Abstract
Tuberculosis (TB) is a major public health burden worldwide, and more effective treatment is sorely needed. Consequently, uncovering causes of resistance to Mycobacterium tuberculosis (Mtb) infection is of special importance for vaccine design. Resistance to Mtb infection can be defined by a persistently negative tuberculin skin test (PTST–) despite living in close and sustained exposure to an active TB case. While susceptibility to Mtb is, in part, genetically determined, relatively little work has been done to uncover genetic factors underlying resistance to Mtb infection. We examined a region on chromosome 2q previously implicated in our genomewide linkage scan by a targeted, high-density association scan for genetic variants enhancing PTST– in two independent Ugandan TB household cohorts (n = 747 and 471). We found association with SNPs in neighboring genes ZEB2 and GTDC1 (peak meta p = 1.9 × 10−5) supported by both samples. Bioinformatic analysis suggests these variants may affect PTST– by regulating the histone deacetylase (HDAC) pathway, supporting previous results from transcriptomic analyses. An apparent protective effect PTST– against body-mass wasting suggests a link between resistance to Mtb infection and healthy body composition. Our results provide insight into how humans may escape latent Mtb infection despite heavy exposure.
Keywords: tuberculosis, genetic association, early clearance of Mycobacterium tuberculosis, genetics of immunity
Introduction
Tuberculosis (TB) is one of the most devastating communicable diseases in world health, with approximately 10.4 million incident cases and 1.4 million deaths from TB in 20151. About nine in ten people initially infected with Mycobacterium tuberculosis (Mtb), however, do not go on to develop active disease. Hence, TB pathogenesis follows a two-step process2, starting with initial infection, (latent Mtb infection, LTBI) diagnosed by the tuberculin skin test (TST) or interferon-gamma release assays (IGRA), and progression to symptomatic disease in a subset of infected persons.
While a role for human genetic susceptibility to TB has been well-established3, 4, genetic susceptibility to Mtb infection has been less studied. Genomewide analyses of TB, whether for genetic linkage5–9 or, more recently, for association10–17, have not provided consistent evidence for major TB susceptibility loci. Difficulty of replication may stem in part from the clinical definitions used for TB18. Few studies have examined latent Mtb infection7, 17, 19–21; yet, while few, these studies reveal consistency in genomic loci identified.
Within the framework of household contact study22, 23, we have focused on the persistent TST negative (PTST–) phenotype, which measures relative resistance to Mtb infection over an extended period of time, despite heavy exposure within the household24. The hypothesis that this resistance may be influenced by innate and adaptive immune factors is an ongoing area of investigation25–27. Studying resistance may reveal genetic insights into mechanisms underlying TB pathogenesis16, 21. Resistance to latent Mtb infection has particular relevance to the design of a preinfection vaccine26, since reducing the pool of latently infected individuals will reduce the incidence of TB. Analysis of PTST– individuals could identify critical biologic mechanisms underlying resistance to Mtb infection.
We previously found evidence for genetic linkage of regions on chromosome 2q and 5p with the PTST– phenotype7. We then found genetic association with the PTST– phenotype at an existing candidate locus, SLC6A3, within the chromosome 5 region28 that had been identified by another group examining TST reactivity17, 19, 20. Now, we present a fine-mapping association scan for PTST– across the other major linkage region, on chromosome 2q, with two independent samples from Ugandan households ascertained through an index case with TB22, 23, 29. Heritability analysis suggested that approximately half of the genetic variation in PTST– is due to loci on chromosome 2q. A meta-analysis combining the two samples identified two loci of interest, one linked to histone de-acetylase regulation, and another linked to body mass composition, uncovering new biologic pathways underlying resistance to Mtb infection.
Results
Sample Description
Our study sample comprises two independently-recruited cohorts of Ugandan households ascertained through a proband with active TB (summarized in Table 1)22, 29. Sample 1 (n = 165 active TB, 501 LTBI, 81 PTST–) was genotyped for a fine-mapping panel with markers spaced approximately every 10 kb across the chromosome 2 linkage peak observed in an overlapping sample (overlap n = 103 TB, 277 LTBI and 45 PTST–)7, and subsequently imputed to the Illumina HumanOmni5 panel. In addition, genotyping for haplotype tagging SNPs from several candidate loci that may affect resistance to Mtb infection28 was performed. Sample 2 (n = 201 TB, 237 LTBI, 33 PTST–), genotyped for the Illumina HumanOmni5 BeadChip (average marker distance ~ 600 bp), includes a greater proportion of participants with active TB and a smaller proportion of PTST–. In both samples, PTST– are, on average, much younger than non-PTST–, but did not differ significantly in sex ratio or in proportion of HIV+ individuals.
Table 1.
Characteristics of the two Ugandan PTST– samples.
| PTST– | Non-PTST– | Total | p | |
|---|---|---|---|---|
| Sample 1 | ||||
| n | 81 | 666 | 747 | — |
| Active TB | 0 (0.0%) | 165 (24.8%) | 165 (22.1%) | — |
| Female | 38 (46.9%) | 276 (41.4%) | 314 (42.0%) | 0.41 |
| Age, y | 9.3 ± 8.8 | 17.8 ± 13.5 | 16.9 ± 13.3 | < 0.001 |
| HIV+ | 4 (5.4%) | 80 (13.0%) | 84 (12.2%) | 0.061 |
| Sample 2 | ||||
| n | 33 | 438 | 471 | — |
| Active TB | 0 (0.0%) | 201 (45.9%) | 201 (42.7%) | — |
| Female | 16 (48.5%) | 215 (49.1%) | 231 (49.0%) | 0.99 |
| Age, y | 11.5 ± 12.5 | 21.6 ± 13.4 | 20.9 ± 13.5 | < 0.001 |
| HIV+ | 3 (9.4%) | 56 (12.9%) | 59 (12.7%) | 0.78 |
Values are presented either as n (% of total sample) or as mean ± SD. Non-PTST–, LTBI plus active TB; p, p value for test of differences between PTST– and non-PTST– individuals, by 2 × 2 χ2 test for sex, Wilcoxon rank-sum test for age, and Fisher’s exact test for HIV status.
Genetic Association Analysis
We focused on SNPs that showed association in both samples, thus demonstrating internal replication. We tested genetic association in both samples by means of logistic regression, with adjustment for relatedness, and combined results for markers tested in both cohorts, after correction for population structure by genomic control (Supplementary Figure 1; see Supplementary Table 1 for complete results; the most significantly associated SNPs within each sample are listed in Supplementary Tables 2 and 3).
