Abstract
The one-cell C. elegans embryo offers many advantages for mechanistic analysis of cell division processes. Conservation of key genes and pathways involved in cell division makes findings in C. elegans broadly relevant. A key technical advantage of this system is the ability to penetrantly deplete essential gene products by RNA interference (RNAi) and replace them with wildtype or mutant versions expressed at endogenous levels from single copy RNAi-resistant transgene insertions. This ability to precisely perturb essential genes is complemented by the inherently highly reproducible nature of the zygotic division that facilitates development of quantitative imaging assays. Here, we detail approaches to generate targeted single copy transgene insertions that are RNAi-resistant, to engineer variants of individual genes employing transgene insertions as well as at the endogenous locus, and to in situ-tag genes with fluorophores/purification tags. We also describe imaging assays and common image analysis tools employed to quantitatively monitor phenotypic effects of specific perturbations on meiotic and mitotic chromosome segregation, centrosome assembly/function, and cortical dynamics/cytokinesis.
Introduction
The Caenorhabditis elegans one-cell embryo is a powerful system for the mechanistic analysis of cell division (Fig. 1). Studies in this model system combine the ability to perform precise genetic manipulations with quantitative live-cell imaging. In addition, the stereotypical nature of early embryonic divisions facilitates the detection and characterization of even subtle mutant phenotypes. The primary technical advantage of this system is that RNAi-mediated depletion is both specific and highly penetrant. >95% protein depletion can be routinely achieved independently of the target’s intrinsic turnover rate because of the combination of robust mRNA degradation and removal of pre-existing protein from the germline by continued embryo production (Oegema and Hyman 2006). In addition, as fertilization triggers oocyte meiotic divisions and subsequent embryonic mitoses, one is able to visualize the first divisions that occur after penetrant depletion of any essential cell division component. This feature of the system was central to early phenotypic screens that led to the discovery and characterization of the cellular pathways that participate in diverse processes including cell polarity, centriole duplication, nuclear assembly, bipolar spindle formation, kinetochore assembly, kinetochore-microtubule interactions, and cytokinesis (Fraser et al. 2000; Gonczy et al. 2000; Piano et al. 2000; Maeda et al. 2001; Kamath and Ahringer 2003; Simmer et al. 2003; Rual et al. 2004; Fernandez et al. 2005; Sonnichsen et al. 2005) (Fig. 1). Following these initial pathway discovery efforts, two technologies greatly expanded the ability to conduct detailed mechanistic cell biological analysis employing this system. The first is targeted single copy transgene insertions, which enable expression of RNAi-resistant versions of essential genes at endogenous levels, and the second is CRISPR/Cas9, which enables in situ-tagging as well as introduction of non-essential mutations into the endogenous locus.
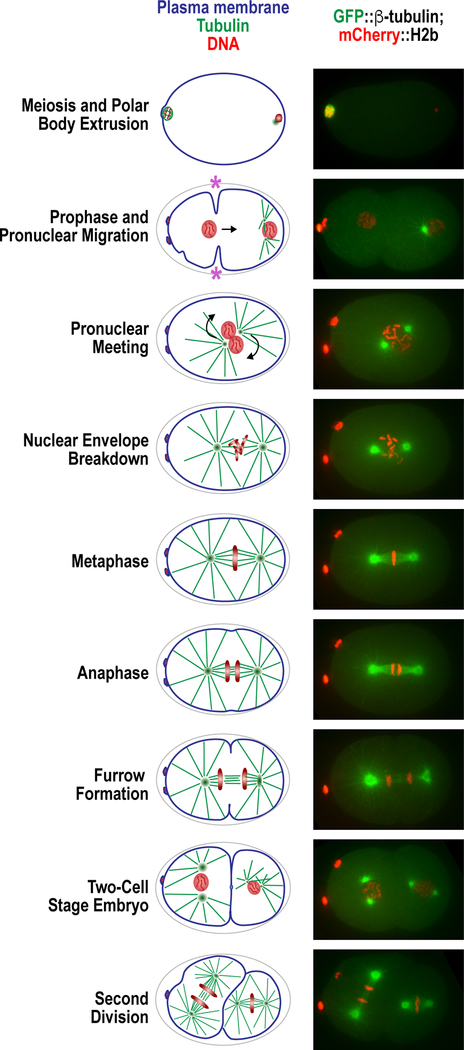
Figure 1. The one-cell embryo processes.
(left) Schematic of the early divisions in the C. elegans embryo, from meiosis I to the second embryonic division. (right) Confocal microscopy images of embryos matching the schematics of the left, expressing GFP::β-tubulin and mCherry::H2b. Pseudo-cleavage furrow is indicated with a purple asterisk.
In this chapter, we describe methods for studying cell division processes in the one-cell C. elegans embryo. We detail approaches for engineering essential cell division genes, visualizing their products, and conducting quantitative imaging based assays to define phenotypes. The ease of culturing C. elegans also enables proteomic analyses, methods for which have been described previously and will not be presented here (e.g. Zanin et al. 2011).
I. Engineering Cell Division Genes
To make use of C. elegans one-cell embryo as a model for mechanistic dissection of cell division genes, it is necessary to manipulate the endogenous locus to introduce tags for visualizing and purifying the gene product, to define the consequences of loss of gene function (employing RNAi and, when not essential for viability, null mutants), and to engineer precise changes in specific genes guided by sequence/structural information. This first section describes methods for achieving these goals.
A. In Situ-Tagging for Imaging & Affinity Purification
For visualization of specific gene products during cell division, two methods are used: immunofluorescence and fluorescent protein tagging. Immunofluorescence is well-suited for high resolution localization in the C. elegans embryo (e.g. Gonczy et al. 1999; Oegema et al. 2001) but the protocol is complicated by the presence of the eggshell making it difficult to quantify; immunofluorescence is also limited by the requirement that specific antibodies be generated and validated for each target. While antibodies are valuable and their generation is highly recommended (we have in hand a library of antibodies targeting cell division components that are available upon request), we routinely rely on fluorescent tags to visualize and quantify localization dynamics. In addition, fluorescent tags can be used for affinity purification and identification of interacting partners using mass spectrometry (Cheeseman et al. 2004; Zanin et al. 2011).
Two methods can be used to generate fluorescent protein fusions: transgenes and in situ tags; the latter is the method of choice in the CRISPR/Cas9 era. However, for genes that cannot be functionally tagged, as well as for general probes that label subcellular structures such as the plasma membrane, the microtubule cytoskeleton, and the chromatin, transgenes are necessary. In this section, we describe methods for functionally tagging genes in situ using CRISPR/Cas9. Transgene insertions are described below in Section I.C.2.
1. Choice of fluorophore
Since the pioneering use of green fluorescent protein (GFP) in C. elegans (Chalfie et al. 1994), a number of fluorescent protein tags have been developed (for a comparative analysis of some of the currently available fluorescent probes, see Heppert et al. 2016). Both intron insertion and codon optimization are important for robust fluorescent protein expression, especially in the germline (Green et al. 2008). As the growth temperature of C. elegans is 20°C and many fluorescent protein properties were optimized for use in mammalian cells (which grow at 37°C), there remains a need to optimize different wavelength and photoconvertible fluorescent proteins specifically for use in C. elegans.
GFP (and it’s fast-folding version sGFP), which absorbs blue light and emits green light, is the historically most widely used fluorophore (note that in C. elegans the S65C variant of GFP, not used in other systems, is commonplace because of its distribution in the early vector sets by Andrew Fire’s lab). In the last few years mNeonGreen, isolated from a lancelet, has emerged as a compelling alternative to green fluorescent protein (Shaner et al. 2013). In our hands, mNeonGreen has improved the signal-to-noise ratio when fused to proteins such as cyclin B (CYB-1) (Kim et al. 2017), although any potential improvement will depend on the target protein and/or the tissue/subcellular compartment in which it is expressed (Heppert et al. 2016).
Red fluorescent proteins such as mCherry, mKate2 and TagRFP have also been adapted for use in C. elegans, although lower intensity and photostability of these proteins make them second choices for visualizing low abundance proteins (Heppert et al. 2016). A novel mCherry variant called wrmScarlet (a codon-optimized version of mScarlet, Bindels et al. 2017) was recently shown to be 6-fold brighter than TagRFP when fused to a muscle-expressed gene (El Mouridi et al. 2017). It will be interesting to assess whether wrmScarlet improves imaging in the red channel in the germline and embryo. Photoconvertible proteins (mEos2 and mMaple3; McKinney et al. 2009; Wang et al. 2014) have also been adapted for used in C. elegans for super-resolution imaging (Kohler et al. 2017; Rog et al. 2017).
To engineer in-situ fluorescently tagged proteins, tags are generally placed at the N- or C-terminus; for some proteins, such as the Argonaute CSR-1, an internal tag proved necessary to maintain function (Gerson-Gurwitz et al. 2016). Addition of flexible linkers between the fluorophore and the gene of interest is also important in the generation of functional fusion proteins. The effect of a fluorescent protein tag on function is not possible to predict but if the gene or its orthologue has been functionally tagged in other systems, such as budding yeast or mammalian cells, this provides a useful starting point.
2. CRISPR-Cas9 – mediated in situ tagging
CRISPR-Cas9 generates a double-stranded DNA break at a specific location, using RNA as a guide (Doudna and Charpentier 2014; Hsu et al. 2014) (Fig. 2A). If provided with a DNA donor template, this break can then be repaired by the homologous recombination machinery to insert specific sequences such as the coding sequence for a fluorescent protein. The endonuclease, Cas9 is directed to specific genomic regions by forming a complex with an RNA duplex containing a tracrRNA and a crRNA; the latter contains a 20-nucleotide sequence that is complementary to the target DNA. The target sequence must be followed by a “protospacer adjacent motif” or PAM, which contains a NGG sequence (where N is any nucleotide). The tracrRNA and crRNA are most often expressed as a single guide (sgRNA), with the sequence of the crRNA-equivalent region varied to target different genomic locations.
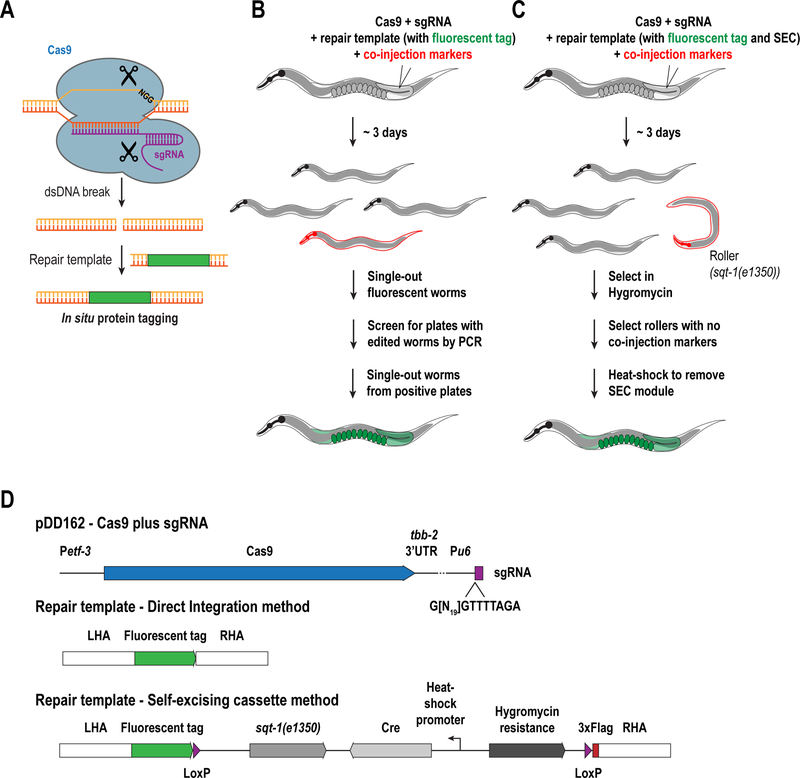
Figure 2. Methods for fluorescently tagging proteins in situ using CRISPRCas9.
(A) Schematic illustrating the CRISPR-Cas9 method (see text for details). Illustrations describing two methods for in situ tagging, using either direct integration (B) or the self-excising cassette method (C). (D) Plasmids to be generated for CRISPR/Cas9-mediated in situ tagging.
There are many approaches for CRISPR/Cas-9 based genome editing in C. elegans and the method of choice will depend on the type of modification that is desired (for a detailed review of currently available methods, see Dickinson and Goldstein 2016). In general, all CRISPR/Cas9 editing protocols require three steps: 1) injection of a mix into the gonad of young adult worms, 2) selection of transformants in the F1 generation, and 3) isolation of genome-edited animals among the F2 or F3 generations (Fig. 2B&C). The injection mix includes selection markers to help identify transformants. Below are two protocols that we have successfully adapted to tag endogenous loci with fluorescent proteins: the self-excising cassette (SEC) method (Dickinson et al. 2015) and the direct integration (DI) method (Waaijers et al. 2013; Waaijers and Boxem 2014). Both methods rely on plasmid-based expression of Cas9 and a single-guide RNA or sgRNA (which has both tracrRNA and crRNA functions; Fig. 2A), followed by homologous recombination–based insertion of the sequence encoding the fluorescent tag, although they differ on the method used to select for insertions.
