Abstract
Cervical cancer is the fourth most common cancer in women. Although cure rates are high for early stage disease, clinical outcomes for advanced, metastatic, or recurrent disease remain poor. To change this panorama, a deeper understanding of cervical cancer biology and novel study models are needed. Immortalized human cancer cell lines such as HeLa constitute crucial scientific tools, but there are few other cervical cancer cell lines available, limiting our understanding of a disease known for its molecular heterogeneity. This study aimed to establish novel cervical cancer cell lines derived from Brazilian patients. We successfully established one (HCB-514) out of 35 cervical tumors biopsied. We confirmed the phenotype of HCB-514 by verifying its’ epithelial and tumor origin through cytokeratins, EpCAM and p16 staining. It was also HPV-16 positive. Whole-exome sequencing (WES) showed relevant somatic mutations in several genes including BRCA2, TGFBR1 and IRX2. A copy number variation (CNV) analysis by nanostring and WES revealed amplification of genes mainly related to kinases proteins involved in proliferation, migration and cell differentiation, such as EGFR, PIK3CA, and MAPK7. Overexpression of EGFR was confirmed by phospho RTK-array and validated by western blot analysis. Furthermore, the HCB-514 cell line was sensitive to cisplatin. In summary, this novel Brazilian cervical cancer cell line exhibits relevant key molecular features and constitutes a new biological model for pre-clinical studies.
Introduction
Cervical cancer is a major public health problem worldwide, making it the fourth most common type of cancer among women. In 2018, there were 570,000 new cases reported and 311,000 related deaths1. Women between the ages of 50 and 60 years-old are most affected by cervical cancer2. Persistent infection of the basal layer of cervical epithelium with high-risk human papillomavirus (HPV), such as 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68, is considered the main risk factor for the development of cervical cancer precursors, known as cervical intraepithelial neoplasia (CIN 1, 2, and 3), and invasive cervical cancer3.
Cervical cancer can be histologically classified as squamous cell carcinoma (SCC), adenocarcinoma and adenosquamous carcinoma, with a prevalence of 77%, 17% and 6%, respectively4. Many efforts have being undertaken to determine the molecular profile of this heterogeneous disease. Recently, The Cancer Genome Atlas (TCGA) integrated information from CNV, methylation, mRNA, and miRNA profiles. Through clustering, the TCGA revealed three molecular subtypes of cervical cancer: SCC keratin-high, SCC keratin-low and adenocarcinoma. Differences observed in the three major subtypes included the enriched expression of some genes such as PIK3CA, ADH7 and SPRR3 in the SCC keratin-high compared with the SCC keratin-low cluster; more frequent CNVs including common EGFR amplifications in SCCs; a high number of aberrations in tumor-suppressor genes related with TGF-β pathway in adenocarcinomas including SMAD4 and TGFBR2 deletions, and increased DNA methylation in adenocarcinomas4,5.
Cervical cancer treatment is based on the stage of disease. For early stage disease, surgery is the primary treatment modality, cure rates are high, and 5-year overall survival is up to 92%6. For advanced disease, which includes recurrent or metastatic disease, the mainstay of therapy is chemoradiation with a platinum-based agent and unfortunately, treatment responses are poor7. To improve outcomes for patients with advanced disease, recent findings on the molecular profile of this tumor type is valuable.
To facilitate the discovery of new antineoplastic agents, many research centers and teams have been carrying out screenings with a multitude of compounds, testing them in in vitro models, using immortalized human cancer cell lines8. This approach provides controlled conditions to evaluate the efficacy of drugs, and enables the unrestricted availability of human source material. However, there is a very low number of cervical cancer cell lines commercially available in comparison with other tumors, such as breast and lung tumors, which currently provides a limited representation of known subtypes and tumor heterogeneity. Therefore, the aim of this study was to establish and to characterize a new human cervical tumor cell line derived from a Brazilian patient.
Results
Clinical characterization and establishment of a primary cell culture
From March 2016 to June 2017, 35 cervical tumor biopsies were processed (Suppl. Table 1). Only one (2,9%) of the cell cultures, named HCB-514, survived for more than 12 months and continued to grow after several freeze-thaw cycles. This cell line was derived from a 30 year-old patient diagnosed with stage IIB squamous cell carcinoma of the cervix. The patient was treated with concurrent chemoradiation with cisplatin from October 10 to November 17, 2016, and was disease-free through her most recent follow-up appointment, on April 25, 2018. The cell culture HCB-514 grew attached to the flask, with cells forming an irregular island pattern with a cobblestone morphology, characteristic of epithelial cells (Fig. 1). When the cell line became confluent, cells were frozen in 5% DMSO in fetal bovine serum (FBS) solution in liquid nitrogen for further assays. After the fourth passage, immunophenotypic characterization was performed. The HCB-514 cell line presented stable outgrowing for more than 6 months, reaching 26 passages, and it was HPV-positive, supporting a spontaneous immortalization process. The cell line was negative for mycoplasma, and a short tandem repeat (STR) analysis showed that the HCB-514 cell line, tumor tissue and peripheral blood shared the same markers, confirming cell line identity (Table 1).
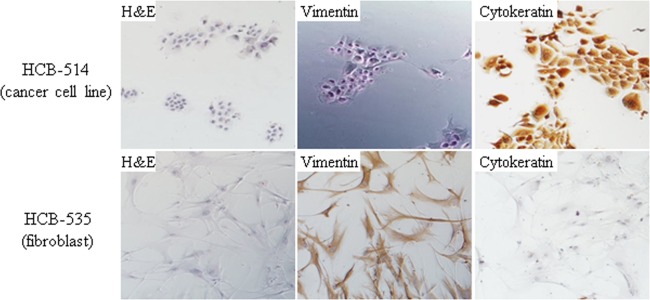
Figure 1.
Representative images of immunocytochemistry of cervical cancer cell line HCB-514 (top images) and the fibroblast cells (HCB-535) (bottom images). All pictures were taken at 100x magnification.
Table 1.
STR profile of cell culture, blood and frozen tissue of the patient.
| Regions | Blood | Tumor | HCB-514 |
|---|---|---|---|
| Amelogenin | X | X | X |
| CSF1PO | 10,12 | 10,12 | 12 |
| D13S317 | 9,11 | 9,11 | 9,11 |
| D16S539 | 9 | 9 | 9 |
| D5S818 | 10,12 | 10,12 | 10 |
| D7S820 | 11,13 | 11,13 | 13 |
| THO1 | 7,9 | 7,9 | 7,9 |
| TPOX | 8,12 | 8,12 | 8,12 |
| WA | 16 | 16 | 16 |
Immunophenotypic characterization
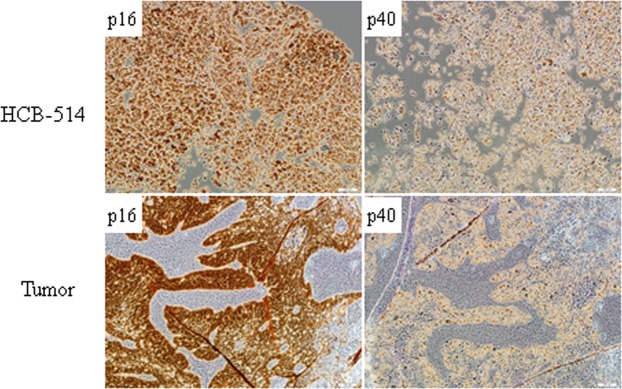
Immunocytochemistry of HCB-514 showed strong cytokeratin expression, but no vimentin expression, similar to SiHa, indicating that HCB-514 is an epithelial cell line (Fig. 1). On the other hand, HCB-535 fibroblast presented the opposite staining pattern, with strong vimentin expression but no cytokeratin expression (Fig. 1). Furthermore, immunohistochemistry from HCB-514 cell block was in accordance with the expression pattern of formalin-fixed paraffin embedded (FFPE) tumor tissue taken in parallel from the patient at the time of biopsy. Both HCB-514 and FFPE tumor tissue were positive for p40, a squamous cell marker, and p16, a marker for cervical cancer (Fig. 2), confirming its squamous cell origin.