ZEB2/GTDC1 association peaks
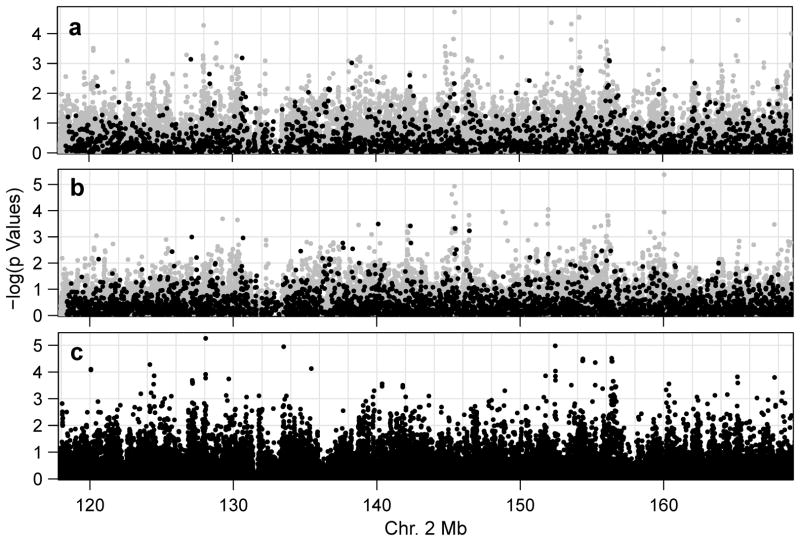
The leading combined association result, for rs7568133 (145.2 Mb; ORmeta = 2.12, 95% CI = (1.50, 3.00) for the A allele; p = 1.9 × 10−5; Figure 1A, Table 2), follows from nominally significant associations in both study samples (p = 0.00062 and 0.0085 in Samples 1 and 2, respectively) (Table 2; Figures 1 and 2). The effect of this variant is consistent between Samples 1 (OR = 2.00, 95% CI = [1.35, 2.98]) and 2 (OR = 2.54, 95% CI = [1.27, 5.11]; p value from Cochran’s Q test for heterogeneity = 0.56), and the minor allele frequencies are similar (0.495 vs. 0.478 in Samples 1 and 2, respectively). rs7568133 falls within the large intron 2 of the DNA-binding transcriptional repressor gene, zinc finger E-box-binding homeobox 2 (ZEB2; Figure 2A). This variant alters several potential DNA-binding motifs, and is an enhancer mark in primary monocytes, but is not listed as an expression quantitative trait locus (eQTL) in the GTEx database (Supplementary Table 4).
Figure 1.
Manhattan plots of association results from (A) the meta-analysis, (B) Sample 1 and (C) Sample 2. Genotyped and imputed markers are represented as black and blue dots, respectively.
Table 2.
Most significant meta-analysis results.
| Sample 1 | Sample 2 | Meta-analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Marker | Gene | Position | Alleles | RAF | Info | OR | 95% CI | p | RAF | OR | 95% CI | p | OR | 95% CI | p |
| rs2028211 | BIN1 | 127,901,657 | C/A | 0.1460 | 0.78 | 1.43 | (0.80, 2.53) | 0.22 | 0.1470 | 3.92 | (2.18, 7.07) | 5.4E-06 | 2.33 | (1.55, 3.52) | 5.3E-05 |
| rs7568133 | ZEB2 | 145,204,976 | A/G | 0.4952 | 0.82 | 2.00 | (1.35, 2.98) | 0.00062 | 0.4775 | 2.55 | (1.27, 5.11) | 0.0085 | 2.12 | (1.50, 3.00) | 1.9E-05 |
| rs10169306 | AC023469.1 | 151,926,472 | A/G | 0.0635 | 0.84 | 1.57 | (0.84, 2.95) | 0.16 | 0.0687 | 4.26 | (2.24, 8.11) | 1.0E-05 | 2.56 | (1.63, 4.02) | 4.3E-05 |
| rs58110523 | FMNL2 | 153,292,235 | C/T | 0.0486 | 0.84 | 2.85 | (1.43, 5.66) | 0.0028 | 0.0483 | 3.71 | (1.48, 9.29) | 0.0052 | 3.13 | (1.80, 5.42) | 4.8E-05 |
| rs74762979 | ARL6IP6 | 153,804,898 | G/A | 0.0957 | 0.88 | 1.88 | (1.07, 3.31) | 0.028 | 0.0763 | 5.25 | (2.39, 11.55) | 3.7E-05 | 2.66 | (1.68, 4.22) | 2.9E-05 |
| rs114101795 | ARL6IP6 | 153,825,388 | A/G | 0.0957 | 0.88 | 1.88 | (1.07, 3.30) | 0.028 | 0.0787 | 5.46 | (2.45, 12.13) | 3.2E-05 | 2.68 | (1.69, 4.25) | 2.8E-05 |
| rs79513402 | ARL6IP6 | 153,829,134 | C/T | 0.0957 | 0.88 | 1.88 | (1.07, 3.30) | 0.029 | 0.0774 | 5.29 | (2.40, 11.66) | 3.6E-05 | 2.66 | (1.68, 4.22) | 2.9E-05 |
| rs78089492 | AC092684.1 | 164,826,751 | G/A | 0.0737 | 0.92 | 2.54 | (1.43, 4.52) | 0.0015 | 0.0655 | 2.56 | (1.27, 5.15) | 0.0082 | 2.55 | (1.64, 3.98) | 3.6E-05 |
Gene, gene that contains or is nearest to marker; Alleles, effect/other allele, where the reference is the minor allele in the sample; RAF, reference allele frequency; Info, IMPUTE2 information quality score.
Figure 2.
LocusZoom plots of the chromosome 2 region surrounding rs7568133, for (A) the meta-analysis, (B) Sample 1 only and (C) Sample 2 only. In A and B, genotyped and imputed markers are represented by squares and circles, respectively. In B, markers selected for haplotype-based analysis (see Online Resource 1, Supplementary Figure 2) are marked with asterisks. The LD structure shown is that of the 1000 Genomes 2014 AFR population.