2.1. Selection of the targeting sequence
To select suitable sgRNA sequence, one can look for an NGG motif or use one of the many available tools online, such as http://genome.sfu.ca/crispr/, http://crispr.mit.edu, https://crispr.cos.uni-heidelberg.de or http://genome.sfu.ca/crispr/). The choice of sgRNA is critical for the success of CRISPR/Cas9 editing and we follow the below recommendation (described in Farboud and Meyer 2015; Paix et al. 2015).
Cleavage site should be as close as possible to the edit site. For knocking in a fluorescent protein-coding region, the cleavage site is typically within 50 bp of the codon to which the fluorescent protein will be fused, generally the Start codon or the penultimate codon.
Guide RNA sequence should have a GC content of 30–75%
Avoid “C” as the nucleotide preceding the PAM, as its presence decreases cleavage efficiency (Doench et al. 2014; Gagnon et al. 2014; Farboud and Meyer 2015).
Off-target cleavage should be minimized. There are many tools available online for this purpose (for instance, http://crispr.mit.edu). If it is difficult to design a highly specific gRNA, we recommend choosing one that has three or more mismatches, preferentially close to the PAM.
Once selected, the 20-nucleotide target sequence can be inserted into an appropriate expression vector through site-directed mutagenesis or the Gibson Assembly DNA cloning method (Gibson 2009). A vector developed by the Goldstein lab, called pDD162 (Dickinson et al. 2013), expresses both codon-optimized Cas9 under the eft-3 germline promoter and the sgRNA under a U6 promoter. Alternatively, this plasmid can been modified to remove the Cas9 coding sequence and express the U6-derived sgRNA on its own, which enables adjusting the ratio of Cas9 to sgRNA plasmids to be injected. Because efficiency of a chosen sgRNA is difficult to predict, one can also use two different sgRNAs simultaneously. This can improve the rate of repair from a template although it can also lead to more off-target cleavage.
2.2. Design of repair template constructs
The repair construct should contain homology regions of 1000 – 1500 bp flanking the coding sequence for the fluorescent tag (Fig. 2D). It is advisable to insert a flexible linker sequence between the fluorescent protein and the gene in order to avoid interference of the tag with protein function (we use either GGGGGG or GGRAGS as linkers). In addition, mutation of the sgRNA recognition site to prevent cleavage of the repair template helps increase editing efficiency (Paix et al. 2015). In the self-excising cassette (SEC) method, the fluorescent tag is followed by a cassette which contains a Hygromycin resistance gene and the sqt-1(e1350) allele that causes a dominant roller phenotype that are used for selection of transformants. In addition, the cassette contains heat-shock–inducible Cre that can be used to remove the entire cassette once successful integration is achieved (Dickinson et al. 2015) (Fig. 2C).
We recommend using Gibson Assembly (Gibson et al. 2009) for construction of repair templates. This technique enables rapid and efficient generation of constructs without the need for restriction enzyme-based digestions and extraneous coding sequences. We first PCR-amplify the homology arms from N2 genomic DNA (genomic sequences are obtained from Wormbase, www.wormbase.org). Concurrently, we PCR-amplify the coding sequence for the fluorescent tag and a backbone vector (e.g. pBlueScript SKII (+)). All of the fragments must contain overlapping ends. The fragments are combined with an Assembly Master Mix (AMM) to assemble the different fragments together and the reaction is transformed into E. coli to screen for clones containing the correct plasmids. For the SEC method, the large, ~5.5 kb sequence of the cassette can be difficult to clone, although efficient ligation-based methods can also be used with these vectors (Dickinson et al. 2015).
2.3. Fluorescent protein tagging using direct integration (DI)
- Prepare 10 µl of a DNA mix containing the following plasmids:
- 100 ng/µL of sgRNA construct. This can be included in the Cas9 construct pDD162 or in a separate plasmid. If two sgRNA constructs are used, each should be 50 ng/µL.
- 30 ng/µL of Peft-3::Cas9 plasmid (pDD162) if the Cas9 and the sgRNA are delivered from separate plasmids.
- 40 ng/µL of repair template plasmid
- Fluorescent co-injection markers (Frokjaer-Jensen et al. 2008; Frokjaer-Jensen et al. 2012); Pmyo-2::mCherry (pCFJ90; 2.5 ng/µL) and Pmyo-3::mCherry (pCFJ104; 5 ng/µL). Green versions of these markers also exist (pCFJ420 (Pmyo-2::GFP::H2b) and pCFJ421 (Peft-3::GFP::H2b)); all are available through Addgene.
Centrifuge the mix at maximum speed for 30 min at 4°C.
Inject the germline of 40 – 50 young adult worms. Germline injection is described in detail in Berkowitz et al. (2008); Kadandale et al. (2009). Recover at 20–22°C.
After three days, pick ~96 L4 larval stage F1 progeny that express the fluorescent co-injection markers and single them onto 60 mm plates. Progeny should be selected from many different parental worms.
After four days, once the F2s have laid their F3 progeny and the plate is close to, or just starved of bacteria, wash worms from half of the plate using 500 µL of M9 buffer and collect the suspended worms into 1.5 mL tubes. The primary goal is to collect enough DNA from the F2 animals, which might contain a population that has the correct genome modification, while leaving enough worms on the plate for propagation.
Spin down worms at 1000 g for 3 min and remove as much M9 as possible.
Resuspend the worm pellet in 75 µL of PCR lysis buffer containing 1 mg/mL proteinase K and transfer into an 8-strip PCR tube.
Subject the tubes to three freeze-thaw cycles by dipping the strips into liquid nitrogen and immediately thawing in a water bath set to 55°C.
Lyse the worms at 65°C for 1 hour, followed by heat-inactivation of proteinase K at 95°C.
Use 4 µL of the lysate as a template for a 25 µL PCR reaction. Use a set of two primers, in which one is complementary outside the homology arm (either left or right) and the other is complementary to the coding sequence of the fluorescent tag, such that a band of 1–2 kb is present in the case of a successful integration event. The choice of the polymerase is important for the success of the PCR; we recommend using either Bio-X-Act (Bioline) or DreamTaq (Thermo Fisher).
If positive plates are identified, single out 16–24 worms from the remaining population onto 35 mm plates. Once these worms have laid progeny, lyse them in 5 µL of PCR lysis buffer and use 1–2 µL as template for a PCR reaction, using the same conditions as described above.
Single progeny from positive plates and determine homozygosity using primers that flank the fluorescent protein insertion. We usually design primers such that a wild-type allele gives a ~500 bp band and an integrant gives a ~1500 bp product.
Note: For tagging of essential genes we have observed that a significant proportion of the F1 worms exhibit developmental defects and/or embryonic lethality. This may indicate that the modification attempted is lethal, but may also be the result of efficient cutting of an essential gene locus without successful repair. In such cases, the amount of Cas9 plasmid (if separate from the sgRNA plasmid) can be decreased by half in the injection mix. Even if significant lethality or developmental defects are observed, selection and verification should be done as there is still a significant chance that the fluorescent protein coding region was integrated successfully in a small number of progeny.
2.4. Fluorescent protein tagging using the SEC method (Dickinson et al. 2015)
Prepare a similar DNA mix as described in the previous section; centrifuge the mix at maximum speed for 30 min at 4°C and inject into the germline of 40 – 50 young adults.
Place three worms per 60 mm plate and recover at 18°C.
If injections were successful, three days later, roller worms should be visible (note that the phenotype does not appear until after the L3 larval stage). Prepare a 5 mg/ml solution of Hygromycin B (Sigma) in water, filter-sterilize and add 500 µL to each plate. Dry plates in a laminar flow hood.
After four days, when the F2s are at the L4/adult stage, select for roller worms that lack any of the fluorescent co-injection markers and single them into 35 mm plates. If no non-fluorescent rollers are identified, check the plates three days later when F3 progeny are present.
Once the progeny of the singled-out rollers are at the L4/adult stage, check for homozygosity. If any normal moving worms are present on the plate, it is an indication that the mother was not homozygous and more progeny will have to be singled-out until all their progeny are rollers. In the case of N-terminal tags, the insertion of the SEC will generate a null allele and therefore, for essential genes, a balancer chromosome will need to be crossed in before proceeding with the next step (Edgley et al. 2006).
Once the SEC insertion is homozygous/balanced, pick ~10–15 L1/L2 worms, transfer them to a 60 mm plate and heat-shock them at 34°C for 4 hours. This will induce expression of the Cre recombinase and subsequent removal of the self-excising cassette.
After five days, look for normal moving worms. Confirm insertion of the fluorescent tag by PCR and microscopy.
B. Disrupting gene function in the C. elegans embryo
In the C. elegans one-cell embryo, there are four methods employed to disrupt gene function: RNA interference (RNAi), temperature-sensitive mutations, null mutations and auxin-inducible degradation (AID, Zhang et al. 2015). While null mutations are useful to study non-essential genes, for essential cell division genes they must be balanced and can only be analyzed in one-cell embryos in the rare cases where the null mutant is maternal effect lethal (i.e. maternal load from the balanced heterozygote is sufficient to support development of a homozygous mutant fertile adult whose progeny embryos lack the gene product and are inviable). Temperature-sensitive alleles are only available for a handful of genes and are not easy to engineer, and while the AID method is highly efficient, it requires functional tagging of the gene to be degraded, which is not possible in some cases. Therefore, to date, the most common method employed to disrupt gene function in the C. elegans embryo is RNAi.
1. RNAi-mediated protein depletion
In C. elegans embryos, RNAi-mediated protein depletion is very efficient. The germline is constantly synthesizing mRNA and proteins that flow into the oocytes, as they are being compartmentalized prior to ovulation and fertilization. Therefore, after destroying a specific mRNA using RNAi, its protein product is depleted from the germline and embryos at a constant rate that does not depend on protein half-life (Oegema and Hyman 2006). In comparison to genetic mutants, this approach is straightforward to use for studying function of essential cell division genes, due to its acute nature.
Although there are technically facile methods for delivery of the double-stranded RNA (dsRNA) that triggers RNAi, such as feeding and soaking, we find that direct injection of dsRNAi into worms results in the most penetrant depletions, typically >95% (Oegema and Hyman 2006) (Fig. 3B). Below we describe our standard RNAi protocol that works reliably to inhibit the function of the many genes and pathways that we have studied over the years.
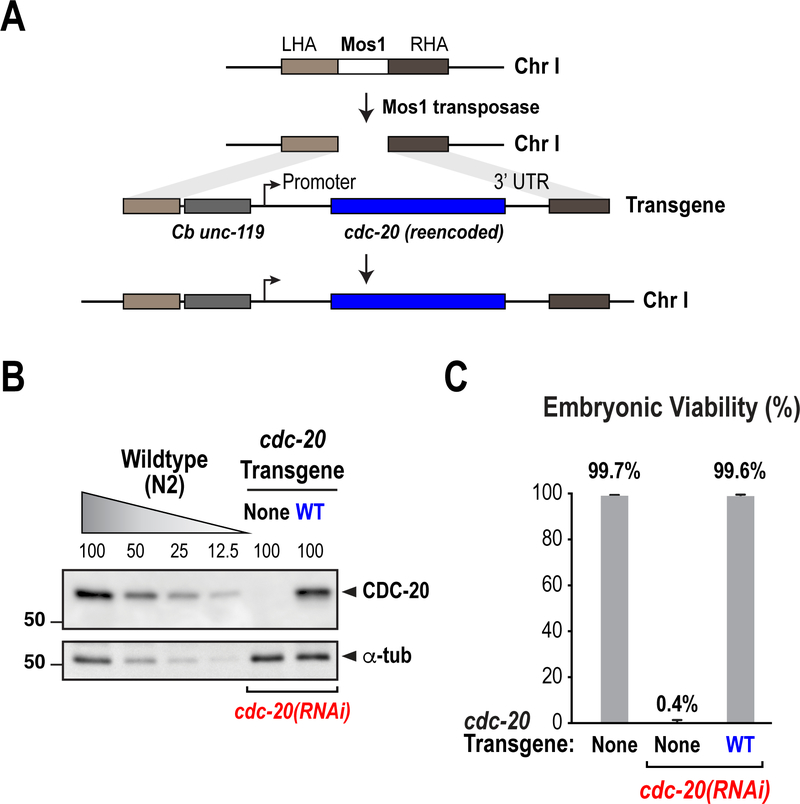
Figure 3. Generation of gene replacement systems using RNAi-resistant transgenes.
Described here is the strategy used for the generation of an RNAi-resistant transgene for CDC-20 (also known as FZY-1). (A) Schematic illustrating the strategy for single-copy transgene insertions using MosSCI. The CDC-20 transgene was inserted in chromosome I and rendered resistant to an RNAi targeting its coding sequence (B) Analysis of transgene expression through immunoblot. A control (N2) sample was titrated in order to assess depletion efficiency and rescue of CDC-20 expression. (C) Rescue of embryonic lethality by the RNAi-resistant CDC-20 transgene. (Adapted from Kim et al. 2017)
1.1. Generation of dsRNA for injection
To target a gene for depletion by RNAi, we first generate a double-stranded RNA (dsRNA) that targets a particular exon of the gene. We typically choose exons in the middle of the gene or towards the 3’ end that are between 400 to 800 bp in length. If only short exons exist in the gene, it is also possible to combine two exons with a short (<50 bp) intron sequence in between them or use cDNA as a PCR template.
Design primers to amplify the target exon(s) by PCR. Primers must contain sites for the T7 (taatacgactcactatagg) or T3 (aattaaccctcactaaagg) RNA polymerases on their 5’ end.
PCR-amplify the target exon using N2 genomic DNA (or, in specific cases, cDNA) as a template.
