Abstract
Alzheimer’s disease (AD) is a progressive neurodegenerative condition marked by a decline in cognitive functions with no validated disease modifying treatment. It is critical for timely treatment to detect AD in its earlier stage before clinical manifestation. Mild cognitive impairment (MCI) is an intermediate stage between cognitively normal older adults and AD. To predict conversion from MCI to probable AD, we applied a deep learning approach, multimodal recurrent neural network. We developed an integrative framework that combines not only cross-sectional neuroimaging biomarkers at baseline but also longitudinal cerebrospinal fluid (CSF) and cognitive performance biomarkers obtained from the Alzheimer’s Disease Neuroimaging Initiative cohort (ADNI). The proposed framework integrated longitudinal multi-domain data. Our results showed that 1) our prediction model for MCI conversion to AD yielded up to 75% accuracy (area under the curve (AUC) = 0.83) when using only single modality of data separately; and 2) our prediction model achieved the best performance with 81% accuracy (AUC = 0.86) when incorporating longitudinal multi-domain data. A multi-modal deep learning approach has potential to identify persons at risk of developing AD who might benefit most from a clinical trial or as a stratification approach within clinical trials.
Subject terms: Data integration, Machine learning
Introduction
Alzheimer’s disease (AD) is an irreversible, progressive neurodegenerative disorder characterized by abnormal accumulation of amyloid plaques and neurofibrillary tangles in the brain, causing problems with memory, thinking, and behavior. AD is the most common form of dementia with no validated disease modifying treatment. An estimated 5.7 million Americans are living with AD in 2018. By 2050, this number is projected to rise to nearly 14 million1. Current available treatments decelerate only the progression of AD and no treatment developed so far can cure a patient who is already in AD. Thus, it is of fundamental importance for timely treatment and progression delay to develop strategies for detection of AD at early stages before clinical manifestation. As a result, the concept of mild cognitive impairment (MCI) was introduced. MCI, a prodromal form of AD, is defined to describe people who have mild symptoms of brain malfunction but can still perform everyday tasks. Patients in the phase of MCI have an increased risk of progressing to dementia1–4. Some patients in their MCI stages are converted to AD within a limit of the time window after baseline, while some are not. It has been reported that MCI patients progress to AD at a rate of 10% to 15% per year and 80% of these MCI patients will have converted to AD after approximately six years of follow-up5,6. It is an ongoing topic among AD-related researches to identify biomarkers that classify patients with MCI who later progress to AD (MCI converter) from those with MCI who do not progress to AD (MCI non-converter).
Various machine learning methods have been applied to identify biomarkers for MCI conversion prediction and improve their performances. Support vector machine (SVM) is one of methods frequently used for solving classification problem. A lot of studies applied SVM for MCI conversion prediction7–12. A multi-task learning along with SVM was used to identify AD-relevant features, showing 73.9% accuracy, 68.6% sensitivity, and 73.6% specificity7. For the use of additional subjects, a domain transfer learning method to use auxiliary samples such as AD and cognitively normal older adults (CN) subjects as well as MCI subjects showed 79.4% accuracy, 84.5% sensitivity, and 72.7% specificity8. A linear discriminant analysis (LDA) was used based on cortex thickness data showing 63% sensitivity and 76% specificity13. Furthermore, the integration of multi-modality data improves the performance for MCI conversion prediction by extracting complementary AD-related biomarkers from each modality. Cerebrospinal fluid (CSF), MRI, and cognitive performance biomarkers were combined, resulting in 68.5% accuracy 53.4% sensitivity, and 77% specificity14,15. Along with MRI and CSF biomarkers, APOE ε4 status were integrated16.
In this study, in order to predict MCI to AD conversion, we proposed a multimodal recurrent neural network method, a deep learning approach, based on the integration of demographic information, longitudinal CSF biomarkers, longitudinal cognitive performance, and cross-sectional neuroimaging biomarkers at baseline obtained from the Alzheimer’s Disease Neuroimaging Initiative cohort (ADNI). Our proposed deep learning method can incorporate longitudinal multiple domain data and take variable-length longitudinal data to capture temporal features at multiple time points. In particular, non-overlapping samples as well as overlapping samples from each data can be used to build a prediction model.