The overall region of association at ZEB2 is primarily driven by strong associations in Sample 1 (Figure 2B), and extends about 200 kb to overlap with the glycosyltransferase-like domain-containing 1 (GTDC1) gene (Figure 2). Three markers within the association peak are among the five most significantly associated markers for Sample 1 (Supplementary Table 2). Of these, rs13390689 and rs79319398 are in introns: GTDC1 intron 3 and ZEB2 intron 2, respectively. Of particular interest is rs79319398, which lies within a DNA region associated with numerous enhancer histone-modification sites in monocytes, neutrophils, and B- and T-lymphocytes, and is predicted to affect three regulatory motifs (Supplementary Table 4). This variant also has a CADD score of 14.59, placing it in the top 3.5% of all possible genetic variants for deleteriousness. Though not within a gene, rs7580080, 7.8 kb downstream of ZEB2, is also connected in epigenomic studies to histone modification marks in monocytes, neutrophils and hematopoietic stem cells, and has a CADD score of 17.66 (Supplementary Table 4).
The association peaks in ZEB2 and GTDC1 appear to be independent. Associated variants in this region (Figure 2B, marked with asterisks) mostly lie within a single LD block within GTDC1 (Supplementary Figure 2); however, rs7568133, is independent of this block and in only weak LD with one other associated SNP in ZEB2, rs79319398. A conditional association analysis of variants in the GTDC1/ZEB2 region, in which allele dosages of rs7568133 were included as a covariate, confirmed the independence of this SNP from the associated SNPs in GTDC1, in that adjusting for the rs7568133 genotype did not reduce the significance of association of the GTDC1 markers by more than an order of magnitude (data not shown). Haplotypes of the 11 SNPs in the LD block, which are highly correlated (r2 near 1.0), and of two SNPs in ZEB2, rs7568133 and rs79319338, which are in complete LD (D′ = 1) but not strongly correlated (Supplementary Figure 2), were tested for association with PTST– in Samples 1 and 2, but were not found to be more significantly associated than the best single markers (data not shown).
Because HIV status is a potential confounder for TST results, we conducted a sensitivity analysis in which HIV+ individuals were omitted. Although the p values for some of the highly associated SNPs in Table 2 were slightly less significant, most likely on account of the smaller number of available individuals, the ORs were very similar (data not shown), indicating that HIV status is not a major cause of misclassification.
FMNL2/ARL6IP6 association peak
In addition, several SNPs in ADP ribosylation factor-like 6 interacting protein 6 (ARL6IP6) were associated in both samples and had meta p-values < 3x10−5 (Table 2), although evidence of association was stronger from Sample 2 (Supplementary Table 3). However, the CADD scores for these SNPs were not notable, and none of them was listed in the GTEx database as an eQTL (Phred-scaled score < 3; Supplementary Table 4). Because the SNPs were not potentially pathogenic, and also because this gene did not have a potential connection to Mtb biology, it was not considered further as a candidate gene. Other strongly associated markers (p < 10−4) from the meta-analysis have greatest support from Sample 2 (Table 2). Only one, rs58110523, is within a gene (intronic to FMNL2 p < 0.01 in both individual samples), but resides in a region with very little association in Sample 1 (Figure 1). These variants, like the index variant rs7568133, change multiple binding motifs but are not linked to many epigenetic marks or transcription factor binding sites (Supplementary Table 4).
Regional heritability analysis
From our variance components analysis for region-specific heritability, we estimated that the chromosome 2 linkage interval explains 9.22% of PTST– risk in Sample 2 (SE = 5.59%; one-sided p = 0.044). The remainder of the autosomal genome explains 12.5% of the overall risk (SE = 6.17%; one-sided p = 0.0084); thus, the whole genome accounts for 21.7% of the risk.
Body mass composition
Because body-mass wasting is correlated to Mtb infection susceptibility30–34, and because GTDC1 has been associated with obesity-related phenotypes in a previous genomewide methylation scan35, we examined the relationship between body mass parameters and PTST– status in our cohort. A smaller proportion of PTST– participants displayed evidence of body-mass wasting than non-PTST– participants by all three measured criteria: BMI, lean mass and fat mass (Table 3). However, only the lean-mass measurement showed a statistically significant difference (5.1% of PTST–s vs. 17.9% of non-PTST–s; p = 0.039). The non-PTST– subjects have a higher prevalence of lean-mass wasting than the PTST– subjects.
Table 3.
Body mass wasting in PTST– and non-PTST– Ugandan individuals.
| PTST– | Non-PTST– | p | |
|---|---|---|---|
| BMI | 6 (12.2%) | 322 (21.0%) | 0.14 |
| Lean mass | 2 (5.1%) | 218 (17.9%) | 0.039 |
| Fat mass | 8 (20.0%) | 301 (24.6%) | 0.51 |
Criteria for wasting were body-mass index (BMI) < 18.5 kg/m2, fat mass index < 1.8 kg/m2 for men and < 3.9 kg/m2 for women, and lean mass index < 16.7 kg/m2 for men and < 14.6 kg/m2 for women30. All tested participants were HIV-negative aged 15 years or older. Values are shown as N (%). p, p value by Fisher’s exact test; values below 0.05 are italicized.
Discussion
We conducted a fine-mapping study exploring genetic variation associated with the PTST– phenotype in two Ugandan household samples over a segment of chromosome 2q with previous evidence for genetic linkage7. We measured disproportionate overall heritability attributable to the region, and more specifically, associated markers in both cohorts and through meta-analysis with the PTST– phenotype. Even though the risk for PTST– attributable to the chromosome 2 linkage region was only borderline significant (p = 0.044), this 51-Mb segment accounted for approximately 9% of the overall risk for, PTST– and more than 40% of the total genomic risk. These results support the hypothesis that at least one major locus underlying PTST– lies in this chromosomal region.
The most significant association result from the meta-analysis, rs7568133, implicates the genes ZEB2 and GTDC1 on 2q22.3. Though this result falls just short of regionwide significance (ca. 8 × 10−6), this marker is associated in both samples with p < 0.01 and has good agreement in effect size. rs7568133 alters five potential regulatory motifs, and thus, although it is not upstream of either gene, it may function in regulation of gene expression. ZEB2 contains a binding motif that potentially disrupts histone deacetylase 2 (HDAC2)36, a gene implicated by gene-set enrichment analysis of differences in transcriptional response to Mtb infection by monocyte-derived macrophages from PTST– and non-PTST– individuals37. The macrophage has a central role in Mtb pathogenesis, from recognition to killing, a key component of the innate immune response thought to influence PTST–25–27. Thus, our results suggest that genetic variation in macrophage response may influence resistance to Mtb infection. Several SNPs with strong association in Sample 1 occur within enhancer histone marks and DNaseI-hypersensitivity sites found in numerous types of immune-system cells, implying that ZEB2 may be under active transcription in these cell types. Moreover, three of these variants have CADD scores greater than 14 (Supplementary Table 1), suggesting that these variants may truly be pathogenic. Together, these findings support a role for the HDAC innate immunity pathway in relative resistance to Mtb infection that may be genetically regulated.