Figure 2.
Representative immunohistochemistry images of HCB-514 cell block slices (top images) and the tumor patient tissue (bottom images) depicting expression of p16 (cervical cancer marker) and p40 (squamous cell carcinoma marker). All pictures were taken at 100x magnification.
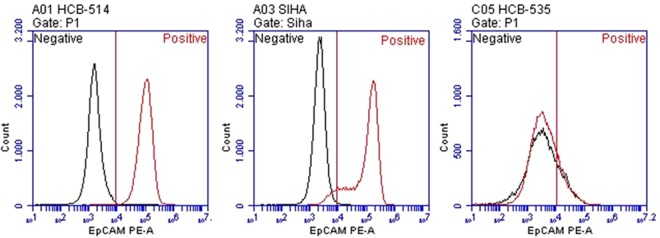
In addition, flow cytometry analysis showed high expression of EpCAM in HCB-514, with 100% of cells staining positive. Expression of EpCAM was present in 92% of SiHa cells and in 19% of the fibroblast cell line (Fig. 3).
Figure 3.
Evaluation of epithelial cellular adhesion molecule (EpCAM) expression in HCB-514. HCB-514 (left), SiHa (center) and HCB-535 (right) cell lines were stained with antibody anti-EpCAM and evaluated in flow cytometry. SiHa, and a fibroblast cell line (HCB-535) were used as positive and negative controls, respectively. Black graphs represent unstained cells; red graphs represent stained cells.
Doubling-time
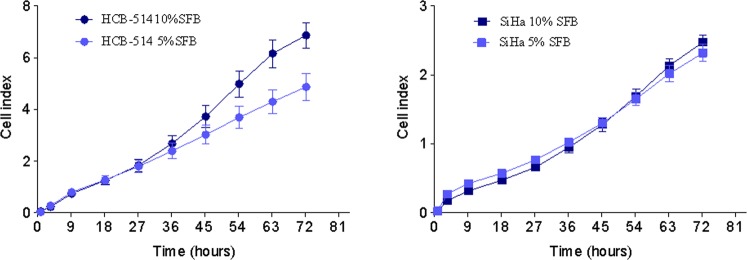
The growth curve for HCB-514 during exponential cell growth showed a doubling time of 18 h in 10% FBS media, and 24 h in 5% FBS media. To evaluate whether this growth was faster or slower than other cell lines largely used for in vitro assays, SiHa was also evaluated and showed a doubling-time of 17 h in 10% FBS media and 21 h in 5% FBS. Thus, the time was similar among cell lines, with a faster doubling-time in 10% than in 5% media (Fig. 4).
Figure 4.
Growth curves of HCB-514 obtained from real time impedance-based technology cell analyzer system (xCELLigence). Different media conditions were assessed. Data represent the mean of 3 independent experiments done in duplicate.
HPV status and genotyping
HPV infection is present in almost all cervical tumors, therefore we evaluated the presence of the virus in the HCB-514 cell line. For this purpose, GP5+/GP6+ primers were used to amplify the highly conserved region of the HPV L1 gene by PCR. The band correspondent to this region was found in HCB-514, confirming the presence of HPV (Suppl. Fig. 1a). To identify which high-risk HPV type was present, a genotyping test was performed with the COBAS® HPV assay, confirming HPV type 16. Furthermore, to confirm that HPV16 infection was present, we evaluated and confirmed the presence of E6 oncoprotein, an important protein responsible for HPV oncogenicity (Suppl. Fig. 1b).
Furthermore, WES allowed us to determine whether the HPV genome integrated into host genomic DNA of HCB-514 and in primary tumor. The results showed two integration sites, one into the MICU2 and the other into CDH13 genes, the last being also found in the WES of patient’s primary tumor.
Moreover, it is hypothesized that HPV genetic variation co-evolves with hosts of certain ethnic groups, which influences disease development and outcome8. Therefore, to identify the variant of HPV in HCB-514 and in the primary cervical cancer, phylogenetic analysis was performed. Both the HCB-514 cell line and the patient’s primary tumor showed the presence of the sublineage HPV16 A1 (Suppl. Fig. 2).
HCB-514 and tumor mutational profile
The WES analysis of paired cell line/blood revealed 520 somatic mutations. The analysis of the most significant non-silent mutations identified 93 mutated genes (Suppl. Table 3), such as HPS-3, IRX2, XPO5, CBWD1, TGFBR1, CUBN, TRHDE, BRCA2, THBS1, TGM5, CCDC22, which were previously described as pathogenic, or likely pathogenic in tumors (Suppl. Table 3).
Regarding the analysis of paired primary tumor/blood, 863 mutations were found, with 68 non-silent mutations identified (Suppl. Table 4). Among them, 18 mutations were shared with HCB-514 (Table 2), including mutations in the BRCA2, TGFBR1, CCDC22 and SHKBP1 genes.
Table 2.
Somatic non-silent mutations found in both HCB-514 and tumor.
| Chromosome | Position | Gene | Referrence allele | Variant allele | Aminoacid | Zygosity |
|---|---|---|---|---|---|---|
| 1 | 228335177 | GUK1 | C | T | p.R129W | Heterozygous |
| 2 | 69049563 | ARHGAP25 | G | A | p.G431E | Heterozygous |
| 5 | 140793112 | PCDHGA10 | G | A | p.E124K | Heterozygous |
| 7 | 77408339 | RSBN1L | A | C | p.K799Q | Heterozygous |
| 9 | 101900171 | TGFBR1 | C | T | p.A202V | Heterozygous |
| 10 | 51130414 | PARG | G | A | p.L59F | Heterozygous |
| 12 | 99194813 | ANKS1B | C | G | p.E1053Q | Heterozygous |
| 12 | 109201563 | SSH1 | G | A | p.R193W | Heterozygous |
| 13 | 32945095 | BRCA2 | G | A | p.W2830* | Heterozygous |
| 15 | 39874463 | THBS1 | G | A | p.R46H | Heterozygous |
| 15 | 43545745 | TGM5 | G | A | p.R215W | Heterozygous |
| 19 | 41084418 | SHKBP1 | G | A | p.R129W | Heterozygous |
| 20 | 4163484 | SMOX | G | A | p.G431E | Heterozygous |
| 20 | 23066708 | CD93 | G | A | p.E124K | Heterozygous |
| 21 | 47421214 | COL6A1 | G | C | p.K799Q | Heterozygous |
| 22 | 17590484 | IL17RA | C | G | p.A202V | Heterozygous |
| 22 | 19213831 | CLTCL1 | C | T | p.L59F | Heterozygous |
| X | 49105196 | CCDC22 | G | T | p.E1053Q | Heterozygous |
Several studies have reinforced the putative impact of APOBEC family of cytidine deaminases enzymes in altered nucleotides found in cervical cancer9. Therefore, mutational signatures from cell line and tumor were analyzed. Both samples showed similar mutation signatures, with C to A and C to T being the most predominant (Suppl. Fig. 3).
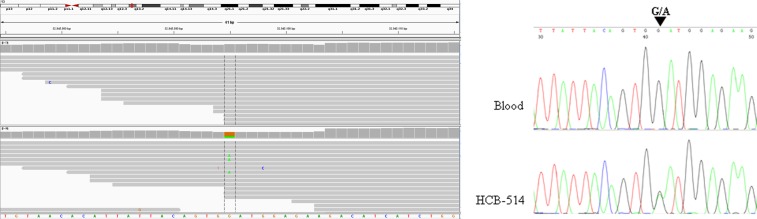
The BRCA2 mutation (p.W2830*) identified was further confirmed by Sanger sequencing (Fig. 5).
