioli L, J. Wound Care 2016, 25, 277. [DOI] [PubMed] [Google Scholar]
- [73].Caley MP, Martins VL, O’Toole EA, Adv. Wound Care (New Rochelle). 2015, 4, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wysocki AB, Staiano-Coico L, Grinnell F, J. Invest. Dermatol 1993, 101, 64. [DOI] [PubMed] [Google Scholar]
- [75].Yager DR, Zhang LY, Liang HX, Diegelmann RF, Cohen IK, J. Invest. Dermatol 1996, 107, 743. [DOI] [PubMed] [Google Scholar]
- [76].Yager DR, Kulina RA, Gilman LA, Int. J. Low. Extrem. Wounds 2007, 6, 262. [DOI] [PubMed] [Google Scholar]
- [77].Tarlton JF, Bailey AJ, Crawford E, Jones D, Moore K, Harding KD, Wound Repair Regen. 1999, 7, 347. [DOI] [PubMed] [Google Scholar]
- [78].Wiegand C, Schonfelder U, Abel M, Ruth P, Kaatz M, Hipler UC, Arch. Dermatol. Res 2010, 302, 419. [DOI] [PubMed] [Google Scholar]
- [79].Wysocki AB, Kusakabe AO, Chang S, Tuan TL, Wound Repair Regen. 1999, 7, 154. [DOI] [PubMed] [Google Scholar]
- [80].Ladwig GP, Robson MC, Liu R, Kuhn MA, Muir DF, Schultz GS, Wound Repair Regen. 2002, 10, 26. [DOI] [PubMed] [Google Scholar]
- [81].Bullen EC, Longaker MT, Updike DL, Benton R, Ladin D, Hou ZZ, Howard EW, J. Invest. Dermatol 1995, 104, 236. [DOI] [PubMed] [Google Scholar]
- [82].Lobmann R, Ambrosch A, Schultz G, Waldmann K, Schiweck S, Lehnert H, Diabetologia 2002, 45, 1011. [DOI] [PubMed] [Google Scholar]
- [83].Weckroth M, Vaheri A, Lauharanta J, Sorsa T, Konttinen YT, J. Invest. Dermatol 1996, 106, 1119. [DOI] [PubMed] [Google Scholar]
- [84].Rayment EA, Upton Z, Shooter GK, Br. J. Dermatol 2008, 158, 951. [DOI] [PubMed] [Google Scholar]
- [85].Liu Y, Min DQ, Bolton T, Nube V, Twigg SM, Yue DK, McLennan SV, Diabetes Care 2009, 32, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Thom SR, Hampton M, Troiano MA, Mirza Z, Malay DS, Shannon S, Jennato NB, Donohue CM, Hoffstad O, Woltereck D, Yang M, Yu K, Bhopale VM, Kovtun S, Margolis DJ, Diabetes 2016, 65, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Margolis DJ, Hampton M, Hoffstad O, Mala DS, Mirza Z, Woltereck D, Shannon S, Trojano MA, Mitra N, Yang M, Bhopale VM, Thom SR, Wound Repair Regen. 2017, 25, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Singer AJ, Clark RA, N. Engl. J. Med 1999, 341, 738. [DOI] [PubMed] [Google Scholar]
- [89].Pastar I, Khan AA, Stojadinovic O, Lebrun EA, Medina MC, Brem H, Kirsner RS, Jimenez JJ, Leslie C, Tomic-Canic M, J. Biol. Chem 2012, 287, 29324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nuutila K, Siltanen A, Peura M, Bizik J, Kaartinen I, Kuokkanen H, Nieminen T, Harjula A, Aarnio P, Vuola J, Kankuri E, Wound Repair Regen. 2012, 20, 830. [DOI] [PubMed] [Google Scholar]
- [91].Pastar I, Ramirez H, Stojadinovic O, Brem H, Kirsner RS, Tomic-Canic M, Surg. Technol. Int 2011, 21, 51. [PubMed] [Google Scholar]
- [92].Li X, Li D, Wang A, Chu T, Lohcharoenkal W, Zheng X, Grunler J, Narayanan S, Eliasson S, Herter EK, Wang Y, Ma Y, Ehrstrom M, Eidsmo L, Kasper M, Pivarcsi A, Sonkoly E, Catrina SB, Stahle M, Xu Landen N, J. Invest. Dermatol 2017, 137, 2630. [DOI] [PubMed] [Google Scholar]