The nearby gene, GTDC1, is involved with obesity and lipid metabolism. This gene may be of interest for TB pathology because resistance to Mtb infection is correlated with maintenance of body weight. Several previous studies reported that body mass composition is both a risk factor for development of active TB as well as for the speed of recovery from active TB30–34. Here, we examined for the first time whether body composition was associated with resistance to Mtb infection. Body composition results (Table 3) show a significant decrease in lean mass body-mass wasting in PTST– vs. non-PTST–, despite the modest sample size. This leads to the hypothesis that GTDC1 is a risk locus for lean mass wasting which in turn influences risk for Mtb infection. The ideal way to explore such a hypothesis is through Mendelian randomization analysis, which we are unable to perform in this dataset because there is not good overlap in the data with individuals having both genotype and bioelectrical impedance data. This will be the subject of future research. Hypocholesterolemia, a consequence of body-mass wasting, may increase susceptibility to Mtb infection through reduced activity of macrophages38. Moreover, previous studies in mice suggest that hypercholesterolemia, whether induced by a high-cholesterol diet or by knockout of apolipoprotein E (ApoE), impairs the immune response to Mtb infection, with much greater susceptibility in ApoE−/− mice39, 40. In contrast, hypercholesterolemic mice lacking LDL-R did mount a robust immune response to Mtb, although, like the ApoE−/− mice, the inflammatory response to Mtb was destructively exaggerated40, 41, and statin drugs appear to increase resistance of human macrophages to Mtb infection42. Finally, methylation of GTDC1 was found to be associated with waist circumference in a European American cohort, but the result was not successfully replicated35.
The chromosome 2 region featured in the present study has also been recently replicated in its association with Mtb infection in a cohort of HIV-infected individuals21. There, a different extreme phenotype approach was taken, by focusing on individuals that were especially susceptible to Mtb infection because they were immunosuppressed and living in TB-endemic settings. In addition, the associated SNPs from this analysis explained the original linkage result. This, in combination with our region-specific heritability estimate, provides evidence for at least one associated locus in this region. rs7568133 is 14 Mb from the major 2q linkage peak for the PTST– phenotype reported earlier7, with greatest LOD score at microsatellite marker ATA27H09 (D2S1353, 2q24.1 at chr2:159,558,931–159,559,082), and a secondary LOD score peak at GATA4E11 (D2S410, 2q14.1 at chr2:116,240,929–116,241,085). Six of the eight most significantly associated markers from the meta-analysis are within 10 Mb of ATA27H09, suggesting that the linkage signal was not caused by a single genetic variant of large effect.
This investigation has several limitations and strengths. First, the power is restricted by the number of available PTST– individuals. Family relationships within the sample reduce the number of effective independent individuals and family-based association requires a more complex association test. Together, these constraints prevent detection of causal variants with uncommon alleles (frequency < 0.05) unless they are of large (quasi-Mendelian) effect. Next, the use of Sample 2 HumanOmni5 genotypes as a reference for imputation of Sample 1 untyped variants potentially compromises the independence of the two samples. However, the differences in the major results from the individual samples (Supplementary Table 1; Supplementary Table 2) suggests that the induced correlation, if any, was slight. Our earlier report23 shows a difference between level of exposure to TB index cases and PTST– vs. LTBI; however, this association is limited to children aged 5 to 15. Finally, the study samples are different in two respects: the average age for both PTST– and non-PTST– individuals is greater in Sample 2, and more non-PTST– have active TB in the Sample 2. These limitations attributable to small sample size are partly due to the observational nature of the study, whereby some subjects were lost to follow-up prior to the end of the 2-year observation period, therefore excluding them from analysis.
In conclusion, we observed an association between PTST- and ZEB2, further supporting a role for differential regulation of the HDAC pathway in individuals resistant to Mtb infection. These variants are likely functional based on high CADD score and presence of enhancer histone marks. Evidence for GTDC1 was weaker, but further suggests a role for body composition in differential trajectories in TB pathogenesis. Deep resequencing, replication, and functional studies are needed to clarify the roles of these genes in Mtb infection.
Materials and methods
Study samples and phenotypes
All procedures performed in studies involving human participants were in accordance with the principles of the Declaration of Helsinki. All study protocols were reviewed and approved by the National HIV/AIDS Research Committee, the Uganda National Council of Science and Technology, and the institutional review board at the University Hospitals Case Medical Center, Cleveland, OH, USA. Informed consent was obtained from all participants.
Participants in Sample 1, the initial sample for fine mapping, were recruited as reported previously7, 23, 28. Briefly, index cases with culture-positive pulmonary TB and their household members were enrolled and evaluated for TB symptoms and reactivity to TST. Participants were classified as PTST– if they tested TST– at recruitment and remained TST– over 24 months of follow-up. Because the TSTs were at least 3 months apart, boosting was unlikely to increase the chances of observing a TST conversion, as we demonstrate elsewhere23. The sample after quality control totalled 747 individuals with a PTST– phenotype28. This sample overlapped with the sample previously studied by linkage analysis7: 360 individuals belonged to both samples.
Individuals in Sample 2, the follow-up sample, were recruited later in the same study, but are independent from Sample 1. A total of 471 individuals from Sample 2 passed quality controls (see below).
HIV-negative individuals at least 15 years of age were measured for body-mass wasting by three related measures: body-mass index (BMI) and its two components, fat mass index (FMI) and lean mass index (LMI)43. The overall sample for body-mass composition comprised 232 PTST– and 1553 non-PTST– individuals from the household contact study22, 23, including 236 individuals genotyped in Sample 1 (41 PTST–, 195 non-) and 253 individuals in Sample 2 (23 PTST–, 230 non-) Not all individuals had available measurements for all three measures. FMI and LMI were estimated by means of bioeletrical impedance analysis30. Criteria for wasting were body-mass index (BMI) < 18.5 kg/m2, fat mass index < 1.8 kg/m2 for men and < 3.9 kg/m2 for women, and lean mass index < 16.7 kg/m2 for men and < 14.6 kg/m2 for women30.