Figure 5.
IGV image from the whole-exome sequencing (WES) result of BRCA2 gene (left) and electropherogram of exon 20 from BRCA2 gene that harbors mutation (G → A) (right).
Copy number variation
Nanostring analysis for CNV was performed for both the cell line and constitutive DNA. Among all evaluated genes, 44 somatic alterations were discovered, including 30 regions with amplification, containing genes such as MYC, TP53, EGFR, TERT, AKT3, MAPK7, BIRC2, YAP1 and 14 losses, containing genes including KIT, RB1, BRCA2 and APC (Tables 3 and 4).
Table 3.
Copy number amplification regions in HCB-514 cell line.
| Chromosome Region | CNV | Cytoband | Cancer genes |
|---|---|---|---|
| chr1 | Amplification | p36.33; q32.1 | |
| chr1 | High amplification | q32.1 - q43 | H3F3A, FH |
| chr1 | Amplification | q43 - q44 | AKT3 |
| chr2 | High amplification | p25.3 - p22.3 | MYCN, C2orf44, NCOA1, DNMT3A, ALK |
| chr2 | High amplification | p22.3 - p21 | EML4 |
| chr2 | Amplification | p21 - q33.1 | MSH2, MSH6, FBXO11, BCL11A, REL, XPO1, IGL@, TTL, PAX8, ERCC3, CHN1, HOXD13, HOXD11, NFE2L2, PMS1, SF3B1 |
| chr2 | Amplification | q37.3 | |
| chr3 | High amplification | q11.1 - q26.1 | TFG, CBLB, GATA2, RPN1, FOXL2, WWTR1, GMPS, MLF1 |
| chr3 | High amplification | q26.1 - q29 | EVI1, PIK3CA, SOX2, ETV5, EIF4A2, BCL6, LPP, DCUN1D1, PRKCI |
| chr3 | High amplification | q29 | TFRC |
| chr5 | Amplification | p15.33 - p11 | IL7R, LIFR, SKP2, TERT |
| chr6 | Amplification | q12 | |
| chr7 | Amplification | p22.3 - q11.21 | CARD11, PMS2, ETV1, HNRNPA2B1, HOXA9, HOXA11, HOXA13, JAZF1, IKZF1, EGFR |
| chr8 | Amplification | p23.3 - q24.3 | PCM1, WRN, WHSC1L1, FGFR1, HOOK3, TCEA1, PLAG1, CHCHD7, NCOA2, HEY1, COX6C, EXT1, MYC, NDRG1, RECQL4, C8orf33, C8orf4 |
| chr9 | Amplification | q32 | |
| chr11 | Amplification | q12.3 - q13.4 | MEN1, CCND1, ORAOV1, FADD |
| chr11 | Amplification | q13.4 - q22.1 | NUMA1, PICALM, MAML2, GAB2, YAP1 |
| chr11 | High amplification | q22.1 - q22.3 | BIRC2, BIRC3 |
| chr12 | Amplification | p13.33 | |
| chr12 | Amplification | q23.1 - q24.33 | ALDH2, PTPN11, BCL7A |
| chr14 | Amplification | q11.1 - q32.33 | BCL2L2, CCNB1IP1, TRA@, NKX2-1, NIN, KTN1, GPHN, TSHR, TRIP11, GOLGA5, DICER1, TCL6, TCL1A, BCL11B, AKT1, IGH@ |
| chr15 | Amplification | q21.3 - q26.3 | IGF1R, FLJ27352, TCF12, PML, NTRK3, IDH2, CRTC3, BLM |
| chr16 | Amplification | p13.3 | |
| chr16 | High amplification | p13.3 - p11.2 | TSC2, CREBBP, CIITA, SOCS1, TNFRSF17, ERCC4, MYH11, PALB2, IL21R, FUS |
| chr17 | Amplification | p13.3 - q11.2 | MAPK7, YWHAE, USP6, TP53, PER1, GAS7, MAP2K4, NF1, SUZ12 |
| chr18 | High amplification | p11.31 | |
| chr18 | Amplification | p11.31 - p11.23 | |
| chr18 | Amplification | p11.1 - q23 | DCC, ZNF521, SS18, MALT1, BCL2 |
| chr19 | Amplification | p13.2 | |
| chr20 | Amplification | p13 - q13.33 | BCL2L1, NCOA3, AURKA, ZNF217, EEF1A2, ASXL1, MAFB, TOP1, SDC4, GNAS, SS18L1 |
Table 4.
Copy number deletion regions in HCB-514 cell line.
| Chromosome Region | Event | Cytoband | Cancer genes |
|---|---|---|---|
| chr1 | Deletion | p11.2 - q21.1 | |
| chr2 | Deletion | q33.1 - q37.3 | CREB1, IDH1, ATIC, FEV, PAX3, ACSL3 |
| chr3 | Deletion | p26.3 - p12.3 | SRGAP3, FANCD2, VHL, PPARG, RAF1, XPC, MLH1, MYD88, CTNNB1, SETD2, BAP1, PBRM1, FHIT, MITF, FOXP1 |
| chr3 | Deletion | q26.1 | |
| chr4 | Deletion | p16.3 - p16.1 | FGFR3, WHSC1 |
| chr4 | Deletion | p16.1 - q22.1 | SLC34A2, PHOX2B, FIP1L1, PDGFRA, CHIC2, KIT, KDR |
| chr4 | Homozygous deletion | q22.1 | |
| chr4 | Deletion | q22.1 - q26 | RAP1GDS1, TET2 |
| chr4 | Deletion | q26 - q35.2 | IL2, FBXW7 |
| chr5 | Deletion | p15.33 | |
| chr5 | Deletion | q11.1 - q13.2 | IL6ST, PIK3R1 |
| chr5 | Homozygous deletion | q13.2 | |
| chr5 | Deletion | q13.2 - q31.3 | APC |
| chr5 | Deletion | q31.3 - q35.3 | PDGFRB, CD74, ITK, EBF1, RANBP17, TLX3, NPM1, NSD1 |
| chr7 | Deletion | q11.21 - q22.1 | SBDS, ELN, HIP1, AKAP9, CDK6 |
| chr7 | Homozygous deletion | q22.1 | |
| chr7 | Deletion | q22.1 - q35 | MET, SMO, CREB3L2, KIAA1549, BRAF |
| chr7 | Deletion | q35 - q36.3 | EZH2, MLL3, SHH |
| chr9 | Homozygous deletion | p13.2 - p13.1 | |
| chr9 | Deletion | p13.1 - p12 | |
| chr9 | Homozygous deletion | p12 - p11.2 | |
| chr9 | Deletion | p11.2 - q13 | |
| chr9 | Deletion | q13 | |
| chr9 | Deletion | q21.11 | |
| chr9 | Deletion | q21.32 | |
| chr11 | Deletion | q22.3 - q25 | ATM, DDX10, POU2AF1, SDHD, PAFAH1B2, PCSK7, MLL, DDX6, CBL, ARHGEF12, FLI1 |
| chr13 | Deletion | q11 - q34 | CDX2, FLT3, BRCA2, LHFP, LCP1, RB1, ERCC5, FOXO1, GPC5, IRS2 |
| chr18 | Deletion | p11.32 - p11.31 | |
| chr18 | Deletion | p11.23 - p11.21 | |
| chr18 | Deletion | p11.21 | |
| chr19 | Deletion | q13.42 | |
| chr21 | Deletion | q11.2 - q22.12 | OLIG2, RUNX1 |
| chr21 | Deletion | q22.2 - q22.3 | TMPRSS2, U2AF1 |
| chrX | Deletion | q24 |
To expand the analysis beyond the 87 genes of the NanoString panel, CNV was also investigated from WES data. The WES showed 64 chromosomal regions harboring a large number of genes with CNV. There were 30 amplified regions, with 9 of them presenting high amplification, harboring genes such as MYNC, ALK, PIK3CA, BIRC3, ERCC4 and PALB2 (Table 3). Deletions were found in 34 regions, harboring genes such as MLH1, PDGFRA, APC, MET and RB1. Homozygous deletions were found in 5 regions (Table 4).
