Abstract
Here we report the biological synthesis of CdS fluorescent nanoparticles (Quantum Dots, QDs) by polyextremophile halophilic bacteria isolated from Atacama Salt Flat (Chile), Uyuni Salt Flat (Bolivia) and the Dead Sea (Israel). In particular, a Halobacillus sp. DS2, a strain presenting high resistance to NaCl (3–22%), acidic pH (1–4) and cadmium (CdCl2 MIC: 1,375 mM) was used for QDs biosynthesis studies. Halobacillus sp. synthesize CdS QDs in presence of high NaCl concentrations in a process related with their capacity to generate S2− in these conditions. Biosynthesized QDs were purified, characterized and their stability at different NaCl concentrations determined. Hexagonal nanoparticles with highly defined structures (hexagonal phase), monodisperse size distribution (2–5 nm) and composed by CdS, NaCl and cysteine were determined by TEM, EDX, HRXPS and FTIR. In addition, QDs biosynthesized by Halobacillus sp. DS2 displayed increased tolerance to NaCl when compared to QDs produced chemically or biosynthesized by non-halophilic bacteria. This is the first report of biological synthesis of salt-stable QDs and confirms the potential of using extremophile microorganisms to produce novel nanoparticles. Obtained results constitute a new alternative to improve QDs properties, and as consequence, to increase their industrial and biomedical applications.
Introduction
Semiconductor nanocrystals or Quantum Dots (QDs) exhibit unique optical and electronic properties with fluorescence emission wavelengths depending on nanoparticle (NPs) size and composition1–3. Nowadays, the interest in studying QDs has increased due to their numerous applications in photovoltaics, optoelectronics, transistors, oil exploration, biomedicine and imaging, among others3–7.
To date, QDs are mainly obtained through chemical synthesis; however, most of these methods involve the use of toxic reagents, high temperatures and anaerobic conditions. In addition, most QDs synthesized by chemical methods present low biocompatibility and poor stability at high osmolarity conditions, thus affecting their potential applications5,8–11. In this context, there is a growing interest in the generation of green and eco friendly methods for QDs production to be used in different technological applications.
During the last years, the industrial and scientific interest for developing eco-friendly and sustainable methods to synthesize CdS, CdSe and CdTe QDs has grown. The addition to chemical synthesis procedures of bidentate thiols [e.g. dithiothreitol (DTT), mercaptosuccinic acid (MSA), mercaptopropionic acid (MPA)] and ligands with different functional groups (amino, hydroxyl, carboxylic acid, among others) has been used to improve NPs biocompatibility and stability12. Nevertheless, these methods produce NPs that still display low biocompatibility, sensitivity to pH and high ionic strength, as well as elevated production costs11,13–15.
Several chemical methods based on the use of biological reagents and mild conditions have been developed during the last years16–19. These methods, denominated biomimetic, have allowed the production of nanomaterials in a simpler and eco-friendly way. In addition, biomimetic procedures have contributed to improve the properties of NPs, being the increase on biocompatibility one of the most significant improvements since it has a direct impact on the range of nanoparticles applications14,15.
Most biomimetic methods described to date involve the use of mild temperatures (90 °C), aerobic conditions, and biological molecules, such as biological thiols17–19. Other biological components such as peptides, nucleotides, fusion proteins and phosphorylated molecules have also been used for the chemical synthesis of QDs14,16,18–21.
In this context, biological synthesis of nanoparticles has emerged as a sustainable and green alternative to classical production methods. Several protocols reporting the production of NPs using cell extracts or living cells have been developed during the last decade. These biologically produced nanoparticles display high stability, water solubility, biocompatibility, low costs and high production rates, among others22–24. From these protocols, the biosynthesis of NPs using microorganisms has been implemented as a cost effective and eco-friendly alternative.
Several microorganisms, including bacteria and fungi, are used to biosynthesize cadmium-based QDs24–28. Most biosynthesis processes reported to date involve different thiolated biomolecules as relevant precursors of the biosynthetic process29–31. Moreover, the importance of biological thiols such as glutathione and H2S has been recently showed26,27,32,33. All these reports relate the biosynthesis of QDs with the presence of biological thiols promoting the intra- and extracellular generation of QDs. Recently, our group determined the importance of phosphorylated biomolecules in the biological synthesis of CdS QDs. Phosphate groups present in biologically relevant molecules such as nucleotides and metabolic sugars, contribute to the generation of the nanocrystal and constitute the external layer of the QDs16. Despite these advances, the molecular mechanism involved in QDs biosynthesis is still unknown16,23,25,34.
With the aim of finding new methods to biosynthesize QDs with improved properties, our group has focused on using bacteria inhabiting extreme environments (extremophiles) for QDs production. Accordingly, we reported the biological synthesis of QDs at low temperatures using novel psychrotolerant bacterial strains isolated from Antarctica26,27. In addition, our group recently reported the production of fluorescent Cd-nanoparticles by acidophilic bacteria used in biomining operations35,36. Bacteria of the Acidithiobacillus gender (A. ferrooxidans, A. thiooxidans and A. caldus) were used to biosynthesize Cd-QDs under acidic conditions (pH 3,0). Interestingly, QDs produced by acidophilic bacteria were highly stable at low pHs, a characteristic not observed in QDs produced by chemical methods or biosynthesized by the mesophilic bacteria Escherichia coli35. To date, the use of other extremophile microorganisms to produce QDs with novel properties has not been explored.
Most halophilic microorganisms inhabit soil or liquid environments characterized by the presence of high salt concentrations37–39. Halophilic prokaryotes, belonging to Bacteria or Archaea domains, can adapt to a wide range of osmolarities, some of them living near NaCl saturation37. Salt-tolerant bacteria have been isolated from a wide range of environments at all latitudes40–42. To date, most halophilic microorganisms have been isolated with the purpose of obtaining new biomolecules of industrial interest like enzymes, exopolysaccharides and phytohormones, among many others43. However, their use in the biosynthesis of fluorescent nanoparticles has not been explored.
Salt stable nanomaterials are a useful alternative for different applications, particularly for those involving NPs transport in presence of ionic solutions such as real time sensing in oil explorationand bioimaging5,44. The injection of transportable fluorescent nanoparticles would allow spatially resolved measurements of temperature, pressure, rock porosity and permeability of an oil reservoir5. In addition, discovering and characterizing the biomolecules involved in fluorescence salt-stability of biosynthesized nanocrystals would allow the development of new chemical methods to produce NPs with improved properties.
The objective of the present work was to determine if halophilic microorganisms isolated from extreme environments with high salt concentrations (Atacama Desert, Uyuni Salt Flat and Dead Sea) can biosynthesize Cd-QDs under high osmolarity conditions. In addition, we characterized the QDs produced by halophilic bacterium and we tested the ability of QDs to retain their fluorescence intensity in presence of elevated salt concentrations (fluorescence stability), a condition affecting most QDs described to date.
Results
Isolation and characterization of halophilic bacteria
15 bacterial isolates able to grow in Luria Bertani (LB) medium supplemented with NaCl 8 or 15% at different temperatures (28 to 37 °C) were obtained from Atacama Salt Flat (ASF), Uyuni Salt Flat (USF) and Dead Sea (DS) samples, as described in methods.
The ability of all isolates to grow at different NaCl concentrations was determined (Table 1). Obtained results indicate that bacterial isolates are able to grow in a wide range of NaCl concentrations (1 to 22%). All isolates were also capable to grow at 28 and 37 °C. Based on these results, these microorganisms can be classified as halophiles. Therefore, they present a unique opportunity to study the process of QDs biosynthesis given the extreme conditions in which they can survive.
Table 1.