- [93].Saravanan R, Adav SS, Choong YK, van der Plas MJA, Petrlova J, Kjellstrom S, Sze SK, Schmidtchen A, Sci. Rep 2017, 7, 13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Gardiner M, Vicaretti M, Sparks J, Bansal S, Bush S, Liu M, Darling A, Harry E, Burke CM, PeerJ. 2017, 5, e3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD, BMC Microbiol. 2008, 8, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D, PLoS ONE 2008, 3, e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kalan L, Loesche M, Hodkinson BP, Heilmann K, Ruthel G, Gardner SE, Grice EA, MBio 2016, 7, e01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bonassi S, Ugolini D, Kirsch-Volders M, Stromberg U, Vermeulen R, Tucker JD, Environ. Mol. Mutagen 2005, 45, 258. [DOI] [PubMed] [Google Scholar]
- [99].Stojadinovic O, Brem H, Vouthounis C, Lee B, Fallon J, Stallcup M, Merchant A, Galiano RD, Tomic-Canic M, Am. J. Pathol 2005, 167, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Amini-Nik S, Cambridge E, Yu W, Guo A, Whetstone H, Nadesan P, Poon R, Hinz B, Alman BA, J. Clin. Invest 2014, 124, 2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Poon R, Nik SA, Ahn J, Slade L, Alman BA, BMC Cell Biol. 2009, 10, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Krisp C, Jacobsen F, McKay MJ, Molloy MP, Steinstraesser L, Wolters DA, Proteomics 2013, 13, 2670. [DOI] [PubMed] [Google Scholar]
- [103].Hanakawa Y, Amagai M, Shirakata Y, Sayama K, Hashimoto K, J. Cell Sci 2000, 113, 1803. [DOI] [PubMed] [Google Scholar]
- [104].Trengove NJ, Stacey MC, Macauley S, Bennett N, Gibson J, Burslem F, Murphy G, Schultz G, Wound Repair Regen. 1999, 7, 442. [DOI] [PubMed] [Google Scholar]
- [105].Junka A, Wojtowicz W, Zabek A, Krasowski G, Smutnicka D, Bakalorz B, Boruta A, Dziadas M, Mlynarz P, Sedghizadeh PP, Bartoszewicz M, J. Pharm. Biomed. Anal 2017, 137, 13. [DOI] [PubMed] [Google Scholar]
- [106].Moues CM, van Toorenenbergen AW, Heule F, Hop WC, Hovius SE, Wound Repair Regen. 2008, 16, 488. [DOI] [PubMed] [Google Scholar]
- [107].Serena TE, Adv. Wound Care (New Rochelle). 2014, 3, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Serena TE, Cullen BM, Bayliff SW, Gibson MC, Carter MJ, Chen L, Yaakov RA, Samies J, Sabo M, DeMarco D, Le N, Galbraith J, Wound Repair Regen. 2016, 24, 589. [DOI] [PubMed] [Google Scholar]
- [109].Falanga V, Sabolinski M, Wound Repair Regen. 1999, 7, 201. [DOI] [PubMed] [Google Scholar]
- [110].Karim RB, Brito BL, Dutrieux RP, Lassance FP, Hage JJ, Adv. Skin Wound Care 2006, 19, 324. [DOI] [PubMed] [Google Scholar]
- [111].Brem H, Tomic-Canic M, J. Clin. Invest 2007, 117, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Gallagher KA, Liu ZJ, Xiao M, Chen H, Goldstein LJ, Buerk DG, Nedeau A, Thom SR, Velazquez OC, J. Clin. Invest 2007, 117, 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Dangwal S, Stratmann B, Bang C, Lorenzen JM, Kumarswamy R, Fiedler J, Falk CS, Scholz CJ, Thum T, Tschoepe D, Arterioscler. Thromb. Vasc. Biol 2015, 35, 1480. [DOI] [PubMed] [Google Scholar]
- [114].Nunan R, Harding KG, Martin P, Dis. Models Mech 2014, 7, 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Panuncialman J, Hammerman S, Carson P, Falanga V, Wound Repair Regen. 2010, 18, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Lebrun E, Kirsner RS, JAMA Dermatol. 2013, 149, 1059. [DOI] [PubMed] [Google Scholar]