Many of the amplified genes identified are related with PI3K-AKT and MAPK signaling pathways. Other altered regions harbored genes mostly related with the DNA repair process, apoptosis and transcriptional factors. Moreover, the cell line presented alterations in tyrosine-kinase receptors, such as EGFR and ALK amplification and PDGFRA and MET deletion.
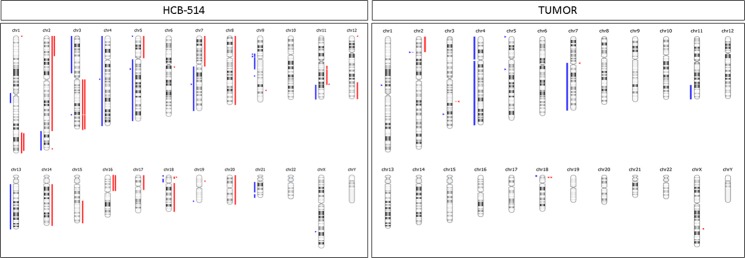
Regarding the CNV present in patient’s primary tumor, the number of CNVs was reduced compared with the HCB-514 cell line, with 4 amplification regions and 10 deletion regions. Again, both primary tumor and cell line shared some of these alterations, such as MYCN and ALK amplification and PDGFRA, KIT, IL2, FBXW7, MET and ATM deletion (Suppl. Table 4). Figure 6 summarizes CNV in the HCB-514 cell line and primary tumor.
Figure 6.
Copy number variations from whole-exome sequencing (WES) of HCB-514 (left) and tumor (right) were compared with patient’s blood DNA. Amplifications are shown in red markers and deletions in blue markers.
Phospho-RTK expression
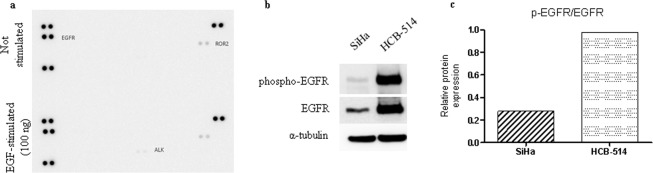
The characterization of RTK expression in HCB-514 revealed basal protein levels of activated EGFR and ROR2 (Fig. 7a, top). Upon EGF stimulation, the only new receptor activated was ALK (Fig. 7a, bottom). Activation of EGFR was also confirmed by the presence of phosphorylated EGFR in Western blot analysis (Fig. 7b), with 3.5-fold more expression in HCB-514 than in SiHa (Fig. 7c).
Figure 7.
(a) Phospho-RTK arrays of HCB-514 were done at basal conditions (up) and upon EGF-stimulation (down). Each RTK is represented in duplicate in the arrays (two spots side-by-side), and three pairs of phospho-tyrosine positive controls are located in the corners of each array. (b) Western blot of EGFR phosphorylation, showing the different amount of phospho-EGFR and total EGFR compared to the SiHa cell line. Alpha-tubulin was used as endogenous control. (c) Quantification of phospho-EGFR normalized with total EGFR.
Response to Cisplatin
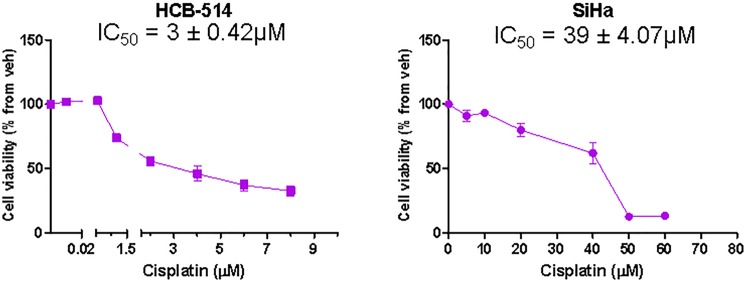
To evaluate the sensitivity of HCB-514 to the chemotherapeutic agent cisplatin, commonly used in clinical practice for cervical cancer treatment, HCB-514 was treated for 72 hours and a MTS assay was performed. The response to cisplatin was dose-dependent and the inhibitory concentration for 50% cells (IC50) value for HCB-514 was 3 ± 0.42 µM, while IC50 value for SiHa was 39 ± 4.07 µM, a 13-fold increase (Fig. 8).
Figure 8.
Cell viability of HCB-514 and SiHa after 72 h of cisplatin treatment. Results were obtained using the MTS assay. Data represent a mean of 3 independent experiments done in triplicate.
Discussion
Cancer cell lines are unique tools for understanding biology and tumor response. At present, immortalized cervical cancer cell lines available in major cell repositories, including ATCC or ECACC, represents only six cervical cancers (not considering HeLa-derived cell lines), not reflecting the whole spectrum of cervical cancer biological heterogeneity, neither representing patients from South America.
In the present study, we successfully established a cervical cancer cell line, HCB-514, out of 35 attempts - a successful rate of 3%. It is difficult to establish cervical cell lines, with success rates ranging from 11–30%, which may explain the reduced number of new cell lines9–12.
Not all mechanisms of continuous cell growth in vitro are known, but, at least for HPV-positive cervical cancers, the presence of papillomavirus E6/E7 expression is an important requirement12. Importantly, the HCB-514 cell line was HPV-16 positive and expressed E6 oncoprotein. Moreover, in our cell line and respective primary tumor, we showed that HPV integrates into the CDH13 gene. This gene negatively regulates keratinocyte proliferation and has been reported as one of the integration-targeted cellular genes (ITGs) in cervical cancer13. Although the role of these ITGs in carcinogenesis is not completely understood, it is suggested that ITGs modulate the expression of nearby genes through a long-distance chromatin interaction mechanism13. In addition, we used phylogenetic analysis to confirm the HPV16 A1 variant in both HCB-514 and primary tumor. This variant has been correlated with a high risk of precancer and cancer in Caucasian women14.
The molecular characterization of the HCB-514 cell line showed mutations in important cancer-related genes, yet exhibited a distinct profile compared to the profile for cervical squamous cell carcinomas recently reported by TCGA4. One of the mutated genes, BRCA2, is a well-known hereditary breast cancer gene that encodes a protein related to DNA damage repair during homologous recombination15. Mutations in BRCA2 occur in approximately 3% of cervical cancers according to the TCGA dataset www.cbioportal.org. Notably, there is limited knowledge of BRCA2 mutational status in cervical cancer, and current literature focuses on germline mutations and its role on non-HPV related cervical cancers, mostly associated with hereditary syndromes16,17. There are no functional studies about BRCA2 mutation in cervical cancer. Interestingly, the pathogenic p.W2830* BRCA2 mutation identified in HCB-514 has been identified in other cancers as a predictive marker for targeted therapies, such as PARP inhibitors; its role in cervical cancers remains unclear.
We also observed a mutation in the TGFBR1 gene, which encodes a receptor that acts on the TGF-β signaling pathway. This pathway is also related to the TGFBR2 gene, which is one of the most commonly mutated genes in cervical cancer4. When TGBR2 is activated by a TGF-β ligand, TGBR1, a serine/threonine kinase, is dimerized, leading to the cell cycle arrest of epithelial cells, control of mesenchymal cell proliferation and differentiation, and other cell processes related to growth suppression18. In this sense, TGF-β inhibitors, such as galunisertib, may emerge as a new approach for the treatment of cervical cancer in TGFBR1-mutated tumors19.