Cadmium and NaCl tolerance of halophilic isolates.
| Isolate | % NaCl Range | CdCl2 MIC (mM) |
|---|---|---|
| ASF1 | 2–15 | 1.375 |
| ASF2 | 2–15 | 0.66 |
| ASF3 | 2–15 | 1.375 |
| ASF4 | 2–15 | 0.66 |
| ASF5 | 2–15 | 1.375 |
| UYSF1 | 2–22 | 1.375 |
| UYSF2 | 2–22 | 0.66 |
| UYSF3 | 2–22 | 0.66 |
| UYSF4 | 1–25 | 1.375 |
| UYSF5 | 1–25 | 1.375 |
| DS1 | 4–25 | 0.66 |
| DS2 | 3–22 | 1.375 |
| DS3 | 3–22 | 0.66 |
| DS4 | 3–22 | 0.66 |
| DS5 | 3–22 | 1.375 |
Cadmium MICs and ranges of NaCl concentrations allowing bacterial growth are shown.
Most CdS QDs biosynthetic methods described to date involve the use of cadmium salts such as CdCl2. Based on this, CdCl2 resistance of isolated halophiles was determined. Minimal inhibitory concentration (MIC) values ranging from 0.66 to 1.375 mM (121 to 251.15 μg/mL) were obtained (Table 1). These values are similar to other strains widely used for QDs biosynthesis belonging to different species such as P. aeruginosa PAO1 (0.55 mM or 100 μg/mL) and E. coli (1.265 mM or 230 μg/mL)27. Obtained results indicate that eight halophilic isolates are resistant to cadmium (1.375 mM).
Biosynthesis of CdS QDs by halophilic bacteria
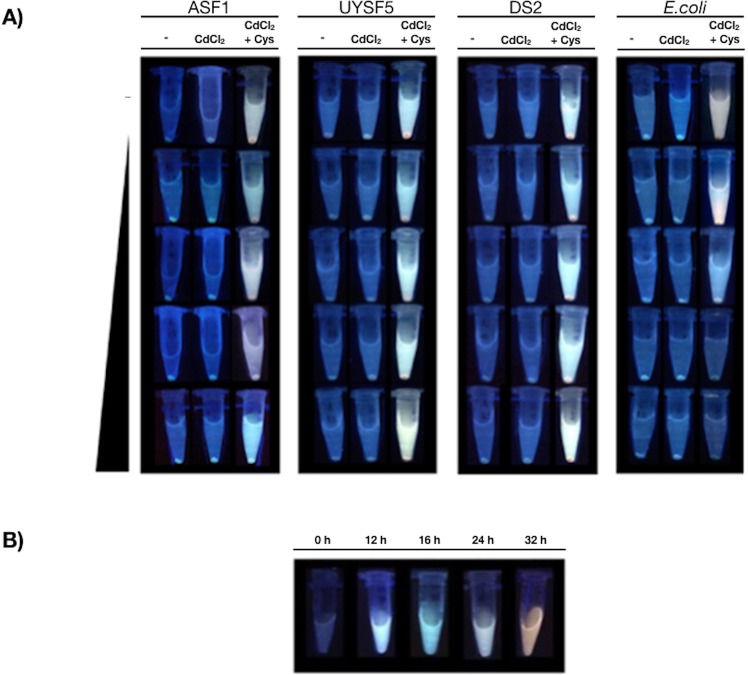
Based on the characteristics of the halophilic strains isolated, we decided to evaluate CdS QDs biosynthesis in culture media supplemented with NaCl 8%. We mainly focus in extracellular biosynthesis since this method facilitates NPs purification and favors future applications. The capacity of all obtained isolates to biosynthesize QDs was evaluated following the protocol described before4,16,26,27,36. Only three cadmium-resistant isolates were able to biosynthesize cadmium-based QDs in the presence of NaCl up to 8%, as determined by fluorescence of cell supernatants (Fig. 1A). The strains with the capacity to biosynthesize QDs were ASF1, UYSF5 and DS2, isolated from Atacama Salt Flat, Uyuni Salt Flat and Dead Sea, respectively. The three isolates capable of biosynthesizing QDs were those that also displayed the highest resistance to Cd (1.375 mM) and NaCl (15% or more) (Table 1) among all isolates. E. coli was used as control, since it has been widely used to study biosynthesis of fluorescent NPs16,25,32. As expected, E. coli was unable to biosynthesize QDs in presence of NaCl 6% or higher.
Figure 1.
Biosynthesis of QDs in presence of different NaCl concentrations by halophilic isolates. (A) The biosynthesis of QDs was evaluated in the halophilic isolates ASF1, UYSF5, DS2 and the control bacteria E. coli. The effect of NaCl (0, 2, 4, 6 and 8%) on QDs biosynthesis in presence or absence of cysteine (2 mM) was evaluated. (B) DS2 was exposed to QDs-biosynthesis conditions and QDs fluorescence after UV exposure was evaluated at different times of biosynthesis (0, 12, 16, 24 and 32 h).
The extremophile strain DS2 was selected for the following experiments since it was able to synthesize QDs, grew at high NaCl concentrations (3–22%) and exhibited resistance to CdCl2 (1.375 mM). In addition, this isolate was able to grow in a wide range of pHs (pH 1–9). The 16S rRNA gene of DS2 was amplified and sequenced to determine the identity of this strain. The obtained sequence showed that DS2 display high identity with Halobacillus sp. (sequence identity: 96–98%), a halophilic spore forming bacterium previously isolated from salt environments45.
To confirm that the fluorescence observed during biosynthesis corresponds to QDs, time dependent changes in the fluorescence of Halobacillus DS2 cells exposed to biosynthetic conditions were evaluated (cysteine 1 mM, NaCl 8% and CdCl2 0.33 mM). Supernatant aliquots were collected at several times and analyzed to evaluate extracellular synthesis of NPs. Fluorescence emission colors changed at different times, moving from blue to red after 32 h (Fig. 1B). This behaviour is characteristic of QDs formation and it has been related to increasing nanocrystal size, which changes the band gap of nanoparticles and their spectroscopic properties16,26,27,32,35. In this context, obtained results are very novel since QDs biosynthesis in the presence of high concentrations of NaCl or using halophilic strains has not been reported to date.
H2S production at high osmolarity is required for QDs biosynthesis
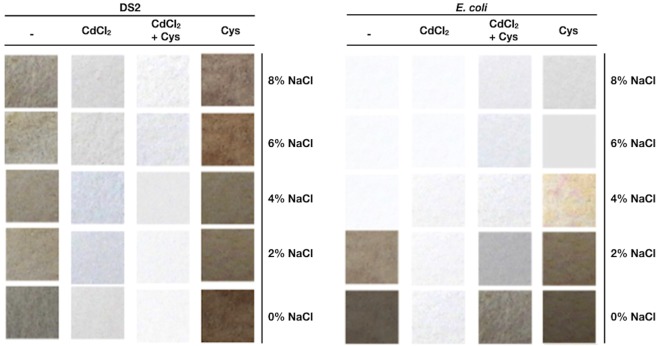
Hydrogen sulfide has been described as a promoter molecule for the biosynthesis process of sulfur-containing nanoparticles16,26,27,35. Based on this, H2S production was assessed in LB media supplemented with several NaCl concentrations. Since bacterial sulfide production has been associated with the activity of cysteine desulfhydrase46, we determined sulfide generation in the presence of cysteine (substrate of cysteine desulfhydrase enzyme). The generation of black precipitates on lead acetate papers exposed to cultures growing in LB media was positive for E. coli at 0 and 2% NaCl, and at all NaCl concentrations tested for the Halobacillus DS2 isolate (Fig. 2). When cysteine was added to bacterial cultures, an increase in sulfide generation was observed at all NaCl concentrations. In the case of E. coli, cysteine allowed the production of sulfide at 0 and 2% NaCl, and scarce amounts were detected at higher NaCl concentrations (Fig. 2).
Figure 2.