- [117].Stojadinovic O, Landon JN, Gordon KA, Pastar I, Escandon J, Vivas A, Maderal AD, Margolis DJ, Kirsner RS, Tomic-Canic M, Exp. Dermatol 2013, 22, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Dinh T, Tecilazich F, Kafanas A, Doupis J, Gnardellis C, Leal E, Tellechea A, Pradhan L, Lyons TE, Giurini JM, Veves A, Diabetes 2012, 61, 2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Ramirez HA, Liang L, Pastar I, Rosa AM, Stojadinovic O, Zwick TG, Kirsner RS, Maione AG, Garlick JA, Tomic-Canic M, PLoS ONE 2015, 10, e0137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Tellechea A, Kafanas A, Leal EC, Tecilazich F, Kuchibhotla S, Auster ME, Kontoes I, Paolino J, Carvalho E, Nabzdyk LP, Veves A, Int. J. Low. Extrem. Wounds 2013, 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Leal EC, Carvalho E, Tellechea A, Kafanas A, Tecilazich F, Kearney C, Kuchibhotla S, Auster ME, Kokkotou E, Mooney DJ, LoGerfo FW, Pradhan-Nabzdyk L, Veves A, Am. J. Pathol 2015, 185, 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Xu W, Jong Hong S, Jia S, Zhao Y, Galiano RD, Mustoe TA, Lab. Invest 2012, 92, 584. [DOI] [PubMed] [Google Scholar]
- [123].Tomic-Canic M, Mamber SW, Stojadinovic O, Lee B, Radoja N, McMichael J, Wound Repair Regen. 2007, 15, 71. [DOI] [PubMed] [Google Scholar]
- [124].Stojadinovic O, Tomic-Canic M, Methods Mol. Biol 2013, 1037, 255. [DOI] [PubMed] [Google Scholar]
- [125].Glinos GD, Verne SH, Aldahan AS, Liang L, Nouri K, Elliot S, Glassberg M, Cabrera DeBuc D, Koru-Sengul T, Tomic-Canic M, Pastar I, Wound Repair Regen. 2017, 25, 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Stojadinovic O, Yin N, Lehmann J, Pastar I, Kirsner RS, Tomic-Canic M, Immunol. Res 2013, 57, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Heilborn JD, Nilsson MF, Kratz G, Weber G, Sorensen O, Borregaard N, Stahle-Backdahl M, J. Invest. Dermatol 2003, 120, 379. [DOI] [PubMed] [Google Scholar]
- [128].Harding K, Int. Wound J 2016, 13, 1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kirsner RS, Vanscheidt W, Keast DH, Lantis JC 2nd, Dove CR, Cazzell SM, Vartivarian M, Augustin M, Marston WA, McCoy Bs ND, Cargill DD, Lee Mshs TD, Dickerson JE Jr., Slade Md HB, Slade Md HB; HP802–247 Study Group, Wound Repair Regen. 2016, 24, 894. [DOI] [PubMed] [Google Scholar]
- [130].Charles CA, Tomic-Canic M, Vincek V, Nassiri M, Stojadinovic O, Eaglstein WH, Kirsner RS, J. Am. Acad. Dermatol 2008, 59, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Cowin AJ, Hatzirodos N, Holding CA, Dunaiski V, Harries RH, Rayner TE, Fitridge R, Cooter RD, Schultz GS, Belford DA, J. Invest. Dermatol 2001, 117, 1282. [DOI] [PubMed] [Google Scholar]
- [132].Vaalamo M, Weckroth M, Puolakkainen P, Kere J, Saarinen P, Lauharanta J, SaarialhoKere UK, Br. J. Dermatol 1996, 135, 52. [PubMed] [Google Scholar]
- [133].Nassiri S, Zakeri I, Weingarten MS, Spiller KL, J. Invest. Dermatol 2015, 135, 1700. [DOI] [PubMed] [Google Scholar]
- [134].Sundaram GM, Common JEA, Gopal FE, Srikanta S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB, Sampath P, Nature 2013, 495, 103. [DOI] [PubMed] [Google Scholar]
- [135].Thom SR, Milovanova TN, Yang M, Bhopale VM, Sorokina EM, Uzun G, Malay DS, Troiano MA, Hardy KR, Lambert DS, Logue CJ, Margolis DJ, Wound Repair Regen. 2011, 19, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Diegelmann RF, Wound Repair Regen. 2003, 11, 490. [DOI] [PubMed] [Google Scholar]
- [137].Wysocki AB, Bhalla-Regev SK, Tierno PM Jr, Stevens-Riley M, Wiygul RC, Biol. Res. Nurs 2013, 15, 407. [DOI] [PubMed] [Google Scholar]
- [138].Wysocki AB, Wound J Ostomy Continence Nurs. 1996, 23, 283. [DOI] [PubMed] [Google Scholar]