We also observed the presence of a mutation in the IRX2 gene. The IRX2 belongs to the iroquois homeobox gene family and encodes a transcription factor known to play multiple roles during embryo development20. In breast cancer, low expression of IRX2 has been observed and shown to be associated with the presence of disseminated tumor cells21. In this line, cell lines edited to overexpress IRX2 exhibited lower cellular motility and reduced secretion of pro-metastatic chemokines20. Moreover, the methylation of the IRX2 promoter region has been correlated with pleural anthracosis disease in lung adenocarcinomas22. Controversially, in primary human osteosarcoma tissues, IRX2 expression was observed to be significantly increased in comparison with normal tissues, and was significantly associated with tumor progression and prognosis23. In vitro assays showed that IRX2 upregulated MMP-9 and VEGF in a PI3K/AKT-dependent manner, and that knockdown of IRX2 in osteosarcoma cell lines inhibited cell proliferation and invasion23,24.
Another gene of interest that was mutated in both the HCB-514 cells and patient’s tumor tissue was SHKBP1. Recently, SHKBP1 was identified in the cervical cancer TCGA, as a significantly mutated gene in this tumor type4. Overexpression of SHKBP1 in cell lines led to a strong delay in the degradation of EGFR upon EGF activation25. This process occurred due to competitive binding of SHKBP1 with protein c-Cbl, thereby interrupting the complex c-Cbl-CIN85, which is responsible for endocytosis of EGFR containing vesicles25.
According to TCGA, an average of 88 somatic CNV was found per cervical tumor, including 26 amplifications, 37 deletions and 26 alterations in whole chromosome arms4. The HCB-514 cell line presented 30 amplifications and 34 deletions, showing 64 alterations in total. Among these CNVs, EGFR and YAP1 genes were amplified in the HCB-514 cell line. Both amplifications have already been described as recurrent in cervical cancer4. In addition, to confirm the biological impact of EGFR gene amplification we analyzed and observed EGFR protein phosphorylation in the HCB-514 cell line. Interestingly, blocking the EGFR tyrosine-kinase protein is an effective approach to treat specific types of tumors, including colorectal, head and neck and lung cancers26.
In cervical cancer, a phase II clinical study with EGFR inhibitor Erlotinib showed that administering it prior to receiving the standard of treatment (cisplatin concurrent with radiotherapy) was safe and showed significant antitumor activity27. In addition, a recent study demonstrated a promising effect of lapatinib (anti- EGFR and HER-2), in HER-expressing cervical cancer cells by reducing the number and size of blood vessels and preventing increased HIF-1α levels28. Regarding YAP1, the protein encoded by YAP1 gene, it is negatively regulated by the Hippo signaling pathway, and its enhanced activity is believed to induce cell proliferation, migration and survival in several cancer types including cervical cancer29,30. Increased activity of YAP1 can occur after TGFα and amphiregulin inhibit the Hippo signaling via EGFR29. Furthermore, YAP1 seems to play a key role in TGF-β1-induced apoptosis and EMT through EGFR regulation30. In cervical cancer, YAP1 was amplified in the EMT cluster samples from TCGA data highlighting the potential role of this gene in EMT-related tumor progression4. However, more functional studies on YAP1 amplification need to be conducted to elucidate its role in cervical cancer.
Finally, we showed the sensitivity of HCB-514 to cisplatin, in line with the patient’s clinical response to cisplatin (with concurrent radiotherapy).
In conclusion, the molecular profile of the novel Brazilian cervical cancer cell line HCB-514 reveals disruption of several key cellular pathways, such as PI3K-AKT, MAPK signaling, DNA repair, apoptosis, transcriptional factors and RTK. This new HPV-16 positive cervical cancer cell line constitutes an important model to better understand the cancer biology of cervical cancer and offers an additional promising tool for cervical cancer pre-clinical studies.
Materials and Methods
Establishment of cervical cancer cell lines
Blood and tumor tissue samples were collected from participants with cervical cancer during routine visits in the Gynecologic Oncology Department of BCH after informed consent was obtained. The collected tumor tissue was maintained in PBS buffer in the refrigerator until processing, which occurred before 24 hours. The sample was initially placed in a petri dish and fragmented with scalpels and incubated for 30 min at 37 °C in a 15 mL falcon containing 5 mL of an enzymatic solution (0.14% collagenase type I [Sigma, Brazil] and 0.01% DNase [2,000 kilounits/mg] [Sigma]) in RPMI 1640 as previously described31. Then, RPMI medium was supplemented with 1% penicillin and streptomycin (P/S; Sigma) and 10% fetal bovine serum (FBS; Sigma) was added to inactivate the enzyme. After centrifugation, the cell pellet was resuspended in FAD media32 adapted by Dr. Matias Melendez (personal communication), which was comprised by three parts of Dulbecco’s modified Eagle’s media (DMEM; Sigma) to one part Ham’s F12 media (F12; Sigma). The media was then supplemented with 1% P/S, 0.4 µg/mL hydrocortisone, 2.5 µg/mL insulin, 13 ng/mL liothyronine, 5 µg/mL transferrin, 2.5 µg/mL insulin and 0.1 µg/mL cholera enterotoxin and 5% FBS (all supplements were purchased from Sigma). Cells were maintained in a T25 flask at 37 °C, 5% CO2. One week after processing, the media was replaced and cell growth was monitored. Controlled trypsinizations were done to preferentially remove the contaminating fibroblasts. Afterwards, the culture media was replaced every 72 h and the cell culture was routinely subcultured once it was confluent. Stock vials were frozen in FBS+ 5% DMSO solution for storage in liquid nitrogen. All methods were performed in accordance with the relevant guidelines and regulations and were approved by the Ethics and Research Committee from Barretos Cancer Hospital (BCH) (985/2015 - CAAE 1,252,699).
Cell lines
For molecular and functional characterization, the immortalized cervical squamous cell carcinoma cell lines, SiHa, was kindly provided by Dr. Luisa Villa (INCT-HPV, Brazil) and a human short-term primary culture of lung fibroblast, named HCB-535, was obtained from BCH. SiHa was grown in DMEM media supplemented with 1% P/S and 10% FBS and HCB-535 in RPMI media supplemented with 1% P/S and 10% FBS.
Immunophenotypic characterization
To confirm cell origin, immunocytochemistry was performed with antibodies against vimentin (positive for fibroblasts) and cytokeratin (positive for epithelial cells). When cells reached the fourth passage, they were plated in a 24-well plate (5 × 105 in 3 DMEM:1 F12 media + 5% FBS) and, upon reaching 80% confluence, cells were fixed with 4% paraformaldehyde for 15 min, washed 3 times with PBS, permeabilized with Triton X-100 (0.1% in water) for 4 min and washed as described. Endogenous peroxidase activity was inhibited for 10 min with H2O2 (3% in methanol). Blockage for non-specific labeling was performed through incubation with 50 µL of the Ultra V Block reagent (Thermo Scientific detection kit) for 10 min and, after washes, primary antibodies cytokeratin AE1/AE3 (Dako, ready to use) or vimentin (Dako, ready to use), incubation was done for 60 min and then washed. A biotinylated goat polyvalent antibody incubation for 10 min was performed, followed by washing. Streptavidin peroxidase incubation was next performed for 10 min and washed. Afterwards, the DAB chromogen (ThermoFisher Scientific) was applied for 10 min for staining. Finally, hematoxylin was used for counterstaining and the cells were photographed with a photographic camera coupled to a microscope (Olympus). The immortalized cell line SiHa, a grade II squamous cell carcinoma and HPV-16 positive, was used as a positive control for the cytokeratin and HCB-535 was used as positive control for vimentin.