Purify the PCR product using a commercial kit. We recommend pooling the products from multiple PCR reactions to obtain approximately 20 µg of PCR product. If the PCR reaction resulted in more than one amplification product, it will be necessary to gel-purify the band of the correct size.
Set up two independent in vitro transcription reactions using the purified PCR product as a template and either T7 or T3 polymerases (MEGAscript, Ambion), following the manufacturer’s instructions.
Incubate reactions for 5 hours at 37°C.
Remove the PCR template by digestion with DNase I for 15 min at 37°C
Purify the RNAs using a MEGAclear transcription cleanup kit (Ambion).
Quantify RNAs and mix them at an equimolar ratio. Add 3X soaking buffer to a final concentration of 1X.
Denature RNAs for 10 min at 68°C and hybridize them at 37°C for 30 min
Check dsRNA by agarose gel electrophoresis. When compared to single RNAs, the dsRNA should show a mobility shift that indicates successful hybridization.
Quantify dsRNA by absorbance at 260 nm (1 absorbance unit = 40 µg/mL). Aliquot and store at −80°C.
1.2. dsRNA injection
Dilute dsRNA at a final concentration of 1 mg/ml in 1X soaking buffer. If two or three dsRNAs are mixed, make sure that the final concentration for each is higher than 0.7 mg/ml. Individual dsRNAs may need to be titrated to achieve maximum penetrance, however, we do not recommend mixing four or more dsRNAs.
Centrifuge in desktop centrifuge at maximum speed for 15–30 mins at 4°C.
Pick L4 larvae or young adult worms and immobilize onto agarose pads.
Using a glass needle, inject a small volume of dsRNA into the worm intestine.
Recover worms on 60 mm plates with food at 20°C. L4s are more prone to drying out on the agarose pads than adults so make sure to recover them quickly after injection.
Typically, full RNAi penetrance is achieved ~36 or more hours after L4 injection.
2. Temperature-sensitive alleles
While RNAi has many advantages, its usefulness can be limiting for genes that are required for embryo production, such as genes that are essential for germline structure and/or meiotic progression. In addition, RNAi reveals the first severe defect that develops following inhibition of gene function, limiting functional analysis at different temporal stages. Temperature-sensitive (ts) alleles provide a powerful method for the temporal control and extent of gene inhibition (Ward and Miwa 1978; O’Rourke et al. 2011; Mok et al. 2017). Ts mutants typically have singleamino acid changes that weaken protein function. In most cases, ts mutants are viable at 16°C and lethal at 25˚C; intermediate temperatures can be used to partially reduce protein function. For instance, we found that the anaphase promoting complex/cyclosome (APC/C), the E3 ubiquitin ligase responsible for triggering the metaphase-anaphase transition, can be partially inhibited in the 20–22°C range employing mat-3(or344), a temperature-sensitive allele of its essential subunit MAT-3 (Kim et al. 2017). This tuning of gene function can be further exploited by modulating the embryo temperature while imaging, although this is only effective with fast-acting mutants (Davies et al. 2017). In addition, separation-of-function ts alleles can be obtained for some genes. For instance, two different ts alleles for the cytokinesis factor cyk-4 can be used to either disrupt its interaction with its binding partner zen-4 (cyk-4(t1689ts)) or disrupt its GTPase-activation domain (cyk4(or749ts)) (Jantsch-Plunger et al. 2000; Canman et al. 2008).
Care must be taken when choosing a ts allele to study a gene of interest. Some ts alleles are partially penetrant at the restrictive temperature or slow-acting. In addition, as mentioned above, ts alleles are not available for the majority of genes and their engineering is not straightforward.
3. Null alleles
For non-essential genes, null alleles can be used to study their function in one-cell embryos. Several high-throughput mutant screens have isolated and mapped deletions for many genes, some of which result in null alleles. These deletions can be found on Wormbase and are readily available from the Caenorhabditis Genetics Center (CGC) and the National BioResource Project (NBRP). However, because of the random nature of these deletions, they do not always lead to full loss of function and can affect more than one gene. In addition, some deletions can disrupt more than one gene at the same time and therefore are not always specific. Recently, with the advent of CRISPR-Cas9, it is relatively straightforward to generate specific, full gene deletions for any gene.
3.1. Generation of null alleles using CRISPR/Cas9
Originally, CRISPR-based techniques relied on the generation of null alleles through the replacement of the coding sequence of a specific gene with a selection marker, such as unc-119 or the SEC module. However, a simpler method relies on the generation of two cuts within the coding sequence of the gene of interest using two different gRNAs: the first gRNA is homologous to a region downstream of the start codon and the second one has homology near the stop codon. This allows the cellular machinery to repair these cuts through non-homologous end joining, thereby deleting the entire gene in the process. By using purified Cas9 and RNAs (crRNA and tracrRNA) and injecting the complexes directly into the gonad (Fig. 4A), deletions can be generated with high efficiency (Paix et al. 2015; Paix et al. 2017b). Using this approach, we have been able to obtain homozygous deletion mutants within the first generation after injection.
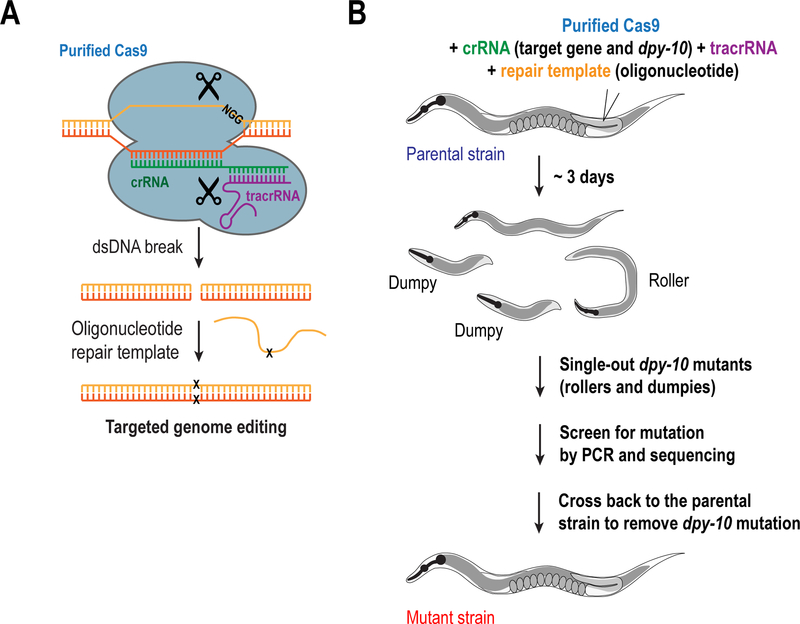
Figure 4. Methods to generate in situ gene modifications through CRISPRCas9.
(A) Schematic illustrating the CRISPR-Cas9 strategy for the generation of in situ modifications using purified Cas9, RNAs and an oligonucleotide as a repairing template. (B) Illustration showing the standard protocol for the generation of in situ mutations or deletions. The method described in the figure utilizes a dpy-10 crRNA as a marker for positively injected worms, which helps in the selection of successfully modified animals, although its use is optional (see text for details).
In the case of essential genes, the limitation is that, given the high efficiency of this method, most worms will give rise to either homozygous deletions or mutants in which both alleles are affected; in either case this results in non-viable progeny. On the other hand, worms with living progeny are likely to be the ones that were not injected properly. Thus, for essential genes, we recommend using a co-CRISPR marker (Arribere et al. 2014; Kim et al. 2014; Ward 2015), such as dpy-10, which allows for the phenotypic selection of homozygous “dumpy” loss-of-function mutants that can be easily distinguished under a stereo microscope for their short morphology. Alternatively, a heterozygous dominant “roller” phenotype can be obtained by mutating Arginine 92 in the dpy-10 sequence to Cysteine using an oligonucleotide as a repairing template (Arribere et al. 2014; Paix et al. 2015) (Fig. 4B) (see section I.C.3). Therefore, worms that get the dpy-10 edit are likely to have also been modified for the gene of interest. Once obtained, these deletions must be balanced to allow their propagation.
Protocol
Use the same guidelines as described above to design the crRNAs targeting the 5’ and 3’ of the coding sequence of interest. For dpy-10, we use the following sequence: gcuaccauaggcaccacgag (Paix et al. 2015).
Order the crRNAs, as well as tracrRNA from a commercial source (e.g. we order from Integrated DNA Technologies (IDT) but many companies synthesize RNAs for CRISPR applications).
Resuspend RNAs in nuclease-free duplex buffer (provided by IDT) at a final concentration of 200 µM each.
Mix 5 µL of tracrRNA with 2.5 µL of each crRNAs to a final volume of 10 µL. If dpy-10 co-CRISPR is used, mix 5 µL of tracrRNA with 2 µL of each targeting crRNA and 1 µL of dpy-10 crRNA.
Incubate RNA mix for 5 min at 95°C, followed by 5 min at room temperature
Incubate the RNA mix with purified Cas9 (can be expressed and purified inhouse or obtained from a commercial source) at a final concentration of 25 µM each, for 5 minutes at room temperature
Centrifuge in desk top centrifuge at maximum speed for 15 min at 4°C
Inject mix into the gonad of ~24 young adults. Recover worms at 20°C.
The following day, transfer the worms into a fresh plate. This is done because a high proportion of the progeny laid within the first 24 hr of injection will not be edited.
After three days, single-out L4s and adults. If using the dpy-10 co-injection marker it may be necessary to wait longer in order to get the mutant worms at the desired stage.
Once the singled-out worms have laid progeny, lyse them in PCR lysis buffer and analyze them by PCR using primers that flank the gene. We normally design oligos that flank the gene of interest; if the gene is deleted, a lower molecular weight band should be present. Alternatively, for genes that are large (>2 kb), we use a three-primer PCR reaction in which two primers flank the gene and a third one binds internally.
Once identified, cross the deleted worms back to the parental strain in order to remove the dpy-10 mutation and/or other potential off-targets.
3.2. Balancing of null alleles
In order to propagate null mutants of essential genes, it is essential to balance them. For this, we cross the mutants to an appropriate balancer chromosome, which itself cannot be propagated as a homozygote and therefore allows for the maintenance of the null allele and the balancer chromosome as heterozygotes (Edgley et al. 2006). The most widely used balancer chromosomes are translocations or inversions, such as hT2(I, III), mIn1(II) and nT1(IV, V). Many of these balancers have been also engineered to express GFP in the pharynx, therefore facilitating the selection of non-fluorescent adults that are homozygous for the null allele (Edgley et al. 2006). A detailed description of available balancers is at http://www.wormbook.org/chapters/www_geneticbalancers/geneticbalancers.html.
We also recommend balancing null mutations of non-essential genes as they often negatively affect worm development and fertility. Therefore, bias during propagation as homozygous mutants may lead to selection for secondary suppressor mutations.
C. Engineering specific mutants for mechanistic studies
Following the initial identification of a loss-of-function perturbation that affects cell division, precise engineering of mutations that affect specific properties becomes necessary to gain mechanistic insight. The C. elegans one-cell embryo is an excellent system for conducting such precise mechanistic analysis of cell division genes. In this system, it is possible to: 1) generate robust gene replacement systems, and 2) evaluate the effect of specific engineered mutations with high precision. Here we describe methods used to design and engineer mutants that disrupt a specific function.
1. Defining functional motifs
The majority, if not all, of the genes that are required for division in the onecell C. elegans embryo have been identified and are well conserved in other species. However, while overall orthology is understood, mechanistic understanding of the function of individual genes is limited. To gain mechanistic insight into how the product of a specific cell division gene functions, a first approach is analysis of the primary protein sequence. Comparing the protein of interest with orthologues in other systems is a good starting point. In some cases, relevant domains have been identified and structurally characterized in other species; in these cases, existing structural and biochemical information and primary sequence alignments can be employed to generate mechanistically informative mutations (e.g. Cheerambathur et al. 2013; Cheerambathur et al. 2017). If no orthologues can be identified, or there is little insight into functional domains, one can analyze secondary and 3D structure prediction; for this purpose there are online prediction programs such as Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) and Jpred (http://www.compbio.dundee.ac.uk/jpred/). This information can then be used to generate truncations that lack specific domains.
Many genes that regulate cell division contain in their sequence so-called Small Linear Motifs or SLiMs, which are short sequences that mediate protein-protein interactions (Diella et al. 2008; Davey et al. 2012b). SLiMs are typically located in disordered regions of proteins and can be evolutionarily conserved. Examples of SLiMs that are frequently found in cell division proteins are:
-
-
Polo-like kinase docking sites: S[S/T]P
-
-
Protein phosphatase 1 (PP1) interaction motifs: e.g. [G/S]ILK and [R/K]VxF (where x is any residue)
-
-
Protein phosphatase 2 – B56 docking motifs: φxxφxE (where φ is a hydrophobic residue)
-
-
Isoprenylation sites: C-terminal Caax (where “a” is an aliphatic residue)
-
-
APC/C degrons: D-box (RxxL) and KEN boxes
Novel SLiMs can also be identified through computational searches (Davey et al. 2011; Davey et al. 2012a); e.g. the ABBA (Acm1, Bub1, BubR1, cyclin A) motif that is present in multiple cell cycle regulators to mediate their interaction with Cdc20 (Di Fiore et al. 2015).