Effect of NaCl on Halobacillus sp. DS2 sulfide production. The production of sulfide by Halobacillus DS2 and E. coli in presence of different NaCl concentrations was determined using papers soaked with lead acetate (Shatalin et al., 2011). (−) Cells in LB. (CdCl2) cells in LB supplemented with CdCl2. (CdCl2 + Cys) Cells in LB supplemented with cysteine and CdCl2. (Cys) Cells in LB supplemented with cysteine.
H2S production under conditions of Cd-based QDs biosynthesis was also evaluated. A decrease in sulfide levels produced by Halobacillus sp. DS2 and E. coli cells was observed at all NaCl concentrations tested, a result that has been associated with the extracellular generation of CdS nanocrystals as consequence of the interaction of sulfide and the metal ion27,47. In summary, H2S assays indicated that the Halobacillus sp. DS2 isolate is capable of synthesizing sulfide even in presence of high salt concentrations, and also that the generation of this compound is associated with CdS generation in culture supernatants.
Spectroscopic characterization of biosynthesized QDs
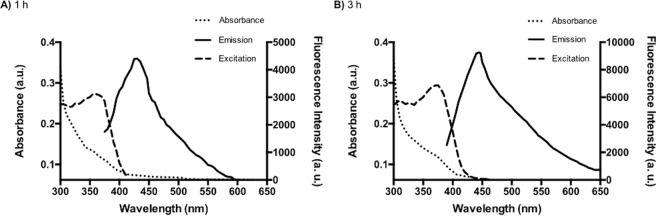
Fluorescent nanoparticles produced by Halobacillus sp. DS2 after 1 and 3 h synthesis were purified from cells following the protocol previously described by Ulloa et al., 2016. Purified nanoparticles were used for all characterization experiments. As shown in Fig. 3A, the absorbance profile of purified QDs presents a small peak at 360 nm characteristic of biosynthesized QDs32. Based on these spectra, we calculated the size of the QDs using the Henglein’s empirical model, which relates the diameter of CdS QDs to the absorption UV-vis spectra48–50. QDs size of 4.4 and 5.0 nm were predicted for NPs biosinthesized at 1 and 3 h, respectively.
Figure 3.
Absorbance, fluorescence and excitation spectra of QDs produced by Halobacillus sp. DS2. (A) Absorbance, emission and excitation spectra of QDs biosynthesized at 1 h. The emission was registered at 450 nm for excitation spectra. (B) Absorbance, fluorescence emission and fluorescence excitation spectra of QDs biosynthesized at 3 h. The emission was registered at 450 nm for excitation spectra.
Also, the fluorescence profile of purified nanoparticles produced by Halobacillus sp. DS2 after 1 or 3 h displayed emission peaks between 420–500 and 440–530 nm, respectively (Fig. 3B). These fluorescence spectra are characteristic of CdS QDs and the differences in emission observed between both QDs are related to differences in the size of the nanocrystal16,17,19,28. Interestingly, a high quantum yield (23.53%) was determined in QDs produced by Halobacillus sp. DS2. This is a desirable property for bioimaging applications that is not commonly observed on biosynthesized QDs16,26,27,32,35. In addition, the excitation spectra of QDs biosynthesized at 1 and 3 h were determined (Fig. 3C). Excitation peaks at 360 and 365 nm, characteristic of CdS QDs, were observed for QDs produced by Halobacillus sp. DS2 after 1 and 3 h synthesis (emission fluorescence was measured at 450 and 500 nm, respectively)32,51,52. In QDs, the fluorescence spectrum is independent of the excitation wavelength due to dissipation of the excitation energy during the excited state (Kasha rules)53.
QDs produced by Halobacillus sp. DS2 are stable at high NaCl concentrations
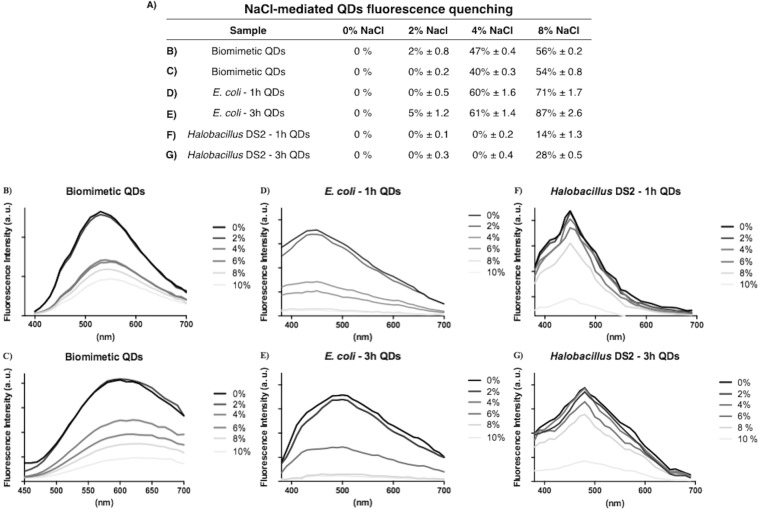
Considering that Halobacillus sp. DS2 is highly resistant to NaCl, and also that this isolate is capable of synthesizing QDs in the presence of different salt concentrations, we decided to evaluate the stability of biosynthesized QDs at high NaCl concentrations. QDs produced by a chemical method and QDs biosynthesized by E. coli were also analyzed as controls. QDs were incubated 30 min with different NaCl concentrations and the effect on QDs-associated fluorescence was determined. As shown in Fig. 4, QDs biosynthesized by the Halobacillus sp. DS2 maintain their fluorescence properties even after exposure to 4% NaCl; no changes on fluorescence intensity were observed at 2 and 4% NaCl (0% fluorescence quenching). In contrast, a quenching in fluorescence higher than 40% was observed when biomimetic QDs or those biosynthesized by E. coli were exposed to 4% NaCl. The exposure to 8% NaCl produces a fluorescence quenching of 14 and 28% on QDs biosynthesized during 1 and 3 h by Halobacillus sp. DS2, respectively. Interestingly, a fluorescence quenching higher than 70% was observed in QDs biosynthesized by E. coli under the same conditions. These results confirm that QDs produced by Halobacillus sp. DS2 are stable at high salt concentrations, a unique characteristic probably related with changes in composition or structure of the nanocrystal produced by the halophilic bacterial isolate.
Figure 4.
CdS QDs biosynthesized by Halobacillus sp. DS2 are stable at high NaCl concentrations. (A) Effect of different NaCl concentrations on the fluorescence emission intensity (percentage of quenching) and spectra of green and red QDs synthesized by a chemical method (B,C), E. coli (D,E) or Halobacillus sp. DS2 (F,G).
Characterization of QDs
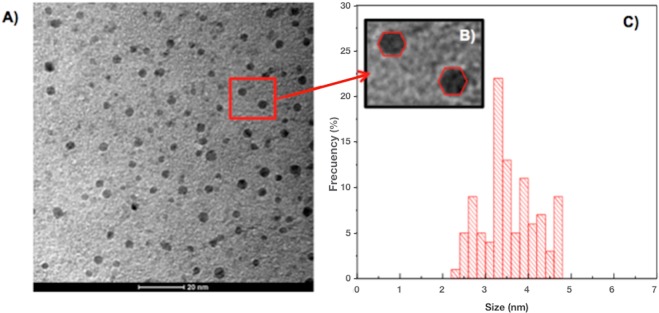
Extracellular CdS-QDs produced after 1 h synthesis were purified and characterized by TEM (Fig. 5A). The electron micrograph shows that NPs are uniformly distributed in an amorphous matrix of low crystallinity. A digital zoom of the TEM image shows that these QDs are regular polyhedra (Fig. 5B), similar to models of hexagonal NPs54. Figure 5C show the size frequency histogram and the average size of biosynthesized NPs (3.56 nm).
Figure 5.