The markers p40 (Biocare Medical, dilution 1:100) and p16 (Roche, Ventana Systems, ready to use), commonly used in the routine pathology for determination of cervical cancer histology subtype33, were evaluated by immunohistochemistry in the cell block of the new cervical cancer cell line established in this work and in FFPE tumor tissue from the cervical cancer patient. Moreover, expression of EpCAM (CD326, BD Biosciences)34, another marker of cervical carcinoma cells, was analyzed in the HCB-514 cell line by flow cytometry. Briefly, 5 × 105 cells were harvested, washed twice in BSA 0,2% in DPBS (centrifugation between the washes was 1500 rpm for 5 min), then 10 µL of EpCAM PE-conjugate were added and incubated for 20 min. Afterwards, cells were washed and analyzed by flow cytometry (Accuri BD). SiHa was used as positive control for EpCAM expression and HCB-535 was used as negative control.
Cell line doubling-time
Growth characteristics of the cell line were analyzed by doubling-time evaluation. Doubling-time was determined through real time impedance-based technology cell analyzer system (RTCA, xCELLigence, Roche), according to manufacturer’s instructions. Firstly, 5,000 cells were plated into E-plate (xCELLigence, Roche) and proliferation rate was evaluated for 72 h. The new cervical cancer was maintained in supplemented media above described in establishment section, with 5% FBS or 10% FBS. SiHa was maintained in DMEM 1% P/S and 5 or 10% FBS. This assay results in a cell index, which considers cell viability, cell number, morphology and adhesion. At the end of assay, a cell growth curve is produced and the doubling-time was calculated from the selected interval, which is basically the interval selected when the cell index doubled inside the log-phase35.
DNA isolation
For molecular characterization, DNA was isolated from 1 × 106 tumor cell pellet in the sixth passage using silica column from Biopur Mini Spin Plus 250 Extraction kit (Biopur), as described by the manufacturer. Blood total DNA was automatically isolated from buffy coat of patient through Mini Kit DNA and Qiasymphony instrument (both from Qiagen) at BCH Biobank Department, according to the manufacturer. This technology combines silica-based extraction with magnetic particles purification. The protocol Blood 200 with an elution volume of 50 µL was used. Tumor DNA was isolated from a biopsy sample obtained from the patient through the Mini Kit DNA and Qiasymphony instrument (QIAgen). For tumor-isolated DNA, macrodissection was performed by an experienced pathologist and up to 25 mg of the sample containing at least 60% tumor area and up to 20% necrosis area were fragmented and submitted to digestion and homogenization procedures, according to the manufacturer’s instructions. The protocol Tissue 200 with an elution volume of 50 µL was used.
All DNA samples were quantified by both NanoDrop® 2000 (Thermo Scientific) and Qubit® 2.0 Fluorometer (Life Technologies) and were then stored at −20 °C for further genetic analysis. Blood DNA was used as reference to distinguish somatic from germline mutations.
Assessment of human papilloma virus (HPV)
The presence of HPV in HCB-514 cell line was evaluated by in house PCR using GP5+ and GP6+ primers, able to amplify a fragment of ~142 pb of the HPV conserved region L136. For this, it was used 5 µL of DNA extracted from HCB-514, 20 mM of Thris-HCl buffer (pH 8.4), 50 mM KCl, 1.25 U of Platinum Taq DNA polymerase, 2 mM of MgCl2, 0.2 mM dNTP mix, 0.6 µM of each primer (GP5+ and GP6+). Amplifications were performed in the equipment Applied Biosystems GeneAmp PCR System 2700 (Applied Biosystems) and the cycle was as described: 94 °C for 4 min followed by 40 cycles of 1 min at 94 °C, 2 min at 40 °C, and 1 min 30 seg at 72 °C. Then, a cycle of 72 °C for 7 min and the PCR products were stored at 4 °C before electrophoresis on a 2% agarose gel. DNA HPV-positive was used as positive control and water was used as negative control. PCR reagents were pursued from Invitrogen.
Moreover, the COBAS® 4800 HPV System (Roche) was used for genotyping HPV, according with manufacturer’s instruction. This assay utilizes DNA target amplification through real time PCR to detect 14 high-risk HPVs in a single assay. It genotypes HPVs 16 and 18 and simultaneously detects other non-16 and 18 high-risk types (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) as a pool result in cases of infection.
In addition, because E6 oncoprotein is required for the oncogenic transformation of HPV-infected cervical epithelial cells and the most common high-risk HPVs associated with cervical cancer are 16 and 18, the presence of their E6 oncoprotein was assessed through an immunochromatographic test, using OncoE6TM Cervical Test (Arbor Vita), according with manufacturer’s instructions. Briefly, the lysate of 1 × 106 HCB-514 cells was incubated with monoclonal antibodies to oncoprotein E6 of HPV types 16 and 18 conjugated with alkaline phosphatase. Then, using a nitrocellulose test strip, this mix migrates up the test strip and if the protein is present, it is possible to observe a purple line in the strip. The position of the line indicates if E6 is from HPV-16 or HPV-18.
Cell line authentication by short tandem repeat (STR) profiling analysis
The STR analysis was carried out in DNA from cell culture, FFPE tumor tissue and blood from the patient as previously described for authenticity confirmation37,38.
Whole exome sequencing (WES) analysis
The DNA from the HCB-514 cell line, the patient’s primary tumor and paired blood sample were used for WES, with input of 50 ng on the Illumina HiSeq2500 ™ System by a commercial company (Mendelics, São Paulo, Brazil). Sequence reads were aligned to the human reference genome build 37 (hs37d5-decoy) using BWA-MEM with Burrows–Wheeler Aligner version 0.7.10-r78939. Duplicate reads were marked with Picard-Tools 1.92 (http://broadinstitute.github.io/picard/). MuTect version 1.1.4; (http://www.broadinstitute.org/cancer/cga/mutect) and Varscan240 were used to call somatic SNVs and indels in tumor-normal pairs, respectively. MuTect was run using default parameters with files from COSMIC version 54 and dbSNP version 132 included as input41,42. We used Ensembl Variant Effect Predictor (VEP)43 to annotate and determine functional consequences of tumor specific variants. The results from SIFT, Polyphen-2, ClinVar were considered. It was also excluded variants that were likely to be germline, i.e., listed in ESP6500 (http://evs.gs.washington.edu/EVS/), 1000Genome or ExAC44,45. The candidate mutations were validated visually using the Integrated Genomics Viewer (IGV)46. Copy number abnormalities (CNA) were identified using Nexus Copy Number version 9.0 (BioDiscovery; El Segundo, CA; https://www.biodiscovery.com/products/Nexus-Copy-Number) with default parameter for BAM ngCGH (matched) input with homozygous frequency threshold and value at 0.97 and 0.8 respectively, hemizygous loss threshold at −0.18, single copy gain at 0.18 and high copy gain at 0.6.
Mutational signature was defined using the package SomaticSignatures of Bioconductor Software, as described by Gehring et al.47.
Integration of HPV viral DNA into the human genome was performed through integration mode of the program HPVDetector, as previously described48. The phylogenetic analysis of HPV sequence was performed using the fasta consensus sequences of HPV16 (one from tumor sample and the other from cell culture) obtained from deep sequencing and a group of HPV16 complete genome sequences retrieved from GenBank were aligned with Muscle49 and edited with Se-Al v2.0a11 (available at http://tree.bio.ed.ac.uk/software/sea/). Phylogenetic reconstructions were performed by maximum likelihood (ML) criterion using RAxML version 8.0.050. The best fit model used for the ML reconstruction was GTR + Γ model without partitions. Node support was evaluated using 1,000 bootstraps cycles.
BRCA2 Sanger sequencing
To validate the mutation identified in WES in BRCA2, the HCB-514 cell and patient’s blood DNAs were submitted to PCR amplification of BRCA2 exon 20, using Hot Start Taq enzyme (Qiagen), as described by Costa et al.51. After amplification, PCR products were purified with the ExoSap enzyme (USB Affymetrix), and submitted to sequencing protocol using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequencing was performed in 3500 XL Genetic Analyzer (Applied Biosystems), as described by Fernandes et al.52.