In addition, the primary sequence of the gene of interest can be scanned for known phosphorylation consensus sequences. This can be done using online resources, such as Group-based Prediction System (GPS; http://gps.biocuckoo.org/). Examples of known consensus phosphorylation sites:
-
-
Cyclin-dependent kinases (Cdks): [S/T]P
-
-
Aurora kinases: [R/K]x[S/T]φ
-
-
Polo-like kinases: [D/E/N/Q]x[T/P]φ
Outside of core catalytic domains of enzymes, functionally critical protein regions often exhibit weak primary sequence conservation across widely divergent species (e.g. between C. elegans and vertebrates). We find that sequence comparisons between more closely related Caenorhabditis species can be very useful for discovery of functionally important regions, SLiMs and post-translational modification sites. Functionally critical residues are, in the majority of cases, conserved across other species of the Caenorhabditis genus such as briggsae, brenneri, remanei and japonica. Such analysis can also reveal critical motifs required for a specific function; for example, analysis of conserved regions within the C-terminus of the nucleoporin MEL-28 revealed a docking site for protein phosphatase 1 that is critical for its function and was subsequently found to also be present in the vertebrate ortholog (Hattersley et al. 2016).
2. Engineering single-copy transgene insertions through the MosSCI system
Prior to 2008, stable insertion of transgenes into the C. elegans genome was achieved primarily using ballistic bombardment, in which the transgene integrated randomly, and often in multiple copies, in the genome (Schweinsberg and Grant 2013). A far superior method which has become the standard for transgene insertions today is Mos1-mediated Single Copy Insertion (MosSCI) (Frokjaer-Jensen et al. 2008; Frokjaer-Jensen et al. 2012) (Fig. 3A). Similar to CRISPR-Cas9, this method consists on the generation of a double-stranded DNA break at a single Mos1 transposon insertion in the genome by Mos transposase (the Mos1 transposon is derived from a Drosophila species). This break can then be repaired, resulting in incorporation of a donor piece of DNA at a single, specific location in the genome. MosSCI strains carry in the background an unc-119(ed3) mutation, which results in an uncoordinated paralysis phenotype. The vectors used for transgene insertion carry a copy of Caenorhabditis briggsae (C.b.) unc-119 that rescues the Unc phenotype, enabling selection of integrants (Frokjaer-Jensen et al. 2008). Subsequent genotyping ensures insertion has occurred at the correct locus. Below we describe the method we use to construct transgenes, how we re-encode them in order to make them RNAi-resistant, and how we generate and validate MosSCI strains.
2.1. Transgene Design
To generate a transgene for a gene of interest, first search for the genomic sequence on Wormbase (http://www.wormbase.org/). It is preferable to build the transgene with its own promoter and 3’ UTR, so that it retains it endogenous regulatory pattern (Fig. 3A). To isolate appropriate UTR sequences the GBrowse tool in wormbase can be used to copy ~2–3 kb of the 5’ upstream region and ~1–1.5 kb of the 3’ downstream region. Although we recommend utilizing 3 kb and 1.5 kb for the 5’ UTR and 3’UTR respectively, we have also been successful with shorter UTR sequences (the test for success being functional rescue as detailed below). For expression specifically in the germline and early embryo, we use control regions that support strong expression in the germline, such as pie-1 and mex-5 promoters and the tbb-2 3’-UTR (Merritt et al. 2008; Merritt and Seydoux 2010; Zeiser et al. 2011).
For the coding sequence, we normally maintain gene introns, as they are important for expression (especially in the germline). Occasionally, introns can be too big and difficult to clone and we have had success after removing such introns from the coding sequence.
2.2. Choice of MosSCI insertion site
There are several available MosSCI insertion strains that contain the Mos1 transposon on different chromosomes (for a list, see http://www.wormbuilder.org/). For most transgenes, we choose to integrate them on either chromosome I (ttTi4348) or chromosome II (ttTi5605); we have had the most success with the latter in terms of integration efficiency and lack of germline silencing. For chromosome I insertions, transgenes are cloned into the pCFJ352 vector, which contains the homology arms for chromosome I, as well as the C.b. unc-119 coding sequence. A similar vector exists for chromosome II, called pCFJ151.
In addition, novel MosSCI strains have been generated, called Universal MosSCI or Uni-MosSCI, in which a region of chromosome II containing the homology arms for pcFJ151 was copied onto other chromosomes (Frokjaer-Jensen et al. 2014). This way, transgenes can be easily transferred to other chromosomes without the need to sub-clone into a different vector. Using this method, we have had significant success with Uni-I (oxTi185) and Uni-V (oxTi365); however, transgenes integrated into Uni-IV (oxTi177) exhibit frequent silencing in the germline (unpublished observations).
2.3. Engineering an RNAi-resistant transgene.
In order to design RNAi-mediated gene replacement systems, it is necessary to alter a coding region in the transgene in order to make it resistant to a specific dsRNA. For this, we re-encode an exon(s) targeted by the dsRNA to generate silent mutations that maintain coding information but prevent RNAi. Two factors are critical for this. First, the C. elegans codon usage must be maintained to prevent recognition of the transgene as foreign by the C. elegans transcriptional silencing machinery. Second, any changes in the coding sequence must be verified to prevent the generation of artificial splicing donor or acceptor sites, which may disrupt gene expression (Green et al. 2008). For this purpose, manual shuffling of the codons in the sequence to be re-encoded, followed by confirmation of the absence of splicing donor or acceptor sites (using online tools such as http://www.cbs.dtu.dk/services/NetGene2/) should suffice. Automated scripts can also be developed for this purpose. Alternatively, a tool known as the C. elegans codon adapter tool online (https://worm.mpi-cbg.de/codons/cgi-bin/optimize.py) (Redemann et al. 2011), which was originally designed to modulate transgene expression levels by modifying the codons in the coding sequence, can be used for re-encoding. For this, make sure that the codon adaptation index (CAI) in your sequence, which is a measurement of the codon weight in the query sequence relative to that of highly expressed genes (Redemann et al. 2011), is maintained in the final product.
Once designed, the re-encoded region is synthesized by a DNA synthesis company. We then use Gibson Assembly to incorporate the fragment into the wildtype transgene.
2.4. Tagged vs untagged transgenes
In addition to re-encoding, transgenes can also be tagged with a fluorescent protein or other tags in order to detect expression of the encoded product. This can be particularly useful when evaluating transgene-encoded mutant proteins that do not rescue removal of the endogenous protein. However, in some cases, addition of a fluorescent tag can partially disrupt gene function and show synergistic effects with other introduced mutations (our unpublished observations). Thus, it is always useful to assess/confirm mutant phenotypes with untagged transgenes, which requires that expression be assessed using immunoblotting or with functional assays (see below).
2.5. Generation of MosSCI strains
This protocol is based on (Frokjaer-Jensen et al. 2008; Frokjaer-Jensen et al. 2012). It requires the injection of a plasmid mix containing the transgene in the appropriate vector and the Mos1 transposase. In addition, a mix containing fluorescent co-injection markers and heat-inducible peel-1 (a negative selection marker) is added; these plasmids will be propagated as extra-chromosomal arrays and their expression can be used to negatively select against animals that have not lost these arrays. Potential integrants are selected on the basis of their rescue of the unc-119(ed3) phenotype and the absence of extra-chromosomal arrays.
We have successfully integrated a number of transgenes of different sizes. Critical factors for a high integration rate are:
-
-
Size of the inserted DNA. Inserts larger than 8 kb are usually more problematic and more worms need to be injected to obtain a strain. However, we have successfully generated integrants of up to 20kb (Wang et al. 2015).
-
-
Integration strain. Chromosome II integrations typically exhibit higher integration efficiency and lower silencing rates than other chromosomes (Frokjaer-Jensen et al. 2012).
-
-
Quality of the injected DNA. We recommend transforming into NovaBlue cells (Millipore) and purifying DNA using a midi-prep kit (Qiagen) (Frokjaer-Jensen et al. 2014).
Protocol
- Generate an injection mix containing
- 50 ng/µL of transgene in targeting vector (pCFJ151 or pCFJ352)
- 100 ng/µL of pCFJ601 (Peft-3::Mos1 transposase)
- 20 ng/µL of pMA122 (Phsp::peel-1) (Optional for negative selection)
- 20 ng/µL of pGH8 (Prab-3::mCherry; pan-neuronal expression)
- 5 ng/µL of pcFJ90 (Pmyo-2::mCherry; pharynx muscle expression)
-
10 ng/µL of pcFJ140 (Pmyo-3::mCherry, body muscle expression)We have stocks of co-injection markers and mix them with the transgene vector at the indicated final concentrations.
Spin injection mixes at max speed on a tabletop centrifuge for 20–30 min.
Inject young adult hermaphrodites of the respective Mos strain with the injection mix. (As mentioned above, integration rates vary depending on the size of the transgene and likely also its sequence composition. We find in practice that injecting 20–30 worms is sufficient to obtain at least one correct integrant although we recommend obtaining at least two independent integrants for initial characterization)
Single injected worms onto 60 mm plates. (we pipette 10–20 µL of M9 on top of the bacterial spot on the plate and transfer 1 injected worm into the drop).
Incubate the injected animals at room temperature until they starve (7–10 days).
Once plates are fully starved, heat-shock them at 34°C in an air incubator for 3 hrs. Heat shock induces expression of the negative selection marker peel-1, which kills worms that contain an extra-chromosomal array.
Leave plates at room temperature after heat-shock and let them recover for 16 – 24 hr. At this point, starved plates can be ‘chunked’ onto a fresh plate to aid identification in the following step. For chunking, use a scalpel to cut a piece of the agar containing moving worms and place it upside down onto a fresh plate with food.
Screen each plate for non-fluorescent moving worms using a fluorescenceequipped stereomicroscope. Worms that contain MosSCI insertions are typically L1 or dauer larvae that move similarly to wild-type worms and lack any of the red fluorescence markers.
Pick 3–5 moving worms that lack red fluorescence and single them on 35 mm plates. In our experience, it is useful to pick more than one worm because a considerable fraction of heat-shocked animals exhibit sterility or growth problems. During this step, it is very important to make sure only single worms are transferred to ensure that any eventual strain represents a single transgene insertion.
Let each singled worm grow and lay embryos, and genotype by PCR. For this purpose, design primers that will give specific products only in the presence of an insertion (Table 1)
Check for homozygosity. If the insertions are homozygous, 100% of the F2 progeny will be non-Unc. If Unc worms are observed in the F2, this means that the transgene is heterozygous and additional non-Unc worms should be singled until Unc worms no longer appear. Homozygosity can also be confirmed using specific primers that give products of different sizes depending on whether an integration is present or not (Table 2).
Table 1.
Oligonucleotides used for the identification of MosSCI integration strains by PCR.
| Oligonucleotides | Expected product |
Notes | ||
|---|---|---|---|---|
| Forward | Reverse | |||
| Mos I (ttTi4348); left integration |
cgtcagagaggagagg aacg |
aaattcaggtctgtacgg aagc |
2093 bp | |
| Mos I (ttTi4348); right integration |
cattcgaagatctgccc act |
gagtccaacgcttatttct cg |
1754 bp | |
| Mos II (ttTi5605); left integration |
cgtcagagaggagagg aacg |
gacatttgagaatggcat tga |
1828 bp | Also for Uni-MosSCI integrations |
| Mos II (ttTi5605); right integration |
cattcgaagatctgccc act |
atcgggaggcgaacct aactg |
1524 bp | Also for Uni-MosSCI integrations |
Table 2.
Oligonucleotides used for the genotyping of MosSCI integration strains by PCR.
| Ooligonucleotides | Expected product | Notes | ||||
|---|---|---|---|---|---|---|
| Forward | Reverse | Internal | Wild-type | Integrant | ||
| Mos I (ttTi4348) |
cccaaaatac ctcccctcat |
tccggagaaa aacttccaaa |
gcgatctttgaa gaagtgacg |
544 bp | 1680 bp | |
| Mos II (ttTi5605) |
cgaaatgtcct cctgattcc |
cgccattgtttc ctgaaaat |
gcgatctttgaa gaagtgacg |
654 bp | 1620 bp | |
| Uni I (oxTi185) |
gttccatagcc accatcacc |
gcatcgctaaa aacgagagg |
tatcgtaaatcg gcgcgagc |
914 bp | 1242 bp | Use a 62°C annealing temperature protocol |
| Uni IV (oxTi177) |
gtcactcaaac cgatgcaga |
gcaatttcggc aatttcagt |
tatcgtaaatcg gcgcgagc |
324 bp | 566 bp | Use a 62°C annealing temperature protocol |
| Uni V (oxTi365) |
ccctcttttggg ctctgcaa |
ttgatgaaagg caagcgtgaa |
tatcgtaaatcg gcgcgagc |
1083 bp | 1316 bp | Use a 62°C annealing temperature protocol |
2.6. Validating parent transgene insertions using null mutants and RNAi-mediated depletion
Designing RNAi-resistant transgenes involves choosing 5’ and 3’ UTR control regions, re-encoding one or more exons, and, in specific cases, inserting a tag for visualization. One or more of these has the potential to negatively impact gene expression and function. Thus, it is extremely important to validate that a transgene insertion designed to express the wild-type gene product (what we refer to as the parent transgene insertion) is fully functional. The gold standard for validating parent transgene insertions is rescue of null mutants (either isolated from forward genetic screens, generated by gene knockout consortia, or made using CRISPR/Cas9). With the advent of CRISPR/Cas9, it is now expected that null mutants will be available or straightforward to generate for all genes and we recommend that these be at hand or engineered in parallel (see I.B.3. above) to generating parent transgene insertions. Of note, all of the parent transgene insertions we have employed for introducing specific mutations have been validated by crossing to mutants and demonstrating rescue (Espeut et al. 2012; Cheerambathur et al. 2013; Lettman et al. 2013; Zanin et al. 2013; Moyle et al. 2014; Shimanovskaya et al. 2014; Kim et al. 2015; Wang et al. 2015; Gerson-Gurwitz et al. 2016; Hattersley et al. 2016; Kim et al. 2017) In the case of non-essential genes, this requires a quantifiable mutant phenotype that can be used to assess functional rescue. If the transgene has been re-encoded for RNAi-resistance and RNAi-mediated depletion is associated with severe defects, transgene functionality can also be assessed using RNAi. We typically use RNAi immediately after first obtaining parent transgene insertions and subsequently confirm rescue of null mutants.