TEM analysis of purified CdS QDs biosynthesized by Halobacillus sp. DS2. (A) Representative TEM image of biosynthesized CdS QDs. (B) Digital zoom of the TEM image of QDs. (C) Frequency size histogram of biosynthesized QDs.
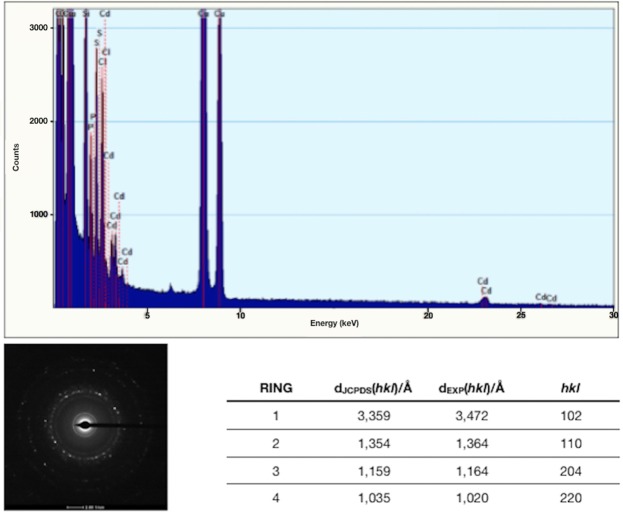
Energy dispersive X-ray spectroscopy (EDS) analysis showed that QDs are composed of cadmium and sulfide, while the amorphous matrix contains carbon, oxygen, phosphorus and chlorine (Fig. 6A,B). Most of these elements are characteristic of biomolecules bound to the nanocrystal. By electron diffraction we obtained patterns with the interplanar distances of biosynthesized QDs (Fig. 6C). The SAED pattern shows that biosynthesized NPs have a crystalline structure with interplanar distances of d1 = 0.245 nm, d2 = 0.207 nm, d3 = 0.123 and d4 = 0,104 nm. This is in agreement with previously reported data indicating that these interplanar distances correspond to the structure of CdS nanocrystals, specifically polycrystalline wurtzite (hexagonal) nanoparticles55. Four diffraction rings consistent with spacings corresponding to a hexagonal phase of cadmium sulfide were observed in the NPs produced by Halobacillus sp. DS2.
Figure 6.
EDS characterization of purified CdS QDs biosynthesized by Halobacillus sp. DS2. (A) EDS of biosynthesized QDs. (B) Electron diffraction pattern of QDs. (C) Interplanar spaces of biosynthesized QDs and those corresponding to hexagonal CdS of the JCPDS 41-1049 file.
The FTIR spectrum for CdS-QDs and cysteine are shown in Fig. 7. In both spectra, a peak close to 1575 cm−1 corresponding to carboxylate group is observed. However, for both spectra, this peak is shifted to lower wavelengths, between 1570 and 1690 cm−1. This shift can be due to the formation of a amino-acid Zwitterion55. In that case, the carboxilate group presents a resonance effect in which both C-O bonds share the electron charge of the double bond of C=O, then the bond multiplicity is lower and the vibration frequency changes. The characteristic band for N–H stretching modes is observed at 3346 and 3250 cm−1 indicating the presence of an NH2 group. Also, the S-H stretching mode is clearly seen at 2550 cm−1, as expected for cysteine. Interestingly, this band is not present in the spectrum of CdS–QDs. This is likely the result of covalent bond formation between the thiol and Cd at the surface of CdS-QDs.
Figure 7.
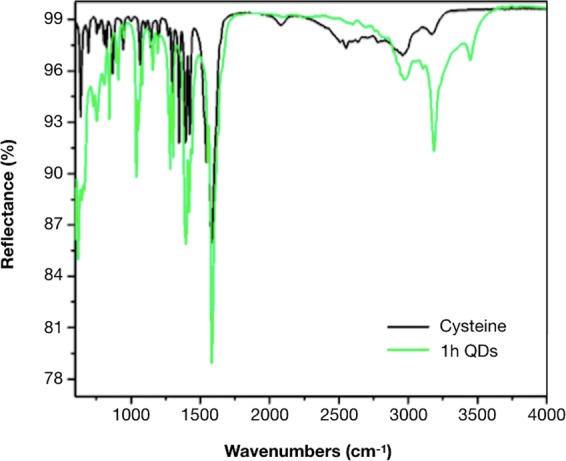
FTIR analysis of CdS QDs biosynthesized by Halobacillus sp. DS2. FTIR spectrum of cysteine (A) and biosynthesized QDs (B).
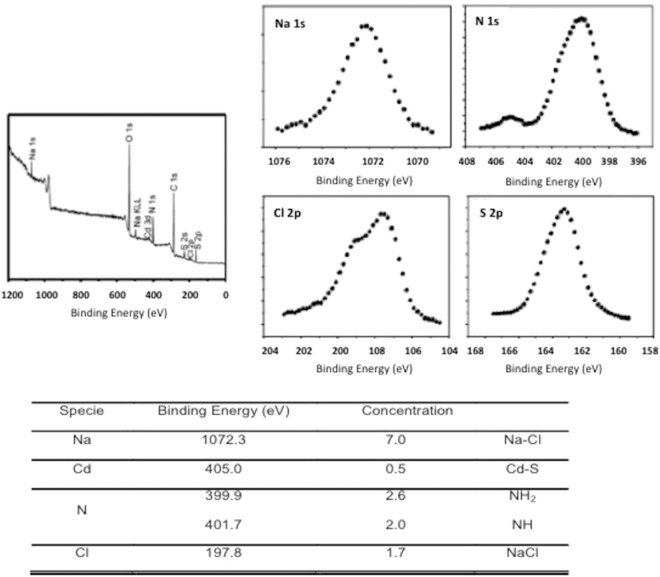
An HRXPS characterization was performed in biosynthesized QDs. The full spectrum shows the presence of several chemical species (Fig. 8). The identification of the binding energies is linked with the different molecules and the NPs in the sample. Sodium and chlorine, Na1s and Cl2p respectively, are mainly attributed to NaCl. The fit of the sulphur S2p revealed two components that can be attributed to CdS NPs and cysteine molecules. Finally, the nitrogen N1s shows two peaks corresponding to NH2 and NH present in cysteine.
Figure 8.
HRXPS characterization of CdS-QDs biosynthesized by Halobacillus sp. DS2.
Discussion
Fifteen salt-resistant bacteria were isolated from three different extreme environments characterized by the presence of high salt concentrations. Growth of bacterial isolates at different salt concentrations and pHs were evaluated, being most of them able to grow in a wide range of pHs (particularly acidic) and salt concentrations (3–25%).
Several reports have described that extreme halophiles belonging to different phylogenetic phyla such as Actinobacteria, Bacteroidetes and Alpha-proteobacteria exhibit unique physiological and nutritional properties56. The halophilic isolates studied here grow in medium supplemented with up to 15% NaCl and given that they are able to grow optimally at NaCl concentrations higher than 10% (1.7 M) they can be categorized as extreme halophiles57. Other halophiles are able to grow optimally at lower salt concentrations, like those present in the sea (3.5%, ~0.6 M)56,58.
The extremophile strain DS2 was able to synthesize QDs, grew at high NaCl concentrations and exhibited resistance to CdCl2 (1.375 mM). The 16S rRNA gene analysis revealed that the DS2 isolate displays high identity with Halobacillus sp. Several Halobacillus isolates have been described to date, most of them displaying high tolerance to NaCl (1–25%) and heavy metals45. Additionally, the capacity to grow at pH ranges between 5–8 was reported on one isolate obtained from a salt mine in Pakistan59.
The Halobacillus sp. DS2 isolate was also able to grow in a wide range of pHs (pH 1–9). Based on these results, the Halobacillus sp. DS2 isolate can be classified as polyextremophile (acidophilic extreme halophile), since it can grow at acidic pHs (pH 1–6), high concentrations of NaCl and can tolerate elevated concentrations of the heavy metal cadmium60.