The datasets analyzed in the current study are not publicly available, because the Ugandan participants did not consent to broad data sharing. However, individual-level data may be requested through a data access committee, chaired by Dr. Sudha Iyengar (ski@case.edu). All genetic association results (summary statistics) are available in Supplementary Table 1 (Online Resource 2).
Genotypes and quality control (QC)
The first phase of the study, conducted on Sample 1, focused on fine mapping a genomic region previously implicated by linkage analysis, 146–176 Kosambi cM on chromosome 2q7. We selected single-nucleotide polymorphisms (SNPs) within map position range chr2: 116,623,530–170,141,754, in GRCh37 coordinates, to cover the 1-LOD support interval (an approximate 95% confidence interval for location) underneath the linkage peak for the PTST– phenotype, at approximately 10-kilobasepair (kb) intervals for genotyping by means of the Illumina (San Diego, CA) iSelect platform. One informative SNP (minor allele frequency (MAF) ≥ 0.1) was selected within each 10-kb window with maximum Illumina assay design score. Of 4,672 SNPs attempted, 3,626 were successfully genotyped on Sample 1 and processed using Illumina Genome Studio, and 3,478 passed marker QC (call rate ≥ 0.9, minor allele frequency ≥ 0.005, p > 10−6 from exact test of deviation from Hardy-Weinberg proportions (HWP) in unrelated subjects, as tested by PLINK 44). Sample QC for the primary analysis has been described28. Samples with call rate < 0.95 over the fine-mapping panel were omitted (total n = 34), as were all Mendelian incompatible genotypes within families.
DNA samples in Sample 2 were typed for 4,310,364 markers on the Illumina HumanOmni5 Beadchip, version 1.0. Genotypes were called using Illumina GenomeStudio. Analysis was restricted to the region of chromosome 2 genotyped for Sample 1. Samples were required to have call rate ≥ 0.98, and samples with 10th percentile of GenCall scores < 0.42 over all markers passing initial QC (call rate ≥ 0.90, p > 10−6 for deviation from HWP) were subject to manual inspection of fluorescence intensity data (B allele frequency) plotted against map position of at least one autosome. Before analysis, markers were subject to a more stringent QC (call rate ≥ 0.98, MAF ≥ 0.01). Genetic sex was verified by means of X-chromosome heterozygosity and percentage of successfully called Y-chromosome genotypes. Relationships and unintentional (non-)duplicates were checked by means of PLINK’s --genome function44 applied to a sparse set (pairwise r2 < 0.1 between markers) of common polymorphisms (MAF ≥ 0.05). Unreported relationships more distant than second-degree were classified as unrelated.
We augmented the fine-mapping marker panel for Sample 1 by imputation45. Because none of the 1000 Genomes Phase 3 populations is respresentative of our Ugandan genomes, we used a set of Ugandan genomes typed for the HumanOmni5 panel, including Sample 2 as a subset, as a reference for imputation. Haplotypes of 44,542 common variants (MAF ≥ 0.5%) spanning the fine-mapping region ± 500kb were determined by means of SHAPEIT246, including the available parent-offspring duos and trios for more accurate phasing. Haplotypes from a subset of 302 unrelated individuals from the HumanOmni5-genotyped sample composed the reference data set for imputation into the discovery cohort (n = 892, including some without a PTST– phenotype). Genotypes from the Sample 1 fine-mapping panel were prephased using SHAPEIT2 before imputation in 5-Mb segments with 500 kb overlap using IMPUTE245. Imputation yielded an augmented panel of 40,335 SNPs with IMPUTE2 imputation quality score47 ≥ 0.5.
We carried out a principal components analysis (PCA) on the 471 Sample 2 individuals passing QC to detect ancestry outliers and to correct for population structure during association analysis (see below). A genome-wide panel of 160,884 common (MAF ≥ 0.05) independent (pairwise r2 < 0.1) variants passing marker QC from the HumanOmni5 panel was chosen, excluding four genomic regions with extensive linkage disequilibrium, which can create artifactual principal components (PCs)48: Chr. 2, 135–137 megabasepairs (Mb) (lactase gene LCT), Chr. 6, 27–35 Mb (HLA region), Chr. 8, 6–16 Mb (inversion polymorphism), and Chr. 17, 40–45 Mb (extensive LD in admixed populations). We calculated principal components (PCs) by two different methods: first, using EIGENSOFT49, which assumes that all individuals are unrelated; and second, using PCAiR50, which performs PCA on an optimal unrelated subset of the sample and which uses genotype loadings to project PCs for relatives. Although the PCAiR approach is more valid for family data, we used EIGENSOFT PCs in association analyses because they resulted in a smaller genomic control (GC) parameter value (see below). To confirm African ancestry, a second PCA was conducted with addition of 119 unrelated individuals from the HapMap CEU, YRI, CHB and JPT samples, using a panel of 130,718 markers in common between the Omni5 PCA panel described above and the 1000 Genomes Phase 1, version 3 data set.
Statistical methods
Association analysis on the imputed Sample 1 genotype data was conducted by means of logistic regression, using the generalized estimating equations (GEE) model implemented in the gee package in R to allow for correlations within families. The number of minor alleles from genotyped markers, or the allele dosage data (expected number of minor alleles) from imputation, were used as a genotype predictor under an additive model (on the logarithmic scale). The “exchangeable” correlation structure was specified, in which all relatives within a family were assumed equally correlated; if this model failed to converge to a stable estimate, the “independence” structure was used, which provides a still valid, albeit less powerful, approach. For this study, because of the limited sample size and the complexity of the statistical model, families were defined by grouping individuals connected by first-degree relationships; within-household correlations owing to common household environment, and correlations between more distant family members, were not modeled. Only imputed SNPs with MAF ≥ 0.03 were tested for association, after it was discovered that the GEE algorithm had difficulty converging with some of the rarer imputed variants, even under the “independence” correlation structure, whereas models on genotyped SNPs with MAF ≥ 0.01 converged well. We chose GEE for association analysis, instead of a generalized linear mixed model (GLMM) adjusting for all relationships, not only because of model complexity but also because there were very few second- and third-degree relative pairs in either sample, and because complex correlations due to sharing the same household, the same bed, etc., are difficult to model by GLMM but are estimated from the data by GEE. With only a regional marker map, we were unable to assess inflation of test statistics by means of the genomic control parameter λ51. However, the value of λ over the region of linkage was only 1.007, and the quantile-quantile plot of the results is consistent with no genome-wide inflation (see Results). Because we expect this region to harbor truly associated variants, we are confident that the type I error was well controlled. Following Sobota et al.52, we estimated the number of effective independent tests by isolating a set of low-dependence markers, using PLINK’s --indep-pairwise function with an r2 threshold of 0.2. This approach yielded 6,246 effective independent tests for a nominal p value threshold of 8.0 × 10−6 for regionwide significance of 0.05; and for the Omni5 marker set, 6,103 independent tests for nominal p = 8.1 × 10−6.