Not all parent transgene insertions will be functional and this can be due to either issues with promoter / 3’ UTR choice, re-encoding for RNAi resistance, and, if present, the tag. As the goal of any project is to obtain a functional parent transgene insertion as soon as possible, when failure is encountered we recommend inserting a genomic region containing the coding region together with upstream and downstream control sequences (without any re-encoding or a tag) and assessing rescue of a mutant. Once a functional genomic region is defined, re-encoding and a tag (if desired) can be added sequentially to generate the desired functional parent transgene insertion.
Once a robust parent transgene insertion is generated, the transgene construct can be modified to introduce any desired mutations, deletions, etc. It is important that expression of mutants be confirmed: if the transgene includes a tag for visualization and the introduced mutation does not affect localization, quantitative analysis of localized fluorescence can be used to evaluate expression. If the transgenes lack a tag or if introduced mutations disrupt localization, immunoblotting should be performed (see I.C.4. below).
3. Generating gene modifications at the endogenous locus
For introducing mutations that do not lead to lethality in essential genes and for non-essential genes, we recommend engineering the endogenous locus of the gene using purified Cas9 and RNAs. The Cas9-mediated cut is repaired using an oligonucleotide that inserts the desired mutation (Arribere et al. 2014; Paix et al. 2014; Zhao et al. 2014; Katic et al. 2015) (Fig. 4A). The efficiency of this method is highly dependent on the distance between the Cas9 cut site and the mutation site; we typically choose a gRNA whose cutting site is within 10–15 bp of the mutation site.
The oligonucleotide used to insert the mutation must contain 30 – 60 base pairs of homology to each side of the cut and must be mutated to prevent continuous cleavage of the DNA once the mutation has been inserted (Arribere et al. 2014; Paix et al. 2014; Zhao et al. 2014; Katic et al. 2015). In addition, if the edit is engineered on the 3’ side of the PAM, then an anti-sense oligo must be used to insert the mutation (Paix et al. 2017a).
In order to screen for the desired mutation, there are three different approaches:
-
-
First, while designing the mutation in the repair template, insert a restriction enzyme site, which allows screening of transformation by amplifying the region by PCR and analyzing it by digestion. Alternatively, the mutation itself may add or remove a restriction site from the genomic sequence. For instance, double alanine mutations can add TseI sites (GC[A/T]GC) to the sequence.
-
-
Second, PCR-amplify with oligos that specifically recognize the mutation (Gaudet et al. 2009).
-
-
Third, PCR-amplify the potentially mutated region and screen by sequencing. Note that, even if the mutation appears to have been successfully introduced using the first two methods of evaluation, it is still important to confirm the presence of the mutation by sequencing.
Protocol
Mix purified Cas9, the crRNA and tracrRNA, just as described above.
Add the repair oligo to the mix at a final concentration of 5 µM (oligo must be ordered either HPLC or PAGE-purified).
Centrifuge mix at maximum speed for 15 min and inject into ~24 worm gonads. Recover worms at 20°C.
The next day transfer worms onto fresh plates.
After three days, single-out L4s and adults. If using the dpy-10 co-injection marker it may be necessary to wait longer in order to get them at the adult stage.
Once the singled-out worms have laid progeny, lyse them and amplify their DNA by PCR, using primers that flank the mutation. Screen for the mutation using one of the methods described above.
Once identified, cross the mutant worms back to the parental strain in order to remove the dpy-10 mutation and other potential off-targets, while keeping track of the mutation through the crosses.
4. Western blotting to validate expression
Once transgene insertions with mutations are generated or mutations introduced into the endogenous locus, it is important that protein expression be assessed using immunoblots (Fig. 3B&C) or by quantification of fluorescence (see section IIB).
To immunoblot the products of RNAi-resistant transgenes, we typically inject L4 larvae with a dsRNA targeting the endogenous mRNA and 36–48 hr later collect adult worms for analysis. In the case of transgenes expressed in the background of null alleles or in situ edits, adults can be collected at any point, although we suggest isolating L4 stage worms 24–48 hr before the generation of lysates in order to avoid collecting old worms. It is crucial for these experiments to generate a good antibody for the protein of interest (Zanin et al. 2011).
Protocol
Add 60 µL of M9 to conical screw cap 1.5 mL tubes and mark the top of the meniscus. Top up with 500 µL of M9.
Pick 60 gravid adult worms and transfer them to tubes with M9
Add 0.5 mL of M9 plus 0.1% Tween-20 and spin down for 2 min at 200 g.
Carefully remove supernatant and wash worms 3X with M9 plus 0.1% Tween-20
Remove buffer until the 60 µL mark
Remove 15 µL of buffer and add 15 µL of 6X sample buffer to a final concentration of 1.5X sample buffer
Sonicate samples in a water bath for 10 minutes at 70°C
Incubate tubes at 100°C for 5 minutes
Repeat step vii
Load 10 µL of worm lysate (equivalent to ~10 worms) on a gel lane, transfer and immunoblot with a validated primary antibody. As a loading control, we routinely use α-tubulin or Actin.
II. Monitoring of Cell Division Processes:
A number of assays have been developed to monitor phenotypic consequences of specific perturbations in the one-cell C. elegans embryo. In this section, we outline techniques that we employ to mount and image embryos, and to analyze events associated with both mitotic and meiotic divisions.
To execute cell division, many subcellular structures must be assembled and disassembled in a cell cycle-coupled manner. These structures include the nucleus, centrioles, centrosomes, the bipolar spindle, kinetochores and the contractile ring. Imaging fluorescently tagged components enriched at these different subcellular structures (generated as described in sections I.A. and I.C.) defines dynamics in wildtype embryos and establishes a baseline for assessing the effects of specific perturbations on the observed dynamics. A first step employing tagged fusions is measurement of fluorescence intensity/imaged structure dimensions over time. In addition to intensity/size changes, methods such as fluorescence recovery after photobleaching and photoactivation provide detailed insights into the dynamics of components at specific cellular locations (e.g. Gallo et al. 2010; Goehring et al. 2011; Olson et al. 2012; Green et al. 2013; Kohler et al. 2017).
Monitoring of chromosome dynamics and spindle morphology/length, using markers such as mCherry::H2b (which labels chromatin), GFP::α-tubulin (which labels microtubules), and GFP::γ-tubulin (which labels centrosomes/spindle poles) can be used to analyze the molecular mechanisms of mitotic and meiotic chromosome segregation. For example, tracking spindle pole separation has been used to analyze the mechanisms underlying load-bearing microtubule attachment formation at kinetochores (Oegema et al. 2001; Desai et al. 2003), while measuring chromosome distribution on the spindle helped address how microtubule attachments are stabilized prior to anaphase onset (Cheerambathur et al. 2017). Generating monopolar spindles with unattached kinetochores, followed by monitoring of the time between NEBD and chromosome decondensation is useful to study spindle assembly checkpoint (SAC) signalling (Essex et al. 2009; Kim et al. 2017) and direct imaging of meiotic chromosome dynamics has helped identify distinct mechanisms driving chromosome alignment and segregation (Monen et al. 2005; Dumont et al. 2010; Hattersley et al. 2016).
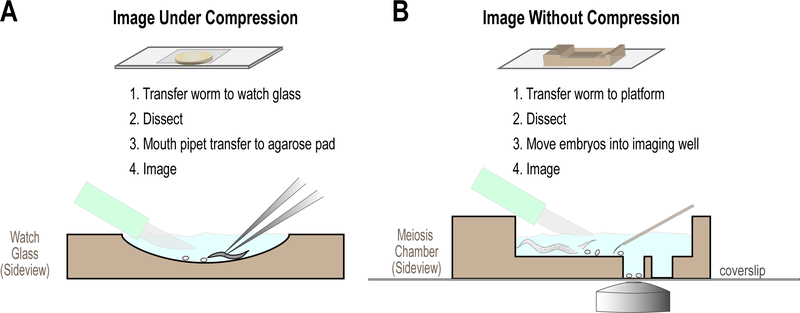
A. Mounting One-Cell Embryos for Imaging
1. Dissection and mounting of embryos on agarose pads
To ensure high quality spatial and temporal resolution of the events that occur in the early embryo, dissection of embryos from gravid adults and mounting on agarose pads provides optimal imaging conditions for many assays (Green et al. 2008). (Fig. 5A). Below we outline the standard method used to mount embryos for live imaging.
Figure 5. Mounting C. elegans embryos for imaging.
Schematics illustrating the standard methods used to mount embryos for live imaging, using either agarose pads (A) or meiotic chambers (B). In both cases, gravid adult worms are dissected and individual embryos visually selected. See text for details.
1.1. Making agarose pads
To create spacers for making an agarose pad, tape two glass slides together. Put two of these spacers on a desk parallel to each other and put a single slide between them.
Use a transfer pipet to place a small drop of agarose (melted by placing an eppendorf tube containing 2% agarose in a heating block at 95˚C) on the center of the slide.
After waiting for a few seconds, place a second slide crosswise over the agarose drop, so that it rests on the spacers and compresses the drop to generate a circular pad of agarose on the center of the slide. iv. After the agarose has cooled, pick up the agarose pad-slide sandwich and twist the two slides until one slide has a thin pad of agarose.
Note: The agarose breaks down over time at high temperatures and a new tube should be placed in the heating block every hour.
1.2. Mounting embryos onto agarose pads
Transfer one or two adult worms to a specimen watch glass (Electron Microscopy Sciences, #71570–01) containing a small amount (4–5 ml) of M9.
Hold the worms at one end with fine forceps (Dumont Swissmade, #0209–5PO) and use a scalpel to cut each worm in half.
Hold each half of the worm with the forceps and scrape the embryos out of the worm with the blunt side of scalpel, like toothpaste from a tube. Using a scalpel with a small blade (Bard-Parker, #371615) can be very helpful for this.
Pick up the embryos with a capillary tube, controlled with a mouth pipette and transfer them to the agarose pad, trying to minimize the amount of liquid transferred. 25 µL capillary tubes can be used for making mouth pipets. Heat the glass over a flame and once molten, rapidly stretch the glass to create a smaller diameter aperture. The ideal diameter for the capillary tube is slightly larger than an embryo.
Transferring the embryos to the pad; use the mouth pipette to remove excess liquid if necessary.
Use an eyelash tool, made by affixing an eyelash to the end of a toothpick with glue or nail polish, to herd the embryos together.
Use the scalpel to cut an arrow or a line in the agarose pad, to facilitate finding them under the microscope. viii. Place a 22 mm x 22 mm coverslip over the embryos and transfer the slide to the microscope for imaging.
An alternative method that can be simpler for beginners is to place a worm in a small drop (2–3 µl of M9) on a 22 mm x 22 mm coverslip and use a pair of fine needles (22 Gauge) to dissect the embryos from the worm on the coverslip. The coverslip can then be placed onto the agarose pad and the embryos imaged. However, mouth pipetting and careful positioning of similarly developmentally staged embryos for simultaneous imaging is advantageous for maximizing image acquisition of multiple samples.
2. Dissection of fertilized oocytes and embryos in osmotically balanced media.
The dissection, mouth pipetting and mounting under compression of C. elegans embryos in M9 media is possible because the eggshell is resistant to osmotic and mechanical stresses. However, to analyze events occurring during with the maternal meiotic divisions that occur after fertilization but prior to completion of eggshell formation (Olson et al. 2012), oocytes must be dissected in osmotically balanced media and mouth pipetting avoided. For the same reasons, analysis of embryos in which RNAi treatment renders the egg shell permeable to small molecule inhibitors (Carvalho et al. 2011), also requires an alternative approach for image analysis. Imaging of the meiotic divisions can be carried out in anaesthetized worms (McCarter et al. 1999; Winter et al. 2017) but is challenging due to the low frequency of events of interest and the negative effects of anesthesia. The depth of the event in the body of the worm and distance from the objective also compromise imaging resolution and signal intensity. For these reasons, we have preferred dissection of the fertilized oocyte to obtain high quality images for analysis (Fig. 5B).
Protocol
Prepare chamber from muwells (Carvalho et al. 2011; www.muwells.com) by rinsing thoroughly with water, wicking away all residual liquid before addition of 0.8x Egg Salts Buffer. The buffer should be filled to the top of the chamber and direct pipetting of the buffer over the wells (Fig. 5B) can be employed to remove air bubbles that would prevent embryos settling into wells.
Mount chamber on a slide (e.g. metal slide with wide aperture for access for objective) and tape into position.
Using a stereomicroscope, select worm to dissect and pick from bacterial lawn and place in media. Avoid transferring an excessive amount of bacteria.