Three halophilic isolates were able to synthesize CdS quantum dots in presence of high NaCl concentrations (6–8%); a result somehow surprising since the biosynthesis of QDs under high salinity concentrations has not been reported before. E. coli (a classical model of QDs biosynthesis) is unable to biosynthesize QDs in presence of NaCl 4% or higher (Fig. 1). This result is related at least in part to the inability of E. coli to produce H2S in presence of high NaCl concentrations as determined in Fig. 2.
A few reports have shown the effect of Na+ over the cellular production of H2S. In cyanobacteria and plants it has been described that sodium toxicity is due in part to the ability of this ion to interact with the 3-phosphoadenosine-5-phosphatase (PAPase) and to inhibit their activity removing the toxic intermediary of S-assimilation, 3-phosphoadenosine-5-phosphate (PAP)61,62. PAP accumulation in E. coli could be related to a reduction of H2S production through the enzyme PAPs reductase (encoded by cysH in E. coli) that catalyzes the production of sulfite. PAP accumulation generates a product inhibition of PAP reductase, thus decreasing the formation of sulfite, a substrate required for the formation of H2S by the sulfite reductase enzyme (encoded by cysJI and cysG in E. coli).
In previous reports it has been described the existence of sodium-tolerant PAPases in halophilic microorganisms, such as the case of halA from the cyanobacterium Arthospira platensis62. Based on the results obtained in cultures amended with cysteine, all isolates analyzed have desulfhydrase enzymes (EC 4.4.1.15), a large family of enzymes that catalyze the conversion of cysteine in sulfide, ammonium and pyruvate, and vice versa. The effect of NaCl on the cysteine desulfhydrase of the halophilic strain Halobacterium salinarum has been previously studied63. It was shown that the purified desulfhydrase enzyme from H. salinarum has a lower sensitivity to NaCl than the homologous enzyme from E. coli63.
QDs produced by Halobacillus sp. DS2 presented high fluorescence stability under osmotic stress conditions, particularly when compared with chemical QDs and biological QDs synthesized by E. coli (Fig. 2). To date, the biological production of QDs with novel or improved properties has been scarcely studied. Our group recently reported the biosynthesis of QDs with increased tolerance to acidic conditions by using acidophilic extremophile bacteria35,36. We also determined that metal resistant psychrotolerant bacteria isolated from Antarctica could synthesize Cd-QDs at low temperatures26,27. Also, the use of Antarctic bacteria to biosynthesize CdS QDs in presence of other toxic metal contaminants (arsenic) was recently reported as a new alternative for bioremediation64. The cellular processes involved in the synthesis of NPs in all these bacteria isolated from extreme environments have not been determined, however, the importance of sulfide on the biosynthesis process has been established26,27,35,64.
The novel properties of QDs produced by extremophile bacteria are most probably related to the generation of nanocrystals with novel compositions and/or structures, probably as consequence of the presence/activity of cellular biomolecules. This last point has been scarcely studied in QDs produced through biological approaches. Because of this, in the present work we performed a detailed chemical and structural characterization of the CdS QDs produced by halophilic bacteria.
The electron microscopy image shows that CdS QDs have an average size of 3.6 ± 0.8 nm with low size polydispersity. Hexagonal NPs structures were observed in QDs biosynthesized by Halobacillus sp. DS2 (Fig. 4B). QDs display a crystalline structure with interplanar distances corresponding to CdS nanocrystals with polycrystalline wurtzite (hexagonal). The high structural and size definition of QDs is not commonly obtained in the biological synthesis of nanocrystals, where most nanostructures present high polydispersity and poorly defined structure models. These structural properties are in agreement with the spectroscopic properties of biosynthesized QDs.
In general, a drawback of CdS QDs biosynthesized by bacteria is the broad emission spectra that they present, a characteristic not desirable for fluorescence-based applications. The emission spectra of QDs produced by Halobacillus sp. DS2 after 1 and 3 h have full width at half maximum (FWHM) values of 85 and 110 nm, respectively. These values are higher than those obtained in QDs produced by some chemical methods (below 40 nm) but are lower than FWHM values reported for CdS QDs synthesized by bacteria. CdS QDs produced by E. coli, Antarctic Pseudomonas strains and acidophilic bacteria present FWHM values of 200, 215–225 and 220 nm, respectively26,35. The emission spectra of QDs biosynthesized by Halobacillus sp. DS2 are most probably consequence of the presence of organic or inorganic components on the surface of the nanocrystal. In this context, emission espectra of QDs produced by Halobacillus sp. DS2 are favored by the presence of NaCl during biosynthesis (Fig. S1). The unusual sharp maximum on the emission spectra of QDs biosynthesized in presence of NaCl was exacerbated at higher salt concentrations (4, 6 and 8%). Altogether, spectroscopic characteristics of biosynthesized QDs do not confirm the excitonic nature of nanoparticles and are probably consequence of trap-states on the QDs surface.
The chemical composition of biosynthesized QDs was determined by EDS and FT-IR. Obtained results indicate that CdS QDs are covered with an organic layer in which elements such as oxygen, carbon and phosphorus, probably come from proteins produced by Halobacillus sp. DS2 or from the cysteine molecule used to stimulate the production of H2S (Fig. 2). All these components have been previously described in other biological methods for QDs production35. The presence of chlorine is also expected in cells exposed to high concentrations of NaCl, mostly because it is a small anion of easy adsorption on reactive surfaces. The FTIR analysis of the organic composition of QDs indicates the presence of groups that most probably correspond to cysteine residues or protein molecules bound to the CdS nanocrystal through an S-Cd linkage (Fig. 7).
The HRXPS confirmed the presence of CdS and cysteine in the nanocrystals produced by halophilic bacteria. Interestingly, the presence of sodium and chlorine, mainly attributed to NaCl was also observed in QDs. NaCl presence is a novel characteristic of biosynthesized QDs, and is most probably consequence of NaCl interaction with biomolecules composing the organic capping of QDs like proteins or peptides.
Altogether, presented results indicate that the polyextremophyle bacterium Halobacillus sp. DS2 can synthesize CdS nanoparticles in presence of high NaCl concentrations in a process that is related, at least in part, to their capacity to generate S2− in these conditions. Interestingly, QDs produced by halophilic bacteria display unique properties when compared with other biological QDs, such as narrow emission spectra (FWHM) and stability at high NaCl concentrations. The novel properties of these NPs are most probably consequence of the presence of NaCl and cysteine. Experiments to confirm this possibility are currently being developed in our lab.
Finally, the results of our work open new possibilities in the field of QDs biosynthesis and confirm the great potential of using microorganisms with the capacity to tolerate extreme conditions (osmolarity, acidity, temperature or metal stress, among many others) to synthesize nanoparticles with properties not easily found in nanomaterials produced by conventional methods (chemical or biological). Understanding the process of biosynthesis and determining the molecules involved will contribute to the development of new methods to produce nanomaterials with the specific properties that different industrial applications require.
Methods
Sample collection
Samples of soil, sediments and water were collected from different sites in the Atacama Salt Flat, Uyuni Salt Flat and the Dead Sea. Samples were stored at 4 °C until their processing.
Isolation of bacterial strains
Samples (100 mg of soil or 100 μL of water) were suspended in 1 mL sterile distilled water (final volume) and vigorously stirred by vortexing (30 s). After that, 10 μL of each solution were used to inoculate 990 μL of LB culture medium and incubated at 28 °C. After 24 h, aliquots were seeded on LB agar plates and incubated at the same temperature for 24–48 h.
Selection of salt-resistant bacteria
Resistant bacteria were selected by plating on LB agar plates supplemented with NaCl (8%). Bacterial growth at 28 °C was monitored after 24 and 48 h.