Copy number variation analysis
HCB-514 chromosomal alterations were also analyzed by the Nanostring plataform, using the nCounter® v2 Cancer CN Assay panel (NanoString Technologies, Seattle, WA, USA). This panel counts the CNV of 87 involved genes commonly amplified or deleted in various cancers (www.nanostring.com/products/CNV). As control, the genomic DNA from the HCB-514 patient’s peripheral blood was used. The DNA input was 600 ng for HCB-514, and 473 ng for peripheral blood.
The raw data was captured by the nSolverAnalysis Software v3.0® program (NanoString Technologies). For the normalization of the results, 54 probes were used for regions of the genome that do not usually present CNV. After the data normalization, the mean of the counts from the 3 probes per each gene was calculated and the number of copies per gene was calculated as previously described53,54.
Human-receptor tyrosine kinase (RTK) array and Western blot
To characterize HCB-514 protein expression, the Proteome Profiler Human Phospho-RTK Array Kit (#ARY001, R&DSystems) was used, according to the manufacturer’s instructions, and basal and epidermal growth factor (EGF)-stimulated protein expression were evaluated. Briefly, cells were grown in two T75 flask until confluence. Then, for EGFR stimulation, EGF diluted in 10 mL of free-serum media (final concentration 10 ng/mL) was firstly added to the cells and incubated for 15 min. After, proteins were collected using lysis buffer (50 mM Tris pH7.6–8, 150 mM, NaCl, 5 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 10 mM,sodium pyrophosphate, 1% NP-40, and protease cocktail inhibitors) and 500 µg of protein lysates were incubated as previously described55. Membranes were revealed with ECL Western Blotting Detection Reagents (RPN2109, GE Healthcare) and the chemiluminescent signal was detected by ImageQuant™ LAS 4000 mini documentation system (GE Healthcare).
For Western blot, protein lysates were separated in 8% SDS-PAGE gel, transferred, incubated in phospho-EGFRY1068 (1:1000), EGFR (1:1000) or α-tubulin (1:2000) primary antibody and revealed, as detailed described by Silva-Oliveira et al.56. Antibodies were purchased from Cell Signaling company.
Viability evaluation
The cell viability of the HCB-514 cell line to cisplatin was evaluated by MTS assay as previously reported38. Briefly, 6,000 cells were plated in 96-wells, allowed to adhere, and then treated with a range from 0.01 to 8 µM of cisplatin (Sigma). After 72 h, the Cell Titer 96 Aqueous Cell Proliferation Assay (MTS reagent, Promega) was added and incubated for 3 h. Absorbance values were normalized with absorbance of DMSO 1%-treated cells and the IC50 (inhibitory concentration for 50% cells) was calculated through non-linear regression.
Statistical analysis
GraphPad Prism was used to determine IC50 in drug-response assays and to plot the cell index data from doubling-time assay and the relative protein expression from Western blot analysis.
Supplementary information
Acknowledgements
The authors would like to thank Barretos Cancer Hospital Research Support Department (NAP) for sample collection, Barretos Cancer Hospital Biobank for sample processing, Dr. Flávia de Paula and Gabriela Fernandes for technical support of STRs and BRCA2 Sanger validation, respectively, and Dr. Laura Musselwhite (Duke University) for revising the manuscript. This study was supported by grants from the FINEP (MCTI/FINEP/MS/SCTIE/DECIT-01/2013 - FPXII- BIOPLAT - Process number 01.13.0469.00) and Barretos Cancer Hospital. PhD scholarship from FINEP (Grant numbers 384088/2014-7 and 380434/2015-6) and Barretos Cancer Hospital to MNR. Post-doctoral scholarship from FINEP to CCM (Grant number 383968/2015-1). Master degree scholarship from CAPES/FAPESP to FFM (Grant number 2015/20077-3). Post-doctoral scholarship from Bill and Melinda Gates Foundation to CMO (Grant number 1151345). LFL is supported by Public Ministry of Labor Campinas (Research, Prevention and Education of Occupational Cancer) in Campinas, Brazil.
Author Contributions
Conceptualization: M.N.R., R.M.R.; Experiments: M.N.R., L.F.L., C.M.O., V.A.O.S., C.C.M., F.F.M.; Data analysis/ Patient material/Clinical information: M.N.R., A.F.E., L.F.L, C.M.O., V.A.O.S., G.M.M., R.R., C.E.A., C.P.S.; Writing original draft: M.N.R., A.F.E., L.F.L., C.M.O., R.M.R.; Review & Editing: M.N.R., A.F.E., L.F.L., C.M.O, V.A.O.S., C.C.M., F.F.M., G.M.M., R.R., C.E.A., C.P.S., R.M.R.
Data Availability
Genetic data has been deposited at the European Genome-phenome Archive (EGA, http://wwwdev.ebi.ac.uk/ega/), which is hosted by the EBI, under accession number EGAS00001003343.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-38315-7.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Instituto Nacional de Câncer José de Alencar Gomes da Silva (INCA). Estimativa2014: Incidência de câncer no Brasil. (Rio de Janeiro, Brasil 2014).
- 3.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijer CJLM, Steenbergen RDM. Gynaecological cancer: Novel molecular subtypes of cervical cancer - potential clinical consequences. Nat. Rev. Clin. Oncol. 2017;14:397–398. doi: 10.1038/nrclinonc.2017.52. [DOI] [PubMed] [Google Scholar]
- 6.American Cancer Society. Cancer Facts & Figures 2018. Atlanta: American Cancer Society; 2018.