Association analysis on Sample 2 was carried out in similar fashion, except that the fourth PC from the EIGENSOFT PCA was included as a predictor to adjust for population structure. The first 20 PCs were evaluated for association with the PTST– phenotype in Sample 2. PCs 3 and 4 were found to be significant when included singly, but in the presence of PC 4, PC 3 had a nonsignificant effect, and thus only PC4 was included in association analysis as a covariate. A sparse genome-wide scan of about 10,000 SNPs from the Sample 2 Omni5 panel, excluding regions implicated in TB susceptibility (the HLA region, but also all genes mentioned in two previous reports28, 53) was conducted to obtain an estimate of the genomic control (GC) parameter λ for genome-wide inflation of test statistics51. Because λ from the final analysis was greater than 1.05, we corrected association p values for genome-wide inflation by the method of Bacanu et al.54. It was uncertain what was causing the overall inflation of test statistics. One possibility was that inflation was caused by including markers with low minor allele counts, but the value of λ was not reduced by increasing the minimum MAF to several values from 0.03 to 0.10. Second, we compared λ from association analyses adjusting for PC 4 from EIGENSOFT, and adjusting for four PCs from PCAiR that were significantly associated with PTST–, and found that adjusting for EIGENSOFT PC4 resulted in a smaller value of λ54.
We conducted meta-analysis by the inverse-variance-weighted fixed-effect method, and calculated Cochran’s Q statistic and I2 to assess effect heterogeneity between the two samples, using a custom script for the statistical software R.
Haplotypes of 13 SNPs in genes GTDC1 and ZEB2 were determined for both Samples 1 and 2 by means of SHAPEIT2, with the --duohmm option to make use of parent/child relationships. Haplotypes from two sets of SNPs, a set of 11 and a set of two, showing linkage disequilibrium were used for haplotype-based association analysis. Best-guess haplotypes from these two sets were counted, and haplotypes with sample frequencies between 3% and 20% were tested for association with PTST– vs. all other pooled haplotypes, under the same GEE regression model used for single-marker association testing.
We used the restricted maximum likelihood (REML) estimation approach in the GCTA software package55 to partition genetic variance in Sample 2 explained by the specific region on Chr. 2 (position 117,911,357 to 168,853,091) and, separately, all other genotyped SNPs across the genome. Briefly, we filtered SNPs (excluding SNPs with call rate < 0.95 and minor allele frequency < 0.05), generated separate genetic relationship matrices for the region on Chr. 2 and the rest of the genome, then performed REML estimation using the expectation maximization fitting method to estimate the proportion of “risk” for being PTST explained by each of the two genetic partitions (i.e., region on Chr. 2 & all other SNPs). REML analysis was adjusted for age, sex, and HIV status.
Differences in proportions in body-mass wasting between PTST– and non-PTST– subsets in the sample measured for body mass composition were evaluated by Fisher’s exact test.
Annotating strongly associated variants
We explored the likely effects of genetic variants with the most significant association results with information from several well-known databases. We obtained Combined Annotation-dependent Depletion (CADD) scores56, a measure of deleteriousness based on evolutionary conservation and on numerous measures of regulatory importance and predicted protein effects, from the CADD Web site (http://cadd.gs.washington.edu/). We report CADD scores as PHRED-like scores, in which a score of 10x indicates pathogenicity within the top 100 × 10−x percent of possible variants genome-wide. We extracted specific information on chromatin structure, effects on DNA regulatory motifs and association results from other GWAS and expression quantitative trait locus (eQTL) studies from the HaploReg v4.157 Web site (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php), specifying the ChromHMM (Core 15-state model) algorithm for chromatin structure determination. We searched the GTEx database58 for prominent PTST– associated markers for evidence of eQTL activity in 53 human tissues. Finally, we acquired a measure of overall evidence for a regulatory role from RegulomeDB (59; http://www.regulomedb.org/).
Supplementary Material
Supplementary Materials (Microsoft Word format, .doc): Supplementary Figure 1. Quantile-quantile plots of analyses for (A) Sample 1 and (B) Sample 2. Supplementary Figure 2. Linkage disequilibrium structure of markers selected for haplotype-based association analysis in the GTDC1/ZEB2 region. Supplementary Figure 3. LocusZoom plots of chromosome 2 regions containing the most significantly associated markers of (A) Sample 1 and (B) Sample 2. Supplementary Table 2. Major association results from Sample 1. Supplementary Table 3: Major association results from Sample 2.
Supplementary Table 1 (Microsoft Excel format, .xls): Supplementary Table 1. Full results from chromosome 2 association analyses and meta-analysis.
Supplementary Table 4 (Microsoft Excel format, .xls): Supplementary Table 4. Summary of annotation data on strongly associated markers.
Acknowledgments
The authors wish to acknowledge the contributions made by senior physicians, medical officers, health visitors, laboratory and data personnel: Dr. Lorna Nshuti, Dr. Roy Mugerwa, Dr. Alphonse Okwera, Dr. Deo Mulindwa, Dr. Christopher Whalen, Denise Johnson, Allan Chiunda, Mark Breda, Dennis Dobbs, Mary Rutaro, Albert Muganda, Richard Bamuhimbisa, Yusuf Mulumba, Deborah Nsamba, Barbara Kyeyune, Faith Kintu, Gladys Mpalanyi, Janet Mukose, Grace Tumusiime, Pierre Peters, Annet Kawuma, Saidah Menya, Joan Nassuna, Keith Chervenak, Karen Morgan, Alfred Etwom, Micheal Angel Mugerwa, and Lisa Kucharski. We would like to acknowledge Dr. Francis Adatu Engwau, former Head of the Uganda National Tuberculosis and Leprosy Program, for supporting this project. We would like to acknowledge the medical officers, nurses and counselors at the National Tuberculosis Treatment Centre, Mulago Hospital, the Ugandan National Tuberculosis and Leprosy Program and the Uganda Tuberculosis Investigation Bacteriological Unit, Wandegeya, for their contributions to this study. Clinical study implementation and data management were supported by the National Institutes of Health, grants N01-AI95383, HHSN266200700022C/N01-AI70022. Genotyping and data analysis was supported by R01HL096811 and analyses were also supported by T32HL007567.This study would not be possible without the generous participation of the Ugandan patients and families.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Author Contributions
EM, MJ, WHB and CMS conceived and designed the study. EM and MJ recruited families and collected the study sample. LLM maintained the database of clinical study data. RPI, NHB, BT, FQ, LT and AS processed the raw genotype data and conducted quality control. TRH, WHB, and CMS developed the conceptual biologic model. RPI, NHB, JBH and WSB performed statistical analyses. RPI and CMS drafted the manuscript. All authors read and approved the final manuscript.