Use forceps and scalpel to dissect the worm. As the object is to capture the oocytes/embryos without the protection of a fully formed egg shell, take care to hold and cut worm away from the vulva where the embryos develop prior to hatching. Practice is required to avoid high frequency of physical damage or death of the recently fertilized oocyte.
Following dissection of the worm and extrusion of the earliest oocytes and embryos from the carcass, visually select the oocytes/embryos at the appropriate developmental stage (Fig. 5B) and use an eyelash tool to move these selected oocytes/embryos to a well. Embryos moved above the well will naturally drift down to settle on the coverslip below the chamber within ~10s. If you desire, multiple embryos can be added to the same well. As the oocytes/embryos drift down randomly it can be hard to position them close together (those too close may land on top of each other, precluding imaging), but as the oocyte/embryo falls, moving the eyelash tool above it can generate a current to guide it in a particular direction.
Imaging can now be carried out on the chamber as described above. Note that as there is no compression of the oocyte/embryo as with mounting on an agarose pad with a coverslip, the distance from the objective to the object of interest may be increased. In the case of the oocyte meiotic spindle, this can be exacerbated by its asymmetrical localization to the cortex. See below for a method to correct for variable intensity due to altered imaging depth (section IIB).
B. Image Acquisition & Analysis Overview with Selected Examples
Many imaging modalities have been employed to analyze one-cell C. elegans embryos, with the most common being spinning disk confocal imaging with acquisition of 5–10 plane z-stacks at specific time intervals. It is important when designing imaging protocols to optimize the conditions (illumination intensity, exposure time, spacing of z-planes, interval between acquisitions, etc.) to best match the question of interest and to minimize photodamage-induced toxicity. As there is much variation in imaging systems, we will not specifically discuss details of image acquisition here but will focus on analysis of acquired images.
Following image acquisition, quantitative analysis is important to define the effects of precisely engineered perturbations generated as described above. We first describe temporal landmarks that are used to align multiple timelapse sequences of one-cell embryos for quantitative analysis and then discuss intensity-based measurements used to assess localized accumulation and loss of components at structures important for cell division. We also briefly discuss automated image analysis approaches, focusing on a specific application to cytokinesis.
1. Temporal landmarks for alignment of timelapse imaging data
One of the major advantages of the C. elegans one-cell embryo as an experimental model is its inherent reproducibility. For both intra and inter-experimental comparisons, many temporal landmarks that are dependent on progression through cell cycle can be used. For example, a common definition of mitotic duration used in many experimental systems is the time from nuclear envelope breakdown (NEBD) to anaphase onset. We define NEBD in C. elegans as the time at which nucleoplasmic histone signal equalizes with the cytoplasmic background and anaphase onset as the first frame when separation of the sister chromatids is visible (Fig. 1). Here we will define many of the temporal landmarks we use to describe the events that occur in the nascent embryo.
Following oocyte NEBD, ovulation and fertilization, the oocyte meiotic spindle forms and aligns the bivalent chromosomes (Fig. 1, first panel). As with mitosis and meiosis II, anaphase I onset is defined when visible separation of the chromosomes occurs, although in the case of meiosis I it is homologous chromosomes and not sister chromatids that are separating.
Following completion of the meiotic programme and the jettisoning of polar bodies, chromosomes decondense to form the maternal and paternal pronuclei (Fig. 1, second panel), which then undergo DNA replication. During this time, visible membrane contractility is observable as first ruffling of the membrane, followed by coalescent into a pseudo-cleavage furrow. Complete regression of this furrow and the meeting of the two pronuclei are both reliable temporal landmarks (Fig. 1, third panel). Following migration of the pronuclei toward the center of the embryo, nuclear envelope breakdown is initiated and defined as above (Fig. 1, fourth panel). The chromosomes then align (Fig. 1, fifth panel) and the continuing cell cycle progression triggers anaphase onset and separation and decondensation of the chromosomes (Fig. 1, sixth panel). Chromosome decondensation (defined as the point at which chromosomes begin to occupy a larger area and decrease in intensity), nuclear reasseambly and initiation of ingression of the cytokinetic furrow are also useful temporal markers (Fig. 1, seventh panel).
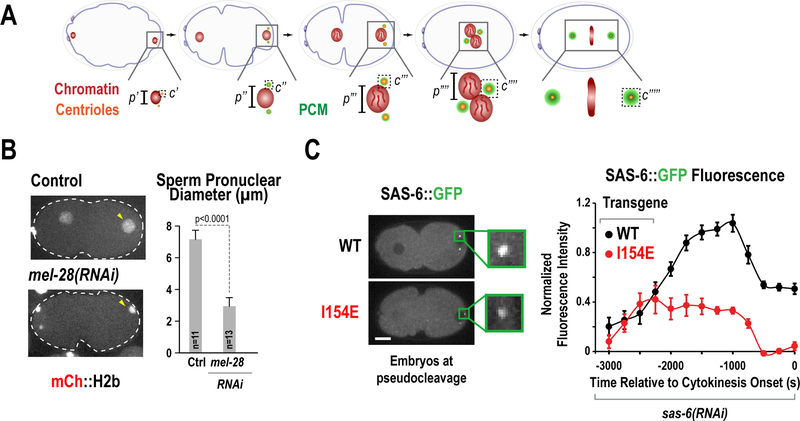
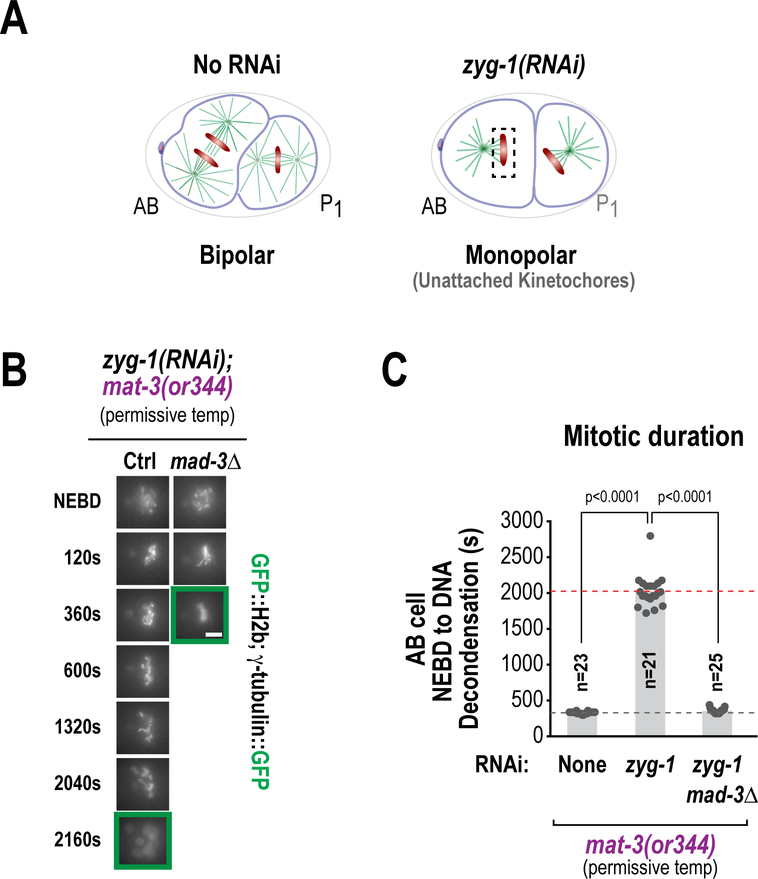
Use of these landmarks underpins many key assays that describe events in the one-cell embryo. For example, the processes of both pronuclei formation and expansion (by monitoring pronuclear size) and centriole assembly / centrosome maturation (by measuring centriole / centrosome size and/or intensity of fluorescent markers) can be described in relation to either anaphase of meiosis II or pseudo-cleavage furrow regression (Fig. 6). Chromosome dynamics during cell division (meiosis and mitosis) can be assessed by employing kymography or a minimal bounding box (Fig. 7 A&B) or measuring rate of separation of chromosomes or centrosomes in anaphase (Fig. 8 A&B) In addition, during the second embryonic division (Fig. 1, eighth and ninth panels), the time from NEBD until chromosome decondensation can be used to assess the function of the spindle assembly checkpoint when monopolar spindles are generated (for instance, through depletion of the centriole duplication factor ZYG-1) (Fig. 9).
Figure 6. Quantification of nuclear reformation and centriole assembly.
(A) Schematics illustrating two early events in the C. elegans embryo: nuclear reformation after exit from meiosis II and centriole assembly. Parameters such as p=pronuclear diameter and c=centriole intensity can be measured over time. (B) Measuring male pronuclear diameter in either control embryos or embryos depleted of the nucleoporin mel-28 (Adapted from Hattersley et al. 2016). (C) Measuring SAS6 accumulation at centrioles upon centriole assembly. Embryos depleted of endogenous SAS-6 by RNAi were expressing a reencoded version of SAS-6::GFP, either wild-type or a mutant deficient in centriole assembly (I154E) and centriole intensities were quantified over time (adapted from Lettman et al. 2013)
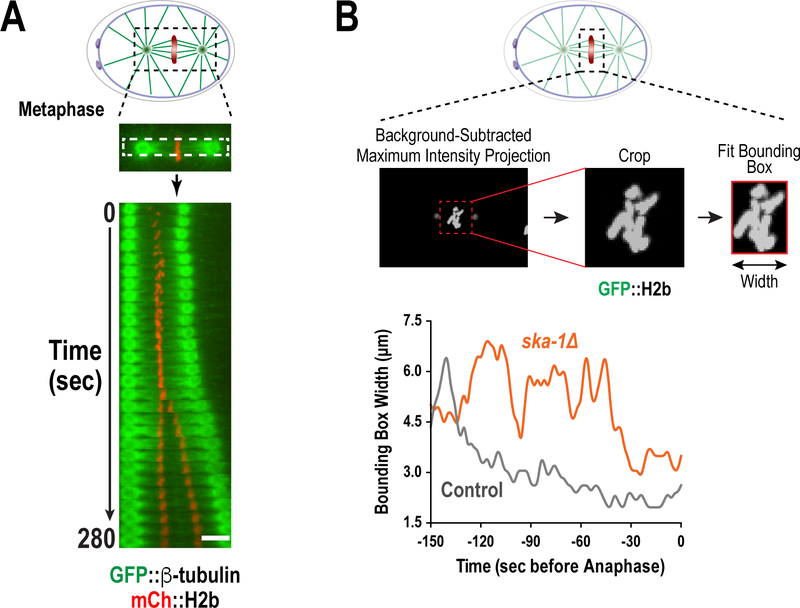
Figure 7. Methods to evaluate kinetochore-microtubule interactions and chromosome alignment during mitosis.
(A) Kymographs illustrating chromosome and spindle dynamics were produced by taking visual sections through the spindle, oriented from spindle pole to spindle pole, during a temporal progression. (B) The minimal bounding box (MMB) method to quantify chromosome dynamics during mitosis. A bounding box was fit to the minimal area of chromosomes, indicated by a maximum-intensity projection of GFP::H2b signal, and width of this box was plotted over time. (All panels adapted from Cheerambathur et al. 2017)
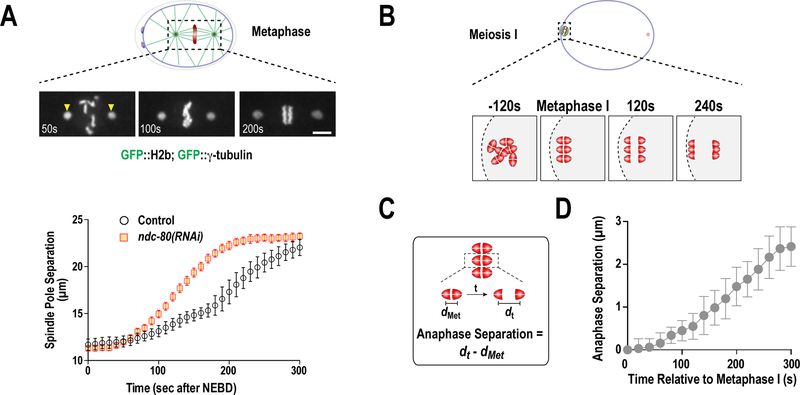
Figure 8. Analysis of mitotic spindle length and meiotic chromosome segregation.
(A) Spindle pole-tracking assay. Distance from the center of spindle-pole to spindle-pole was plotted over time during mitosis to describe changes in spindle length that reflect perturbations in microtubule interactions with chromosomes and/or the cortex (Cheerambathur et al. 2017). (B) Schematic illustrating the process of bivalent chromosome segregation during meiosis I. (C and D) Plot of mean homologous chromosome segregation over time. The distance between the separating homologous chromosomes was measured from the center of each half of the bivalent, positioned in the center of the spindle. The separation of the homologous chromosomes at metaphase (dmet) was subtracted from all values to describe chromosome segregation over time (dt).
Figure 9. Analysis of spindle assembly checkpoint (SAC) signaling.
(A) Schematics of two-cell C. elegans embryos either untreated or depleted of ZYG-1, a component required for centriole duplication. ZYG-1 depletion causes the generation of a monopolar spindle where the kinetochores facing away from the centrosome are devoid of attachment. (B) Representative image sequences of two cell embryos (AB cell) depleted of ZYG-1 in the presence of a temperature-sensitive APC/C allele (mat-3(or344)) at the permissive temperature. This treatment causes cells to arrest in a manner that is dependent on the essential SAC component MAD-3. The addition of the (mat-3(or344)) allele compromises APC/C activity and extends the duration of the mitotic arrest, allowing the detection of subtle perturbations (Bezler/Gonczy 2010, Kim 2017. (C) Quantification of mitotic duration from the experiment in (B). These perturbations are used as a sensitized assay for the SAC (Adapted from Kim et al. 2017).