- 7.Wolford JE, Tewari KS. Rational design for cervical cancer therapeutics: cellular and non-cellular based strategies on the horizon for recurrent, metastatic or refractory cervical cancer. Expert Opinion on Drug Discovery. 2018;13(5):445–457. doi: 10.1080/17460441.2018.1443074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 9.Javed S, et al. Significance of CD133 positive cells in four novel HPV-16 positive cervical cancer-derived cell lines and biopsies of invasive cervical cancer. BMC Cancer. 2018;18:357. doi: 10.1186/s12885-018-4237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelland LR, Burgess L, Steel GG. Characterization of four new cell lines derived from human squamous carcinomas of the uterine cervix. Cancer Res. 1987;47:4947–4952. [PubMed] [Google Scholar]
- 11.Ku JL, Kim WH, Park HS, Kang SB, Park JG. Establishment and characterization of 12 uterine cervical-carcinoma cell lines: common sequence variation in the E7 gene of HPV-16-positive cell lines. Int. J. Cancer. 1997;72:313–320. doi: 10.1002/(SICI)1097-0215(19970717)72:2<313::AID-IJC19>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 12.Magaldi TG, et al. Primary human cervical carcinoma cells require human papillomavirus E6 and E7 expression for ongoing proliferation. Virology. 2012;422:114–124. doi: 10.1016/j.virol.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, et al. Dysregulation of host cellular genes targeted by human papillomavirus (HPV) integration contributes to HPV‐related cervical carcinogenesis. Int. J. Cancer. 2016;138:1163–1174. doi: 10.1002/ijc.29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirabello, L. et al. HPV16 Sublineage Associations With Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J. Natl. Cancer Inst. 108 (2016). [DOI] [PMC free article] [PubMed]
- 15.Nesic K, Wakefield M, Kondrashova O, Scott CL, McNeish IA. Targeting DNA repair: the genome as a potential biomarker. J. Pathol. 2018;244:586–597. doi: 10.1002/path.5025. [DOI] [PubMed] [Google Scholar]
- 16.Constantinou P, Tischkowitz M. Genetics of gynaecological cancers. Best Pract. Res. Clin. Obstet. Gynaecol. 2017;42:114–124. doi: 10.1016/j.bpobgyn.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Mersch J, et al. Cancers Associated with BRCA1 and BRCA2 Mutations other than Breast and Ovarian. Cancer. 2015;121:269–275. doi: 10.1002/cncr.29041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasche B, Pennison MJ, Jimenez H, Wang M. TGFBR1 and cancer susceptibility. Trans. Am. Clin. Climatol. Assoc. 2014;125:300–312. [PMC free article] [PubMed] [Google Scholar]
- 19.Herbertz S, et al. Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-beta signaling pathway. Drug Des. Devel. Ther. 2015;9:4479–4499. doi: 10.2147/DDDT.S86621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner S, et al. Iroquois homeobox 2 suppresses cellular motility and chemokine expression in breast cancer cells. BMC Cancer. 2015;15:896. doi: 10.1186/s12885-015-1907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werner S, et al. Suppression of early hematogenous dissemination of human breast cancer cells to bone marrow by retinoic Acid-induced 2. Cancer Discov. 2015;5:506–519. doi: 10.1158/2159-8290.CD-14-1042. [DOI] [PubMed] [Google Scholar]
- 22.Sato T, et al. Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: Its correlation with smoking history and chronic obstructive pulmonary disease. Int. J. Cancer J. Int. Cancer. 2014;135:319–334. doi: 10.1002/ijc.28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, et al. Knockdown of IRX2 inhibits osteosarcoma cell proliferation and invasion by the AKT/MMP9 signaling pathway. Mol. Med. Rep. 2014;10:169–174. doi: 10.3892/mmr.2014.2215. [DOI] [PubMed] [Google Scholar]
- 24.Liu T, et al. IRX2-mediated upregulation of MMP-9 and VEGF in a PI3K/AKT-dependent manner. Mol. Med. Rep. 2015;12:4346–4351. doi: 10.3892/mmr.2015.3915. [DOI] [PubMed] [Google Scholar]
- 25.Feng L, Wang J-T, Jin H, Qian K, Geng J-G. SH3KBP1 binding protein 1 prevents epidermal growth factor receptor degradation by the interruption of c-Cbl-CIN85 complex SHKBP1 prevents EGFR degradation by the interruption of c-Cbl-CIN85 complex. Cell Biochem. Funct. 2011;29:589–596. doi: 10.1002/cbf.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seshacharyulu P, et al. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin. Ther. Targets. 2012;16:15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogueira-Rodrigues A, et al. Phase 2 trial of erlotinib combined with cisplatin and radiotherapy in patients with locally advanced cervical cancer. Cancer. 2014;120:1187–1193. doi: 10.1002/cncr.28471. [DOI] [PubMed] [Google Scholar]
- 28.Martinho O, et al. HER Family Receptors are Important Theranostic Biomarkers for Cervical Cancer: Blocking Glucose Metabolism Enhances the Therapeutic Effect of HER Inhibitors. Theranostics. 2017;7:717–732. doi: 10.7150/thno.17154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He C, et al. The Hippo/YAP pathway interacts with EGFR signaling and HPV oncoproteins to regulate cervical cancer progression. EMBO Mol. Med. 2015;7:1426–1449. doi: 10.15252/emmm.201404976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, et al. YAP modulates TGF-β1-induced simultaneous apoptosis and EMT through upregulation of the EGFreceptor. Sci. Rep. 2017;7:45523. doi: 10.1038/srep45523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santin AD, et al. Induction of human papillomavirus-specific CD4(+) and CD8(+) lymphocytes by E7-pulsed autologous dendritic cells in patients with human papillomavirus type 16- and 18-positive cervical cancer. J. Virol. 1999;73:5402–5410. doi: 10.1128/jvi.73.7.5402-5410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu YJ, et al. The mesothelial keratins: a new family of cytoskeletal proteins identified in cultured mesothelial cells and nonkeratinizing epithelia. Cell. 1982;31:693–703. doi: 10.1016/0092-8674(82)90324-5. [DOI] [PubMed] [Google Scholar]
- 33.Bergeron C, et al. The clinical impact of usingp16(INK4a) immunochemistry in cervical histopathology and cytology: an update of recent developments. Int. J. Cancer. 2015;136:2741–2751. doi: 10.1002/ijc.28900. [DOI] [PubMed] [Google Scholar]
- 34.Richter CE, et al. Primary cervical carcinoma cell lines overexpress epithelial cell adhesion molecule (EpCAM) and are highly sensitive to immunotherapy with MT201, a fully human monoclonal anti-EpCAM antibody. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2010;20:1440–1447. [PMC free article] [PubMed] [Google Scholar]
- 35.Witzel F, Fritsche-Guenther R, Lehmann N, Sieber A, Blüthgen N. Analysis of impedance-based cellular growth assays. Bioinforma. Oxf. Engl. 2015;31:2705–2712. doi: 10.1093/bioinformatics/btv216. [DOI] [PubMed] [Google Scholar]
- 36.de Roda Husman A-M, Walboomers JMM, van den Brule AJC, Meijer CJLM, Snijders PJF. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 1995;76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 37.Dirks WG, Faehnrich S, Estella IAJ, Drexler HG. Short tandem repeat DNA typing provides an international reference standard for authentication of human cell lines. ALTEX. 2005;22:103–109. [PubMed] [Google Scholar]
- 38.Silva-Oliveira RJ, et al. Cytotoxicity of allitinib, an irreversible anti-EGFR agent, in a large panel of human cancer-derived cell lines: KRAS mutation status as a predictive biomarker. Cell. Oncol. Dordr. 2016;39:253–263. doi: 10.1007/s13402-016-0270-z. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. Curr. Protoc. Bioinforma. 2013;44:15.4.1–17. doi: 10.1002/0471250953.bi1504s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed]
- 45.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson JT, et al. Integrative Genomics Viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–3675. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandrani P, et al. NGS-based approach to determine the presence of HPV and their sites of integration in human cancer genome. Br. J. Cancer. 2015;112:1958–1965. doi: 10.1038/bjc.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinforma. Oxf. Engl. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 51.Costa JL, et al. Nonoptical Massive Parallel DNA Sequencing of BRCA1 and BRCA2 Genes in a Diagnostic Setting. Hum. Mutat. 2013;34:629–635. doi: 10.1002/humu.22272. [DOI] [PubMed] [Google Scholar]
- 52.Fernandes GC, et al. Prevalence of BRCA1/BRCA2 mutations in a Brazilian population sample at-risk for hereditary breast cancer and characterization of its genetic ancestry. Oncotarget. 2016;7:80465–80481. doi: 10.18632/oncotarget.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn S, et al. A nCounter CNV Assay to Detect HER2 Amplification: A Correlation Study with Immunohistochemistry and In Situ Hybridization in Advanced Gastric Cancer. Mol. Diagn. Ther. 2016;20:375–383. doi: 10.1007/s40291-016-0205-4. [DOI] [PubMed] [Google Scholar]
- 54.Norton N, et al. Assessment of Tumor Heterogeneity, as Evidenced by Gene Expression Profiles, Pathway Activation, and Gene Copy Number, in Patients with Multifocal Invasive Lobular Breast Tumors. PloS One. 2016;11:e0153411. doi: 10.1371/journal.pone.0153411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinho O, et al. In Vitro and In Vivo Analysis of RTK Inhibitor Efficacy and Identification of Its Novel Targets in Glioblastomas. Transl. Oncol. 2013;6:187–196. doi: 10.1593/tlo.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva-Oliveira RJ, et al. AKT can modulate the in vitro response of HNSCC cells to irreversible EGFR inhibitors. Oncotarget. 2017;8:53288–53301. doi: 10.18632/oncotarget.18395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genetic data has been deposited at the European Genome-phenome Archive (EGA, http://wwwdev.ebi.ac.uk/ega/), which is hosted by the EBI, under accession number EGAS00001003343.