References
- 1.World Health Organization. Global Tuberculosis Report. WHO Press; Geneva, Switzerland: 2016. [Google Scholar]
- 2.Comstock GW. Epidemiology of tuberculosis. Am Rev Respir Dis. 1982;125:8–15. doi: 10.1164/arrd.1982.125.3P2.8. [DOI] [PubMed] [Google Scholar]
- 3.Stein CM, Sausville L, Wejse C, Sobota RS, Zetola NM, Hill PC, et al. Genomics of human pulmonary tuberculosis: from genes to pathways. Curr Genet Med Reports. 2017;5:149–166. doi: 10.1007/s40142-017-0130-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Möller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90:71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Cooke GS, Campbell SJ, Bennett S, Lienhardt C, McAdam KP, Sirugo G, et al. Mapping of a novel susceptibility locus suggests a role for MC3R and CTSZ in human tuberculosis. Am J Respir Crit Care Med. 2008;178:203–207. doi: 10.1164/rccm.200710-1554OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellamy R. Genetics and pulmonary medicine: 3. Genetic susceptibility to tuberculosis in human populations. Thorax. 1998;53:588–593. doi: 10.1136/thx.53.7.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein CM, Zalwango S, Malone LL, Won S, Mayanja-Kizza H, Mugerwa RD, et al. Genome scan of M. tuberculosis infection and disease in Ugandans. PLoS ONE. 2008;3:e4094. doi: 10.1371/journal.pone.0004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahasirimongkol S, Yanai H, Nishida N, Ridruechai C, Matsushita I, Ohashi J, et al. Genome-wide SNP-based linkage analysis of tuberculosis in Thais. Genes Immun. 2009;10:77–83. doi: 10.1038/gene.2008.81. [DOI] [PubMed] [Google Scholar]
- 9.Miller EN, Jamieson SE, Joberty C, Fakiola M, Hudson D, Peacock CS, et al. Genome-wide scans for leprosy and tuberculosis susceptibility genes in Brazilians. Genes Immun. 2004;5:63–67. doi: 10.1038/sj.gene.6364031. [DOI] [PubMed] [Google Scholar]
- 10.Thye T, Vannberg FO, Wong SH, Owusu-Dabo E, Osei I, Gyapong J, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thye T, Owusu-Dabo E, Vannberg FO, van Crevel R, Curtis J, Sahiratmadja E, et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat Genet. 2012;44:257–259. doi: 10.1038/ng.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chimusa ER, Zaitlen N, Daya M, Möller M, van Helden PD, Mulder NJ, et al. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum Mol Genet. 2014;23:796–809. doi: 10.1093/hmg/ddt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis J, Luo Y, Zenner HL, Cuchet-Lourenço D, Wu C, Lo K, et al. Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat Genet. 2015;47:523–527. doi: 10.1038/ng.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahasirimongkol S, Yanai H, Mushiroda T, Promphittayart W, Wattanapokayakit S, Phromjai J, et al. Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J Hum Genet. 2012;57:363–367. doi: 10.1038/jhg.2012.35. [DOI] [PubMed] [Google Scholar]
- 15.Png E, Alisjahbana B, Sahiratmadja E, Marzuki S, Nelwan R, Balabanova Y, et al. A genomewide association study of pulmonary tuberculosis susceptibility in Indonesians. BMC Med Genet. 2012;13:5. doi: 10.1186/1471-2350-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobota RS, Stein CM, Kodaman N, Scheinfeldt LB, Maro I, Wieland-Alter W, et al. A locus at 5q33. 3 confers resistance to tuberculosis in highly susceptible individuals. Am J Hum Genet. 2016;98:514–524. doi: 10.1016/j.ajhg.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobat A, Poirier C, Hoal E, Boland-Auge A, de La Rocque F, Corrard F, et al. Tuberculin skin test negativity is under tight genetic control of chromosomal region 11p14–15 in settings with different tuberculosis endemicities. J Inf Dis. 2015;211:317–321. doi: 10.1093/infdis/jiu446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein CM. Genetic epidemiology of tuberculosis susceptibility: impact of study design. PLoS Pathog. 2011;7:e1001189. doi: 10.1371/journal.ppat.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobat A, Gallant CJ, Simkin L, Black GF, Stanley K, Hughes J, et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med. 2009;206:2583–2591. doi: 10.1084/jem.20090892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobat A, Barrera LF, Henao H, Arbeláez P, Abel L, García LF, et al. Tuberculin skin test reactivity is dependent on host genetic background in Colombian truberculosis household contacts. Clin Infect Dis. 2012;54:968–971. doi: 10.1093/cid/cir972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobota RS, Stein CM, Kodaman N, Maro I, Wieland-Alter W, Igo RP, Jr, et al. A chromosome 5q31. 1 locus associates with tuberculin skin test reactivity in HIV-positive individuals from tuberculosis hyper-endemic regions in east Africa. PLoS Genet. 2017;13:e1006710. doi: 10.1371/journal.pgen.1006710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein CM, Hall NB, Malone LL, Mupere E. The household contact study design for genetic epidemiological studies of infectious diseases. Front Genet. 2013;4:61. doi: 10.3389/fgene.2013.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein CM, Zalwango S, Malone LL, Thiel BA, Mupere E, Nsereko M, et al. Resistance and susceptibility to Mycobacterium tuberculosis infection and disease in tuberculosis households in Kampala, Uganda. Am J Epidemiol. 2018 doi: 10.1093/aje/kwx380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma N, Zalwango S, Malone LL, Nsereko M, Wampande EM, Thiel BA, et al. Clinical and epidemiological characteristics of individuals resistant to M. tuberculosis infection in a longitudinal TB household contact study in Kampala, Uganda. BMC Infect Dis. 2014;14:352. doi: 10.1186/1471-2334-14-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons J, Stein CM, Seshadri C, Campo M, Alter G, Fortune S, et al. Immunologic mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat Rev Immunol. 2018 doi: 10.1038/s41577-018-0025-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawn TR, Day TA, Scriba TJ, Hatherill M, Hanekom WA, Evans TG, et al. Tuberculosis vaccines and prevention of infection. Microbiol Mol Biol Rev. 2014;78:650–671. doi: 10.1128/MMBR.00021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verrall AJ, Netea MG, Alisjahbana B, Hill PC, Van Crevel R. Early clearance of Mycobacterium tuberculosis: a new frontier in prevention. Immunology. 