Minimal Inhibitory Concentration (MIC)
MICs were determined using the protocol previously described by Elías et al.65. Solutions containing 5.5 mM CdCl2 were prepared in LB medium. Serial dilutions were set in 96-well microplates and inoculated with 5 μL of a previously grown bacterial culture. Microplates were incubated at 28 °C and bacterial growth was assessed after 24 h.
Optimal growth temperature
Bacterial cultures were grown at 28 °C overnight and then diluted to an optic density at 600 nm (OD600) ~0.2. Serial dilutions were prepared and 5 μL of each dilution were used to inoculate LB + NaCl 8%-agar plates. Plates were incubated at 4, 15, 28 and 37 °C and bacterial growth was determined after 24 h.
Bacterial growth in presence of NaCl
Bacterial cultures were grown at 28 °C overnight and then diluted to OD600~0.2. Serial dilutions were prepared and 5 μL of each dilution were used to inoculate LB-agar plates supplemented with 0, 2, 4, 8, 15 and 22% NaCl. Plates were incubated at 28 °C and bacterial growth was evaluated after 24 h.
Effect of pH on bacterial growth
Bacterial cultures were grown overnight at 28 °C and then diluted to an OD600~0.2. 5 μL of each dilution were used to inoculate tubes with LB adjusted at several pHs (1–10). Tubes were incubated at 28 °C and bacterial growth was determined after 24 h.
Sulfide detection assay
The generation of hydrogen sulfide in bacterial cultures was determined using lead acetate soaked papers as described by Shatalin et al.66. The assay was performed using 1 mL of bacterial cultures grown in 2 mL Eppendorf tubes to which a lead acetate paper was attached under the cap. Briefly, bacterial cultures were grown in 990 μL LB supplemented with several NaCl concentrations (0, 2, 4 and 8%) and grown at 28 °C until OD600~0.6. Then, 0.33 mM CdCl2 and 1 mM cysteine were added and the tubes were covered with a paper embedded in lead acetate (100 mM). Tubes were incubated at 28 °C and sulfide generation was determined after 6 h. Controls were performed in the absence of cysteine.
Chemical synthesis of CdS nanoparticles
Chemical QDs were used as control for the assays of CdS stability at high NaCl concentrations. The synthesis at 1 h and 3 h of fluorescent nanoparticles was carried out at 90 °C using the protocol previously described by Pérez-Donoso et al.17.
Intracellular biosynthesis of CdS nanoparticles
A bacterial pre-inoculum was used to inoculate (1:100) LB media supplemented with NaCl 0, 2, 4, 6 or 8% with or without 0.33 mM CdCl2. Then cultures were grown at 28 °C for 24 h. Samples were centrifuged 5 min at 10,000 x g, and the fluorescence of cell pellets and supernatants was evaluated after excitation at 360 nm using a short wave UV-transilluminator. E. coli was used as control.
Extracellular biosynthesis of CdS nanoparticles
Bacteria were grown 24 h at 28 °C in LB media supplemented with NaCl 8%. Samples of 1 mL were then centrifuged 5 min at 10,000 x g and supernatants were discarded. Cells were suspended in buffer Tris-borax citrate 50 mM pH 8.0 amended with different NaCl concentrations (0, 2, 4, 6 and 8%). These solutions were supplemented with 2 mM cysteine in presence of 0.33 mM CdCl2. The fluorescence of supernatants was evaluated after excitation at 360 nm using a short wave UV-transilluminator as described before26,27,32.
Purification of biosynthesized QDs
Extracellular QDs were purified from cell culture supernatants. Supernatants were filtrated using a 0.22 μm filter. Then, the nanoparticles were precipitated with a 1:1 (v/v) ethanol:water solution and mixed by vortexing32. Samples were incubated at 4 °C for 12 h, centrifuged 10 min at 10,000 x g and the supernatants discarded. Obtained NPs were dialyzed for 1 h with Tris-HCl pH 7.0 (100 mM), washed and concentrated using a 3 kDa filter (Millipore).
Fluorescence of purified nanoparticles
Fluorescence and excitation spectra of purified nanoparticles were determined by using a multiplate reader, Synergy H1 (Biotek). Emission spectrum of purified nanoparticles was obtained after excitation at 360 nm. The excitation spectra of purified nanoparticles were obtained by measuring the fluorescence at 450 nm as described previously27,32.
The Quantum Yield of purified CdS QDs was determined. For this purpose, the fluorescence of 4 samples of QDs with different absorbances (between 0.01 and 0.1) was evaluated (excitation at 360 nm). Also, we use Nile Blue dissolved in ethanol (QY = 0.27) for this determination. Fluorescence spectra were measured to obtain the integrated fluorescence intensity (IFI, is defined as the area of the fluorescence spectrum) and this value was plotted versus the absorbance of the solution. The slope of both curves (m) and the refractive index of the solvents (n) (water: 1.333 and ethanol: 1.335) were used to calculate the QY of CdS QDs considering Nile blue as reference (R).The following equation was used16,67:
Transmission electron microscopy (TEM)
TEM measurements were performed using a FEI Tecnai G2 F20 S-Twin microscope, operated at 200 kV. A drop of the dispersed sample was left to dry out on a commercial carbon coated Cu TEM grid. TEM images were processed and analyzed with Digital Micrograph 3.9.0 (Gatan Inc) and The Gimp 2.4.0 software packages. A statistical study of TEM images was carried out to quantify nanoparticles sizes. This study consists of the size measurement (diameter) of about 200 particles per sample. The counts were then plotted as frequency histograms and the mean particle size was calculated. In addition, samples were chemically characterized by Energy-dispersive X-ray spectroscopy (EDX) and electron diffraction (ED).
Fourier Transform Infrared Spectroscopy (FT-IR)
FT-IR was performed to purified QDs. Samples were dried using a vacuum centrifugal concentrator (IR Concentrator Micro-Cenvac NB-503CIR, N-Biotek Inc.) for 24 h. Then FT-IR spectra were determined using a NicoletTM iSTM10 FT-IR Spectrometer (Thermo Scientific Inc.) with a Smart iTRTM Attenuated Total Reflectance (ATR) accessory provided with a single bounce Ge crystal. The scan frequency was 4000 to 700 cm−1.
X-ray photoelectron spectroscopy (XPS)
The chemical composition of biosynthesized NPs was studied by means of chemical binding energy of the elements through X-ray photoelectron spectroscopy technique (XPS). An XPS–Auger PerkinElmer spectrometer model PHI 1257 that includes an ultra-high vacuum chamber, a hemispheric electron energy analyzer and an X-ray source, with Kα radiation unfiltered from an Al (hν = 1486.6 eV) anode were used. The measurements were performed at 200 W using an emission angle of 70 to obtain information from the deep surface.
Supplementary information
Acknowledgements
This work was supported by Erika Elcira Donoso Lopez, FONDECYT 1151255 (JP-D), INACH RT-25_16 (JP-D, DB) and UNAB DI 488-14/R (JP-D).
Author Contributions
B.C., D.B. and J.P.-D. conceived and designed the study. N.B., P.C., N.O.-A., N.D.-S. and B.C. performed Microbiology experiments and analysed data. M.F. performed the HRXRD experiments. R.E.-G. developed the TEM studies. A.T. performed the XRD and FT-IR experiments. B.C. and J.P.-D. wrote the paper. All authors contributed to the scientific discussion and revision of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-38330-8.
References
- 1.Fulekar, M. H. Bioremediation Technology for Hazardous Wastes-Recent Advances. Bioremediation Technology. 135–166 (2010).