2. Intensity-based measurements
The temporal landmarks described above can be combined with quantification of fluorescence intensity to assess dynamic changes in the levels of components in structures such as centrosomes, kinetochores, spindles or the cell cortex. Image quantification can be done in multiple ways, which have relative advantages or disadvantages dependent on the structure and processes of interest. A simple box method can be used to assess discrete structures in a uniform background volume (Hoffman et al. 2001) (Fig. 10A&B). By contrast, it is more challenging to assess intensity of fluorescence in structures such as the cortex that can have different background signals on opposite sides of the plasma membrane. We typically use spinning disk confocal microscopy for image acquisition of discrete structures and perform analysis on the raw images. Here we describe commonly applied techniques using software such as ImageJ/Fiji, to measure fluorescence intensities.
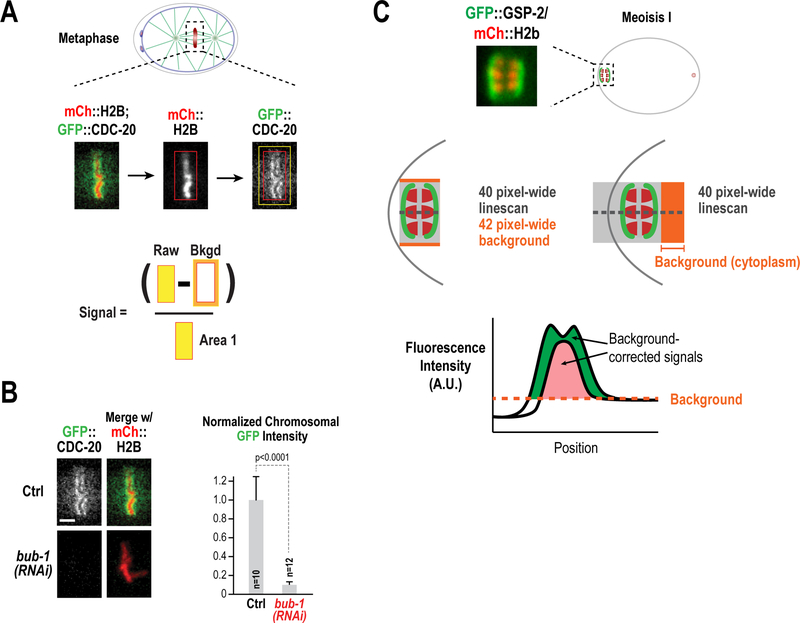
Figure 10. Quantification of signal intensities.
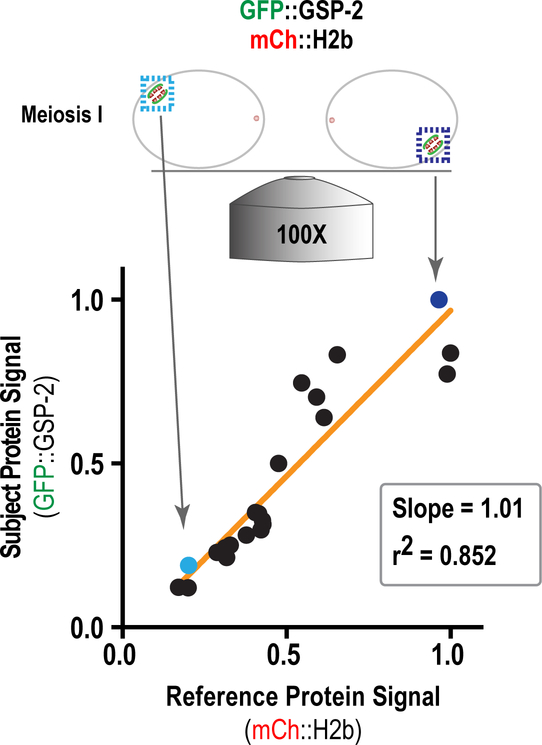
(A) Description of the box method for the quantification of CDC-20 kinetochore localization. A box was drawn around the chromosome area (mCh::H2b) and transferred to the GFP::CDC-20 channel. After recording integrated intensity, the box was expanded in order to calculate background signal. (B) The box method was applied for the quantification of CDC-20 chromosomal intensity in control embryos or embryos depleted of BUB-1 by RNAi (Adapted from Kim et al. 2017). (C) Description of the line-scan method for the quantification of GSP-2 recruitment in anaphase II. In both cases, a 40-pixel linescan was performed perpendicular to the orientation of the meiotic spindle (shown plotted below). In the left example, the line only covers the length of the spindle and background is calculated by expanding the width of the linescan to 42 pixels. This accounts for dynamic change in the background over, for example, a cell membrane or organelle. In the right example, the linescan extends into the embryo and cytoplasmic background was calculated as the mean of this signal.
2.1. Box Method
Depending on the nature of the imaged signal, maximum intensity Z-stack projections, sum intensity Z-stack projections or single Z-plane images should be analyzed at each timepoint of a timelapse sequence. We typically analyze maximum intensity projections or single Z-planes in our experiments. A rectangular box of defined size should be drawn around the object of interest and the integrated intensity (I1) and area of the box (A1) recorded.
To measure background intensity, the box is expanded by 5 pixels on each side, and the integrated intensity (I2) and area (A2) measured.
- The signal and area difference between the expanded box and the original box are used to calculate the average background signal per pixel, using the following formula:
- Total intensity (𝑇) = 𝐼1
- Background signal (B)=
- Corrected intensity = (𝑇−𝐵)𝐴1
This process can be repeated over a time series to measure the relative abundance of a tagged protein over time (for examples of papers from our lab, see Espeut et al. 2012; Moyle et al. 2014; Kim et al. 2015; GersonGurwitz et al. 2016; Kim et al. 2017).
2.2. Line Scan Method
For objects that occur on non-uniform or asymmetric regions of background intensity (for example, in proximity to the plasma membrane where extra-cellular background is lower than that of cytoplasmic background), the box method of quantification is not appropriate (Fig. 10C). Here, use of a line scan is recommended.
Draw a line of defined pixel-width perpendicularly to the direction in which the background is non-uniform. The pixel width of the line should encompass the entirety of the object if possible. This line can extend longitudinally past the object of interest, or solely encompass the object depending on how background intensity is to be calculated.
Plot the pixel intensity along the axis of the line (Value I1). Depending on the structure that is being analyzed, raw signal can be defined as either the integrated intensity at the site of the line scan or as the peak intensity.
Background intensity can be subtracted to reveal the actual signal in two ways (Fig. 10C). First, in a manner comparable to the box method, the pixel width of the line can be expanded to give I2. The formula in the previous section (II.B.2.1) can then be used to calculate the background-subtracted intensity value. Alternatively, given the profile of signal to either side of the object of interest (a co-marker may be used to define this), background intensity on either side of the object of interest may be obtained. Background can then either be defined as the signal present at a specific side of the object of interest (Moyle et al. 2014) or alternatively, as the mean of the intensities at both sides (Hattersley et al. 2016).
2.3. Depth Correction
The relative distance of a fluorescent object from the objective lens can lead to significant variation in detected fluorescence intensity. Thus, to compare measured intensity values, it is important to correct for imaging depth. For this purpose, it is necessary to measure fluorescence intensity of a standard object at different distances from the objective. Consistent and accurate depth correction can be obtained by directly correlating intensity of the signal of interest to that of an invariant signal (Fig. 11). For analysis of chromosome-associated proteins, chromatin visualized using a tagged core histone can be used as the invariant signal. Following measurement of signal of interest and of the invariant probe, the ratio of the two signals can be used to correct for depth-associated changes in intensity (Fig. 11).
Figure 11. Signal depth correction.
Schematic illustrating method used for depth correction of fluorescence signal intensity. GFP::GSP-2 signal correlates with mCh::H2b signal, thus intensity changes in GFP signal due to variation in distance between the objective and object can be corrected ratio to mCh signal.
Dynamic changes in fluorescence intensity distributions can also be analyzed to understand specific cell division processes, e.g. chromosome condensation (Maddox et al. 2006), chromosome alignment (Cheerambathur et al. 2017) (Fig. 7), cortical dynamics (Maddox et al. 2007; Carvalho et al. 2009; Lewellyn et al. 2011), etc. We describe below a specific automated approach that we have employed to analyze cortex dynamics during cytokinesis.
3. Automated Tracking
To facilitate analysis of dynamic imaging data in C. elegans embryos, automated tracking approaches have been used (e.g. Jaensch et al. 2010; Naganathan et al. 2014). The choice of which specific tracking algorithm is best suited for analysis will be dictated by the experimental objective. Python library open CV is one repository of such algorithms.
Here, we present specific information on an automated approach that we have developed to analyze contractile ring closure during cytokinesis in one-cell C. elegans embryos (Fig. 12). This serves to illustrate how a dynamic process in the one-cell embryo can be characterized with detailed quantitative rigor.
Figure 12. Analysis of cytokinesis.
(A) Manual measurement of cytokinetic furrow ingression over time. Membrane ingression is indicated with strains expressing fluorophore-tagged NMY-2 or a plasma membrane marker (PH). Following acquisition of a Z-series during furrow ingression, the Z-slice with maximum ingression was used to measure the amount of cytokinesis ring enclosure. This distance was defined as the distance between the innermost points of the furrow. This can be done manually or using automated software. (B) Parameters for automated analysis of cytokinetic furrow ingression and fluorescence quantification. Following software identification of the cleavage furrow from an image series, the parameters of ring enclosure (orange) were defined. To define the area used for quantification of fluorescence intensity, additional user input is required to define distance from edge of the ring to edge of embryo (drL) and distance from edge of the ring to the center of the embryo (drS). See text for additional details.
3.1. Measuring contractile ring closure
Using a confocal spinning disk microscope, image embryos expressing a marker (e.g. functionally tagged non-muscle myosin II NMY-2) using a 7 × 1.5-µm z-series, acquired every 20s, from around the time of nuclear envelope breakdown until the embryo reaches the four-cell stage. In cases where the first division fails, imaging is terminated when the furrow almost fully regresses (the kinetics of the furrow regression can also be analyzed). As the furrow ingresses asymmetrically and the position of the contractile ring tends to shift along the z-axis as it closes, the z-series should be adjusted such that it is centered on the widest ring diameter.
- Automatic detection of the size and position of the contractile ring can be performed using custom software, written in Python and available from https://github.com/renatkh/cytokinesis. This software package consists of two programs: ‘FindRing.py’ and ‘divPlaneClass.py’. The first, ‘FindRing.py’, detects the contractile ring. The second, ‘divPlaneClass.py’, quantifies intensity values of fluorescently tagged proteins in the cleavage furrow. Below are instructions on use of these automated analysis programs.
- ‘FindRing.py’ requires: i) specification of the first time point when contractile ring intensity becomes is enriched relative to the adjacent cortex, ii) details of imaging conditions, such as the number and pixelspacing of Z-slices per time point. If a chromosome marker is present in the same channel, the appropriate trigger (‘chromosomeMarker’) in the software should be set to ‘True’. If embryos were imaged on agarose pads, the parameter ‘embryoCenterDrift’ should be set to ‘linear’; if the embryos were imaged in meiotic chambers, this parameter value should be set to ‘median’; if the focal position was adjusted during the imaging session or if you are not sure, select the ‘independent’ setting. The software displays a vertically oriented embryo for the user to confirm that the anterior side is at the top of the image. If this is not the case, then the parameter ‘flip’ should be set to ‘True’ and the analysis restarted. Ultimately, the software presents a window with the embryo images that may be scrolled through, which have the position of the detected contractile ring indicated with white circles and the embryo boundary outlined.
- ‘divPlaneClass.py’ quantifies the intensity at the contractile ring and requires specification of imaging conditions. The parameter ‘da’ specifies the step size in degrees for averaging quantification of the ring intensity at angular positions along the ring, relative to the axis of the ring initial movement. The user will need to specify the distance away from the ring edge toward the edge of the embryo (‘drL’) and toward the center (‘drS’); this is used for intensity averaging (Fig. 12B). The output is a list of angular positions and intensity values for every spatial position and total intensity for the contractile ring at each time point. The software also displays reconstructed division planes at every time point with a solid circle indicating the position of the contractile ring and dashed circles indicating inner and outer boundaries used for averaging. If the contractile ring size or position are not accurate, they may be adjusted manually in the *Ring.csv file.
This example highlights one specific case of automated image analysis applied to address a dynamic cell division process. With increasing emphasis on computational approaches in cell biology, such efforts are likely to become widespread in the future.
CONCLUSION/PERSPECTIVE
In the last two decades, the Caenorhabditis elegans one-cell embryo has been at the forefront of mechanistic insight into the events that occur during cell division. This system combines the strengths of both genetic and cell biology approaches as well as widespread gene conservation to continue to inform how cells undergo the process of accurately partitioning DNA content in a developing organism.
Here, we have described both gene engineering and experimental approaches that can be used to drive experimental work. Techniques such as penetrant RNAi, the advent of single-copy insertion transgenes and CRISPR-Cas9, as well as traditional methods such as temperature sensitive and null alleles allows the generation of precise genetic manipulations to interrogate the functions of essential genes of interest. The stereotypical nature of the one-cell embryo combined with high spatio-temporal resolution imaging allows quantitative analysis of the events required to produce the nascent embryo. Here we describe the methods used to engineer C. elegans as well as commonly used techniques used for analysis and definition of phenotypes. The use of these approaches, combined with proteomics, biochemical reconstitutions and structural analysis, will continue to provide important insights in to the mechanistic basis of cell division processes.