2014;141:506–513. doi: 10.1111/imm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall NB, Igo RP, Jr, Malone LL, Truitt B, Schnell A, Tao L, et al. Polymorphisms in TICAM2 and IL1B are associated with TB. Genes Immun. 2015;16:127–133. doi: 10.1038/gene.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guwatudde D, Nakakeeto M, Jones-Lopez EC, Maganda A, Chiunda A, Mugerwa RD, et al. Tuberculosis in household contacts of infections cases in Kampala, Uganda. Am J Epidemiol. 2003;158:887–898. doi: 10.1093/aje/kwg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mupere E, Malone L, Zalwango S, Okwera A, Nsereko M, Tisch DJ, et al. Wasting among Uganda men with pulmonary tuberculosis is associated with linear regain in lean tissue mass during and after treatment in contrast to women with wasting who regain fat tissue mass: prospective cohort study. BMC Infect Dis. 2014;14:24. doi: 10.1186/1471-2334-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mupere E, Malone L, Zalwango S, Chiunda A, Okwera A, Parraga I, et al. Lean tissue mass wasting is associated with increased risk of mortality among women with pulmonary tuberculosis in urban Uganda. Ann Epidemiol. 2012;22:466–473. doi: 10.1016/j.annepidem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mupere E, Zalwango S, Chiunda A, Okwera A, Mugerwa R, Whalen C. Body composition among HIV-seropositive and HIV-seronegative adult patients with pulmonary tuberculosis in Uganda. Ann Epidemiol. 2010;30:210–216. doi: 10.1016/j.annepidem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mupere E, Parraga IM, Tisch DJ, Mayanja HK, Whalen CC. Low nutrient intake among adult women and patients with severe tuberculosis disease in Uganda: a cross-sectional study. BMC Public Health. 2012;12:1050. doi: 10.1186/1471-2458-12-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ezeamama AE, Mupere E, Oloya J, Martinez L, Kakaire R, Yin X, et al. Age, sex and nutritional status modify the CD4+ T-cell recovery rate in HIV-tuberculosis co-infected patients on combination antiretroviral therapy. Int J Infect Dis. 2015;35:73–79. doi: 10.1016/j.ijid.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aslibekyan S, Demerath EW, Mendelson M, Zhi D, Guan W, Liang L, et al. Epigenome-wide study indentifies novel methylation loci associated with body mass index and waist circumference. Obesity. 2015;23:1493–1501. doi: 10.1002/oby.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu LM, Wang J, Conidi A, Zhao C, Wang H, Ford Z, et al. Zeb2 recruits HDAC-NuRD to inhibit Notch and controls Schwann cell differentiation and remyelination. Nat Neurosci. 2016;19:1060–72. doi: 10.1038/nn.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seshadri C, Sedaghat N, Campo M, Peterson G, Wells RD, Olson GS, et al. Transcriptional networks are associated with resistance to Mycobacterium tuberculosis infection. PLoS ONE. 2017;12:e0175844. doi: 10.1371/journal.pone.0175844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Guzmán C, Vargas MH. Hypocholesterolemia: A major risk factor for developing pulmonary tuberculosis? Med Hypotheses. 2006;66:1227–1230. doi: 10.1016/j.mehy.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 39.Han R, Kornfeld H, Martens G. Is hypercholesterolemia a friend or foe of tuberculosis? Infect Immun. 2009;77:3514. doi: 10.1128/IAI.00469-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martens GW, Arikan MC, Lee J, Ren F, Vallerskog T, Kornfeld H. Hypercholesterolemia impairs immunity to tuberculosis. Infect Immun. 2008;76:3464–3472. doi: 10.1128/IAI.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martens GW, Vallerskog T, Kornfeld H. Hypercholesterolemic LDL receptor-deficient mice mount a neutrophilic response to tuberculosis despite the timely expression of protective immunity. J Leukoc Biol. 2012;91:849–857. doi: 10.1189/jlb.0311164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parihar SP, Guler R, Khutlang R, Lang DM, Hurdayal R, Mhlanga MM, et al. Statin therapy reduces the mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. J Inf Dis. 2014;209:754–763. doi: 10.1093/infdis/jit550. [DOI] [PubMed] [Google Scholar]
- 43.VanItallie TB, Yang MU, Heymsfield SB, Funk RC, Boileau RA. Height-normalized indices of the body’s fat-free mass and fat mass: potentially useful indicators of nutritional status. Am J Clin Nutr. 1990;52:953–959. doi: 10.1093/ajcn/52.6.953. [DOI] [PubMed] [Google Scholar]
- 44.Purcell S, Neale B, Todd-Brow K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genomewide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 47.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 48.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 52.Sobota RS, Shriner D, Kodaman N, Goodloe R, Zheng W, Gao Y-T, et al. Addressing population-specific multiple testing burdens in genetic association studies. Ann Hum Genet. 2015;79:136–147. doi: 10.1111/ahg.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein CM. Encyclopedia of Life Sciences (eLS) John Wiley & Sons; Chichester: 2012. Genetics of susceptibility to tuberculosis. [DOI] [Google Scholar]
- 54.Bacanu S-A, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66:1933–1944. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials (Microsoft Word format, .doc): Supplementary Figure 1. Quantile-quantile plots of analyses for (A) Sample 1 and (B) Sample 2. Supplementary Figure 2. Linkage disequilibrium structure of markers selected for haplotype-based association analysis in the GTDC1/ZEB2 region. Supplementary Figure 3. LocusZoom plots of chromosome 2 regions containing the most significantly associated markers of (A) Sample 1 and (B) Sample 2. Supplementary Table 2. Major association results from Sample 1. Supplementary Table 3: Major association results from Sample 2.
Supplementary Table 1 (Microsoft Excel format, .xls): Supplementary Table 1. Full results from chromosome 2 association analyses and meta-analysis.
Supplementary Table 4 (Microsoft Excel format, .xls): Supplementary Table 4. Summary of annotation data on strongly associated markers.