- 2.Zhou M, Ghosh I. Quantum dots and peptides: a bright future together. Biopolymers. 2007;88:325–339. doi: 10.1002/bip.20655. [DOI] [PubMed] [Google Scholar]
- 3.Michalet X, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huynh WU, Dittmer JJ, Alivisatos AP. Hybrid nanorod-polymer solar cells. Science. 2002;295:2425–2427. doi: 10.1126/science.1069156. [DOI] [PubMed] [Google Scholar]
- 5.Kinia G, et al. Salt- and temperature-stable quantum dot nanoparticles for porous media flow. Colloids and Surfaces A: Physicochem. Eng. Aspects. 2014;443:492–500. doi: 10.1016/j.colsurfa.2013.11.042. [DOI] [Google Scholar]
- 6.Ga Gaponik NP, Talapin DV, Rogach AL. A light-emitting device based on a CdTe nanocrystal/polyaniline composite. Phys. Chem. Chem. Phys. 1999;1:1787–1789. doi: 10.1039/a808619b. [DOI] [Google Scholar]
- 7.Órdenes-Aenishanslins N, et al. Use of titanium dioxide nanoparticles biosynthesized by Bacillus mycoides in Quantum Dot Sensitized Solar Cells. Microbial Cell Factories. 2014;13:90. doi: 10.1186/s12934-014-0090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mo D, et al. Cadmium-containing quantum dots: properties, applications, and toxicity. Applied Microbiology and Biotechnology. 2017;101:2713–2733. doi: 10.1007/s00253-017-8140-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Long Y, Zhang Y, Zhong X, Zhu L. Preparation of highly luminescent CdTe/CdS core/shell quantum dots. Chem. Phys. Chem. 2009;9:680–685. doi: 10.1002/cphc.200800672. [DOI] [PubMed] [Google Scholar]
- 10.Peng ZA, Peng X. Formation of high-quality CdTe, CdSe, and CdS nanocrystals using CdO as precursor. J. Am. Chem. Soc. 2001;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- 11.Durán-Toro V, et al. Simple quantum dot-based assay for sensitive and specific quantification of Cu2+ in bacterial cell culture. Analytical Biochemistry. 2014;450:30–36. doi: 10.1016/j.ab.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Clapp A. Overview of stabilizing ligands for biocompatible quantum dot nanocrystals. Sensors. 2011;11:11036–11055. doi: 10.3390/s111211036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh G, et al. Cancer cell targeting using folic Acid/Anti-HER2 antibody conjugated Fluorescent CdSe/CdS/ZnS-MPA and CdTe-MSA Quantum Dots. Journal of Nanoscience and Nanotechnology. 2015;15:9382–9395. doi: 10.1166/jnn.2015.10767. [DOI] [PubMed] [Google Scholar]
- 14.Díaz V, et al. Spectroscopic properties and biocompatibility studies of CdTe Quantum Dots capped with biological thiols. Science of Advanced Materials. 2012;4:609–616. doi: 10.1166/sam.2012.1327. [DOI] [Google Scholar]
- 15.Hardman R. A toxicologic review of quantum dots: toxicity depends on physicochemical and environmental factors. Environmental health perspectives. 2006;114:165. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venegas FA, et al. Biological phosphorylated molecules participate in the biomimetic and biological synthesis of cadmium sulphide quantum dots by promoting H2S release from cellular thiols. RSC Advances. 2017;7:40270–40278. doi: 10.1039/C7RA03578K. [DOI] [Google Scholar]
- 17.Pérez-Donoso JM, et al. Biomimetic, mild chemical synthesis of CdTe-GSH quantum dots with improved biocompatibility. PLoS One. 2012;7:e30741. doi: 10.1371/journal.pone.0030741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aryal BP, Benson DE. Polyhistidine fusion proteins can nucleate the growth of CdSe nanoparticles. Bioconjugate Chem. 2007;18:585–589. doi: 10.1021/bc060277+. [DOI] [PubMed] [Google Scholar]
- 19.Bao H, Wang E, Dong S. One-Pot synthesis of CdTe nanocrystals and shape control of luminescent CdTe–Cystine nanocomposites. Small. 2006;2:476–480. doi: 10.1002/smll.200500346. [DOI] [PubMed] [Google Scholar]
- 20.Li LL, Cui YH, Chen JJ, Yu HQ. Roles of glutathione and L-cysteine in the biomimetic green synthesis of CdSe quantum dots. Frontiers of Environmental Science & Engineering. 2017;11:7. doi: 10.1007/s11783-017-0948-0. [DOI] [Google Scholar]
- 21.He X, Ma N. Biomimetic Synthesis of Fluorogenic Quantum Dots for Ultrasensitive Label-Free Detection of Protease Activities. Small. 2013;9:2527–2531. doi: 10.1002/smll.201202570. [DOI] [PubMed] [Google Scholar]
- 22.Fariq, A., Khan, T. & Yasmin, A. Microbial synthesis of nanoparticles and their potential applications in biomedicine. Journal of Applied Biomedicine (2017).
- 23.Hulkoti NI, Taranath TC. Biosynthesis of nanoparticles using microbes-a review. Colloids and Surfaces B: Biointerfaces. 2014;121:474–483. doi: 10.1016/j.colsurfb.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 24.Bao H, et al. Extracellular microbial synthesis of biocompatible CdTe quantum dots. Acta Biomater. 2010;6:3534–3541. doi: 10.1016/j.actbio.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Jacob JM, Lens PN, Balakrishnan RM. Microbial synthesis of chalcogenide semiconductor nanoparticles: a review. Microbial Biotechnology. 2016;9:11–21. doi: 10.1111/1751-7915.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plaza DO, Gallardo C, Straub YD, Bravo D, Pérez-Donoso JM. Biological synthesis of fluorescent nanoparticles by cadmium and tellurite resistant Antarctic bacteria: exploring novel natural nanofactories. Microbial Cell Factories. 2016;15:1. doi: 10.1186/s12934-016-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallardo C, et al. Low-temperature biosynthesis of fluorescent semiconductor nanoparticles (CdS) by oxidative stress resistant Antarctic bacteria. Journal of Biotechnology. 2014;187:108–115. doi: 10.1016/j.jbiotec.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Mi C, et al. Biosynthesis and characterization of CdS quantum dots in genetically engineered Escherichia coli. Journal of biotechnology. 2011;153:125–132. doi: 10.1016/j.jbiotec.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park TJ, Lee SY, Heo NS, Seo TS. In vivo synthesis of diverse metal nanoparticles by recombinant Escherichia coli. Angewandte Chemie International Edition. 2010;49:7019–7024. doi: 10.1002/anie.201001524. [DOI] [PubMed] [Google Scholar]
- 30.Chen YL, et al. Augmented biosynthesis of cadmium sulfide nanoparticles by genetically engineered Escherichia coli. Biotechnology progress. 2009;25:1260–1266. doi: 10.1002/btpr.199. [DOI] [PubMed] [Google Scholar]
- 31.Kang SH, Bozhilov KN, Myung NV, Mulchandani A, Chen W. Microbial synthesis of CdS nanocrystals in genetically engineered E. Coli Angewandte Chemie International Edition. 2008;47:5186–5189. doi: 10.1002/anie.200705806. [DOI] [PubMed] [Google Scholar]
- 32.Monrás JP, et al. Enhanced glutathione content allows the in vivo synthesis of fluorescent CdTe nanoparticles by Escherichia coli. PLoS One. 2012;7:e48657. doi: 10.1371/journal.pone.0048657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai H, Zhang Z, Guo Y, Jia W. Biological Synthesis of Size-Controlled Cadmium Sulfide Nanoparticles Using ImmobilizedRhodobacter sphaeroides. Nanoscale research letters. 2009;4:717. doi: 10.1007/s11671-009-9303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasula RR, Lim S. Engineering nanoparticles synthesis using microbial. Eng. Biol. 2017;1:12–17. doi: 10.1049/enb.2017.0009. [DOI] [Google Scholar]
- 35.Ulloa G, et al. Use of acidophilic bacteria from the Acidithiobacillus genus to biosynthesize CdS fluorescent nanoparticles (quantum dots) with high tolerance to acidic pH”. Enzymes and Microbial Technology. 2016;95:217–224. doi: 10.1016/j.enzmictec.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Ulloa G, et al. Phosphate favors the biosynthesis of CdS quantum dots in Acidithiobacillus thiooxidans ATCC 19703 by improving metal uptake and tolerance. Frontiers in Microbiology. 2018;9:234. doi: 10.3389/fmicb.2018.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oren, A. Halophilic microorganisms and their environments. Springer Science & Business Media5 (2002).