ACKNOWLEDGEMENTS
P.L-G. acknowledges support provided by a Pew Latin American fellowship. Work in the Desai and Oegema labs is supported by grants from the National Institutes of Health (GM074215 to A.D. and GM074207 to K.O.). A.D. & K.O. receive salary and other support from the Ludwig Institute for Cancer Research.
APPENDIX. Media and supplements
-
NGM agarose plates (1 liter)
3 g NaCl
25 g agarose
2.5 g peptone
1.0 mL cholesterol (5 mg/mL in EtOH)
975 mL ddH2O
*Autoclave 35 min and place in 55°C water bath
*When cooled sterilely add:
1.0 mL 1 M CaCl2
1.0 mL 1 M MgSO4
25 mL 1M KH2PO4 (pH 6.0)
-
M9 buffer (2 liter)
10 g NaCl
12 g Na2HPO4
6 g KH2PO4
0.5 g MgSO4 x 7H2O
Add water to 2 liter
*Autoclave 35 min
-
PCR lysis buffer (0.5 ml)
425 µL H2O
50 µL 10X DreamTaq buffer (ThermoFisher) or any other 10X PCR buffer
25 µL 20 mg/mL proteinase K
* Make fresh every time
-
3X soaking buffer (500 ml)
2.3 g Na2HPO4
1.1 g KH2PO4
630 µL of 5M NaCl
0.38 g NH4Cl
-
6X SDS Sample Buffer
7mL of 0.5M Tris-HCl pH6.8
1 g SDS
0.93 g DTT
* Mix well until completely dissolved
3 mL 100% Glycerol
* Mix well
Add a tiny amount of Bromophenol Blue to give the desired color.
-
1x Egg Salts Media (100 ml)
118mM NaCl
40mM KCl
3.4mM MgCl2
3.4mM CaCl2
5mM HEPES, pH7.4
H2O
(Made up to 0.8x in H2O for use)
Alternatively, Human Cell tissue culture media can also be used for this purpose (Liebovitz’s (1x) Media – Gibco). In addition, the addition of 20% Calf Serum can be used to stabilize small molecule inhibitors in solution.
REFERENCES
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz LA, Knight AL, Caldwell GA, Caldwell KA. 2008. Generation of stable transgenic C. elegans using microinjection. J Vis Exp [DOI] [PMC free article] [PubMed]
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA et al. 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14: 53–56. [DOI] [PubMed] [Google Scholar]
- Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, Oegema K. 2008. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science 322: 1543–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A, Desai A, Oegema K. 2009. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell 137: 926–937. [DOI] [PubMed] [Google Scholar]
- Carvalho A, Olson SK, Gutierrez E, Zhang K, Noble LB, Zanin E, Desai A, Groisman A, Oegema K. 2011. Acute drug treatment in the early C. elegans embryo. PLoS One 6: e24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805. [DOI] [PubMed] [Google Scholar]
- Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A. 2013. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science 342: 1239–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Prevo B, Hattersley N, Lewellyn L, Corbett KD, Oegema K, Desai A. 2017. Dephosphorylation of the Ndc80 Tail Stabilizes Kinetochore-Microtubule Attachments via the Ska Complex. Dev Cell 41: 424–437 e424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR 3rd, Oegema K, Desai A. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18: 2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Cowan JL, Shields DC, Gibson TJ, Coldwell MJ, Edwards RJ. 2012a. SLiMPrints: conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions. Nucleic Acids Res 40: 10628–10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Haslam NJ, Shields DC, Edwards RJ. 2011. SLiMSearch 2.0: biological context for short linear motifs in proteins. Nucleic Acids Res 39: W56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. 2012b. Attributes of short linear motifs. Mol Biosyst 8: 268–281. [DOI] [PubMed] [Google Scholar]
- Davies T, Sundaramoorthy S, Jordan SN, Shirasu-Hiza M, Dumont J, Canman JC. 2017. Using fast-acting temperature-sensitive mutants to study cell division in Caenorhabditis elegans. Methods Cell Biol 137: 283–306. [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev 17: 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Davey NE, Hagting A, Izawa D, Mansfeld J, Gibson TJ, Pines J. 2015. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell 32: 358–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Goldstein B. 2016. CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering. Genetics 202: 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. 2015. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200: 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. 2008. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 13: 6580–6603. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol 12: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley ML, Baillie DL, Riddle DL, Rose AM. 2006. Genetic balancers. WormBook: 1–32. [DOI] [PMC free article] [PubMed]
- El Mouridi S, Lecroisey C, Tardy P, Mercier M, Leclercq-Blondel A, Zariohi N, Boulin T. 2017. Reliable CRISPR/Cas9 Genome Engineering in Caenorhabditis elegans Using a Single Efficient sgRNA and an Easily Recognizable Phenotype. G3 (Bethesda) 7: 1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol 196: 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A, Dammermann A, Lewellyn L, Oegema K, Desai A. 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol Biol Cell 20: 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, Meyer BJ. 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AG, Gunsalus KC, Huang J, Chuang LS, Ying N, Liang HL, Tang C, Schetter AJ, Zegar C, Rual JF et al. 2005. New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res 15: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovsky A, Moerman DG, Jorgensen EM. 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods 11: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9: e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, Seydoux G. 2010. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science 330: 1685–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet M, Fara AG, Beritognolo I, Sabatti M. 2009. Allele-specific PCR in SNP genotyping. Methods Mol Biol 578: 415–424. [DOI] [PubMed] [Google Scholar]
- Gerson-Gurwitz A, Wang S, Sathe S, Green R, Yeo GW, Oegema K, Desai A. 2016. A Small RNA-Catalytic Argonaute Pathway Tunes Germline Transcript Levels to Ensure Embryonic Divisions. Cell 165: 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG. 2009. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res 37: 6984–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Hoege C, Grill SW, Hyman AA. 2011. PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. J Cell Biol 193: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, Schnabel R. 1999. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol 144: 927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Audhya A, Pozniakovsky A, Dammermann A, Pemble H, Monen J, Portier N, Hyman A, Desai A, Oegema K. 2008. Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol 85: 179–218. [DOI] [PubMed] [Google Scholar]
- Green RA, Mayers JR, Wang S, Lewellyn L, Desai A, Audhya A, Oegema K. 2013. The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules. J Cell Biol 203: 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N, Cheerambathur D, Moyle M, Stefanutti M, Richardson A, Lee KY, Dumont J, Oegema K, Desai A. 2016. A Nucleoporin Docks Protein Phosphatase 1 to Direct Meiotic Chromosome Segregation and Nuclear Assembly. Dev Cell 38: 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppert JK, Dickinson DJ, Pani AM, Higgins CD, Steward A, Ahringer J, Kuhn JR, Goldstein B. 2016. Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system. Mol Biol Cell 27: 3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. 2001. Microtubuledependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12: 1995–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaensch S, Decker M, Hyman AA, Myers EW. 2010. Automated tracking and analysis of centrosomes in early Caenorhabditis elegans embryos. Bioinformatics 26: i13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. 2000. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol 149: 1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadandale P, Chatterjee I, Singson A. 2009. Germline transformation of Caenorhabditis elegans by injection. Methods Mol Biol 518: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Katic I, Xu L, Ciosk R. 2015. CRISPR/Cas9 Genome Editing in Caenorhabditis elegans: Evaluation of Templates for Homology-Mediated Repair and Knock-Ins by Homology-Independent DNA Repair. G3 (Bethesda) 5: 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ishidate T, Ghanta KS, Seth M, Conte D Jr., Shirayama M, Mello CC. 2014. A coCRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Lara-Gonzalez P, Prevo B, Meitinger F, Cheerambathur DK, Oegema K, Desai A. 2017. Kinetochores accelerate or delay APC/C activation by directing Cdc20 to opposing fates. Genes Dev 31: 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Moyle MW, Lara-Gonzalez P, De Groot C, Oegema K, Desai A. 2015. Kinetochore-localized BUB-1/BUB-3 complex promotes anaphase onset in C. elegans. J Cell Biol 209: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Wojcik M, Xu K, Dernburg AF. 2017. Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Proc Natl Acad Sci U S A 114: E4734–E4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou H, Desai A, Oegema K. 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev Cell 25: 284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn L, Carvalho A, Desai A, Maddox AS, Oegema K. 2011. The chromosomal passenger complex and centralspindlin independently contribute to contractile ring assembly. J Cell Biol 193: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox AS, Lewellyn L, Desai A, Oegema K. 2007. Anillin and the septins promote asymmetric ingression of the cytokinetic furrow. Dev Cell 12: 827–835. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Portier N, Desai A, Oegema K. 2006. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc Natl Acad Sci U S A 103: 15097–15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol 11: 171–176. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol 205: 111–128. [DOI] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. 2009. A bright and photostable photoconvertible fluorescent protein. Nat Methods 6: 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 2008. 3’ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18: 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Seydoux G. 2010. Transgenic solutions for the germline. WormBook: 1–21. [DOI] [PMC free article] [PubMed]
- Mok CA, Au V, Thompson OA, Edgley ML, Gevirtzman L, Yochem J, Lowry J, Memar N, Wallenfang MR, Rasoloson D et al. 2017. MIP-MAP: High-Throughput Mapping of Caenorhabditis elegans Temperature-Sensitive Mutants via Molecular Inversion Probes. Genetics 207: 447–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol 7: 1248–1255. [DOI] [PubMed] [Google Scholar]
- Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. 2014. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol 204: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganathan SR, Furthauer S, Nishikawa M, Julicher F, Grill SW. 2014. Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. Elife 3: e04165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, Nash B, Price MH, Turnbull DW, Garner AR, Hamill DR et al. 2011. A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One 6: e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 153: 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Hyman AA. 2006. Cell division. WormBook: 1–40. [DOI] [PMC free article] [PubMed]
- Olson SK, Greenan G, Desai A, Muller-Reichert T, Oegema K. 2012. Hierarchical assembly of the eggshell and permeability barrier in C. elegans. J Cell Biol 198: 731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak MJ, Rasoloson D, Paidemarry S, Green R, Reed RR, Seydoux G. 2017a. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Natl Acad Sci U S A 114: E10745–E10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G. 2015. High Efficiency, HomologyDirected Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Seydoux G. 2017b. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122: 86–93. [DOI] [PMC free article] [PubMed]
- Paix A, Wang Y, Smith HE, Lee CY, Calidas D, Lu T, Smith J, Schmidt H, Krause MW, Seydoux G. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol 10: 1619–1622. [DOI] [PubMed] [Google Scholar]
- Redemann S, Schloissnig S, Ernst S, Pozniakowsky A, Ayloo S, Hyman AA, Bringmann H. 2011. Codon adaptation-based control of protein expression in C. elegans. Nat Methods 8: 250–252. [DOI] [PubMed] [Google Scholar]
- Rog O, Kohler S, Dernburg AF. 2017. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S et al. 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14: 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg PJ, Grant BD. 2013. C. elegans gene transformation by microparticle bombardment. WormBook: 1–10. [DOI] [PMC free article] [PubMed]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M et al. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanovskaya E, Viscardi V, Lesigang J, Lettman MM, Qiao R, Svergun DI, Round A, Oegema K, Dong G. 2014. Structure of the C. elegans ZYG-1 cryptic polo box suggests a conserved mechanism for centriolar docking of Plk4 kinases. Structure 22: 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Waaijers S, Boxem M. 2014. Engineering the Caenorhabditis elegans genome with CRISPR/Cas9. Methods 68: 381–388. [DOI] [PubMed] [Google Scholar]
- Waaijers S, Portegijs V, Kerver J, Lemmens BB, Tijsterman M, van den Heuvel S, Boxem M. 2013. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. 2014. Characterization and development of photoactivatable fluorescent proteins for single-moleculebased superresolution imaging. Proc Natl Acad Sci U S A 111: 8452–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wu D, Quintin S, Green RA, Cheerambathur DK, Ochoa SD, Desai A, Oegema K. 2015. NOCA-1 functions with gamma-tubulin and in parallel to Patronin to assemble non-centrosomal microtubule arrays in C. elegans. Elife 4: e08649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JD. 2015. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Miwa J. 1978. Characterization of temperature-sensitive, fertilization-defective mutants of the nematode caenorhabditis elegans. Genetics 88: 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter ES, Schwarz A, Fabig G, Feldman JL, Pires-daSilva A, Muller-Reichert T, Sadler PL, Shakes DC. 2017. Cytoskeletal variations in an asymmetric cell division support diversity in nematode sperm size and sex ratios. Development 144: 3253–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin E, Desai A, Poser I, Toyoda Y, Andree C, Moebius C, Bickle M, Conradt B, Piekny A, Oegema K. 2013. A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev Cell 26: 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin E, Dumont J, Gassmann R, Cheeseman I, Maddox P, Bahmanyar S, Carvalho A, Niessen S, Yates JR 3rd, Oegema K et al. 2011. Affinity purification of protein complexes in C. elegans. Methods Cell Biol 106: 289–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser E, Frokjaer-Jensen C, Jorgensen E, Ahringer J. 2011. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS One 6: e20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142: 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Zhang Z, Ke H, Yue Y, Xue D. 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]