- 38.Kamekura M, Dyall-Smith ML. Taxonomy of the family halobacteriacea and the description of two new genera Halorubrobacterium and Natrialba. J. Gen. Appl. Microbiol. 1995;41:333–350. doi: 10.2323/jgam.41.333. [DOI] [Google Scholar]
- 39.Kushner DJ, Kamekura M. Physiology of halophilic eubacteria. In: Rodriguez-Valera, F (ed.), Halophilic bacteria. CRC Press. Boca Raton, FL. 1988;1:109–138. [Google Scholar]
- 40.Shi W, Takano T, Liu S. Isolation and characterization of novel bacterial taxa from extreme alkali-saline soil. World Journal of Microbiology & Biotechnology. 2012;28:2147–2157. doi: 10.1007/s11274-012-1020-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhou. M, Chen. W, Chen. H, Wei. G. Draft genome sequence of Mesorhizobium alhagi CCNWXJ12–2(Tau), a novel salt-resistant species isolated from the desert of north-western China. Journal of Bacteriology. 2012;194:1261–1262. doi: 10.1128/JB.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abou-Elela SI, Kamel MM, Fawzy ME. Biological treatment of saline wastewater using a salt-tolerant microorganism. Desalination. 2010;250:1–5. doi: 10.1016/j.desal.2009.03.022. [DOI] [Google Scholar]
- 43.Ruppel S, Franken P, Witzel K. Properties of the halophyte microbiome and their implications for plant salt tolerance. Functional Plant Biology. 2013;40:940–951. doi: 10.1071/FP12355. [DOI] [PubMed] [Google Scholar]
- 44.Kostmar C, et al. Stable citrate-coated iron oxide superparamagnetic nanoclusters at high salinity. Ind. Eng. Chem. Res. 2010;49:12435–12443. doi: 10.1021/ie1010965. [DOI] [Google Scholar]
- 45.Desale P, Patel B, Singh S, Malhotra A, Nawani N. Plant growth promoting properties of Halobacillus sp. and Halomonas sp. in presence of salinity and heavy metals. Journal of Basic Microbiology. 2014;54:781–791. doi: 10.1002/jobm.201200778. [DOI] [PubMed] [Google Scholar]
- 46.Awano N, Wada M, Mori H, Nakamori S, Takagi H. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Applied and environmental microbiology. 2005;71:4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunleavy R, Lu L, Kiely CJ, McIntosh S, Berger BW. Single-enzyme biomineralization of cadmium sulfide nanocrystals with controlled optical properties. Proceedings of the National Academy of Sciences. 2016;113:5275–5280. doi: 10.1073/pnas.1523633113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He R, et al. Formation of monodispersed PVP-capped ZnS and CdS nanocrystals under microwave irradiation. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2003;220:151–157. doi: 10.1016/S0927-7757(03)00072-4. [DOI] [Google Scholar]
- 49.Mansur HS, Mansur AA, González JC. Synthesis and characterization of CdS quantum dots with carboxylic-functionalized poly (vinyl alcohol) for bioconjugation. Polymer. 2011;52:1045–1054. doi: 10.1016/j.polymer.2011.01.004. [DOI] [Google Scholar]
- 50.Weller H, et al. Photochemistry of colloidal semiconductors. Onset of light absorption as a function of size of extremely small CdS particles. Chemical physics letters. 1986;124(6):557–560. doi: 10.1016/0009-2614(86)85075-8. [DOI] [Google Scholar]
- 51.Lakowicz JR, Gryczynski I, Piszczek G, Murphy CJ. Emission spectral properties of cadmium sulfide nanoparticles with multiphoton excitation. The Journal of Physical Chemistry B. 2002;106:5365–5370. doi: 10.1021/jp0134953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang SH, Bozhilov KN, Myung NV, Mulchandani A, Chen W. Microbial synthesis of CdS nanocrystals in genetically engineered E. coli. Angewandte Chemie International Edition. 2008;47:5186–5189. doi: 10.1002/anie.200705806. [DOI] [PubMed] [Google Scholar]
- 53.Kasha M. Characterization of electronic transitions in complex molecules. Discussions of the Faraday society. 1950;9:14–19. doi: 10.1039/df9500900014. [DOI] [Google Scholar]
- 54.Quester. K, Avalos-Borja M, Castro-Longoria E. Biosynthesis and microscopic study of metallic nanoparticles. Micron. 2013;54:1–27. doi: 10.1016/j.micron.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 55.Soomro RA, Nafady A, Memon N, Sherazi TH, Kalwar NH. L-cysteine protected copper nanoparticles as colorimetric sensor for mercuric ions. Talanta. 2014;130:415–422. doi: 10.1016/j.talanta.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 56.Bowers KJ, Mesbah NM, Wiegel J. Biodiversity of poly-extremophilic bacteria: Does combining the extremes of high salt, alkaline pH and elevated temperature approach a physico-chemical boundary for life. Saline Systems. 2009;5:9. doi: 10.1186/1746-1448-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qutub. N, Masood Pirzada. B, Umar. K, Sabir. S. Synthesis of CdS nanoparticles using different sulfide ion precursors: Formation mechanism and photocatalytic degradation of Acid Blue-29. Journal of Environmental Chemical Engineering. 2016;4:808–817. doi: 10.1016/j.jece.2015.10.031. [DOI] [Google Scholar]
- 58.Oren, A. The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology and Biochemistry. 2, 113–164 (2006).
- 59.Roohi A, Ahmed I, Iqbal M, Jamil M. Preliminary isolation and characterization of halotolerant and halophilic bacteria from salt mines of Karak, Pakistan. Pak J Bot. 2012;44:365–370. [Google Scholar]
- 60.Bowers, K. J. & Wiegel, J. Temperature and pH optima of xtremely halophilic archaea: a mini-review. Extremophiles, 15, 119–128. [DOI] [PubMed]
- 61.Zhang JY, et al. A lithium-sensitive and sodium-tolerant 3′-phosphoadenosine-5′-phosphatase encoded by halA from the cyanobacterium Arthrospira platensis is closely related to its counterparts from yeasts and plants. Applied and Environmental Microbiology. 2006;72:245–251. doi: 10.1128/AEM.72.1.245-251.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gil-Mascarell R, López-Coronado JM, Bellés JM, Serrano R, Rodríguez PL. The Arabidopsis HAL2-like gene family includes a novel sodium-sensitive phosphatase. The Plant Journal. 1999;17:373–383. doi: 10.1046/j.1365-313X.1999.00385.x. [DOI] [PubMed] [Google Scholar]
- 63.Seckbach, J. Enigmatic microorganisms and life in extreme environments. Springer Science & Business Media1 (2013).
- 64.Glatstein DA, et al. Arsenic and Cadmium Bioremediation by Antarctic Bacteria Capable of Biosynthesizing CdS Fluorescent Nanoparticles. Journal of Environmental Engineering. 2018;144(3):04017107. doi: 10.1061/(ASCE)EE.1943-7870.0001293. [DOI] [Google Scholar]
- 65.Elías AO, et al. Tellurite enters Escherichia coli mainly through the PitA phosphate transporter. Microbiol. 2012;1:259–267. doi: 10.1002/mbo3.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 67.Sens R, Drexhage KH. Fluorescence quantum yield of oxazine and carbazine laser dyes. Journal of Luminescence. 1981;24:709–712. doi: 10.1016/0022-2313(81)90075-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.