Results
Study participants
All individuals used in the analysis were participants of the Alzheimer’s Disease Neuroimaging Initiative (ADNI)17,18. The overall goal of ADNI is to test whether serial magnetic resonance imaging (MRI), position emission tomography (PET), other biological markers, and clinical and neuropsychological assessment could be combined to measure the progression of MCI and early AD. Demographic information, raw neuroimaging scan data, APOE genotype, CSF measurements, neuropsychological test scores, and diagnostic information are publicly available from the ADNI data repository (http://adni.loni.usc.edu). Informed consent was obtained for all subjects, and the study was approved by the relevant institutional review board at each data acquisition site (for up-to-date information, see http://adni.loni.usc.edu/wp-content/themes/freshnews-dev-v2/documents/policy/ADNI_Acknowledgement_List%205-29-18.pdf). All methods were performed in accordance with the relevant guidelines and regulations. In this study, a total of 1,618 ADNI participants aged 55 to 91 were used, which include 415 cognitively normal older adult controls (CN), 865 MCI (307 MCI converter and 558 MCI non-converter), and 338 AD patients (Table 1).
Table 1.
Subject demographics at baseline visit.
| Characteristics | MCI-C (n = 307) |
MCI-NC (n = 558) | CN (n = 415) |
AD (n = 338) |
|---|---|---|---|---|
| Sex (Female/Male) | 117/190 | 236/322 | 206/209 | 152 /186 |
| Memory score (mean ± sd) | −0.26 ± 0.5 | 0.39 ± 0.64 | 1.01 ± 0.56 | −0.86 ± 0.54 |
| Education (mean ± sd) | 15.89 ± 2.75 | 15.94 ± 2.88 | 16.28 ± 2.72 | 15.15 ± 2.99 |
| APOE ε4 (0/1/2) | 105/153/49 | 325/188/45 | 301/103/11 | 113/160/65 |
We used four different types, or modalities of data: demographic information, neuroimaging phenotypes measured by MRI, cognitive performance, and CSF measurements. Demographic information includes age, sex, years of education, and APOE ε4 status. Cognitive performance includes composite scores for executive functioning (ADNI-EF) and memory (ADNI-MEM) derived from the ADNI neuropsychological battery using item response theory as described in detail elsewhere19. CSF biomarkers for AD include amyloid-β 1–42 peptide (Aβ1–42), total tau (t-tau), and tau phosphorylated at the threonine 181 (p-tau). AD-related neuroimaging biomarkers measured by MRI include hippocampal volume and entorhinal cortical thickness.
Experimental setting
To evaluate the performance and effectiveness of our proposed longitudinal multi-modal deep learning method, we used three schemes and compared their performances (Table 2). In the experiment named “baseline”, 4 modalities data at baseline visit (cognitive performance, CSF, demographic information, and MRI) were incorporated. In “single modal”, only longitudinal cognitive performance data was used for the predictor (we tried all other single modality, and the performance with cognitive scores was the best). Finally, the four modalities of longitudinal data were combined and used for training the classifier in the experiment marked as “proposed”. Table 3 shows summary statistics of each modalities of data and hyperparameters used for training GRUs.
Table 2.
Three experimental schemes depending on training dataset composition.
| Longitudinal data | Multimodal data | Representation |
|---|---|---|
| ✓ | baseline | |
| ✓ | single modal | |
| ✓ | ✓ | proposed |
Table 3.
Summary statistics for data and hyperparameters.
| #Features | Hidden Dimension | Time length (Average) | Time length (sd) | |
|---|---|---|---|---|
| Cognitive performance | 2 | 3 | 3.7 | 1.32 |
| Demographic Information | 4 | 5 | 1 | 0 |
| CSF | 5 | 6 | 1.4 | 0.5 |
| MRI | 3 | 4 | 1 | 0 |
For training our models, subjects in CN and AD groups are used as well as MCI-C and MCI-NC. This approach is motivated by8,10,11,20–22. They use CN and AD subjects for training a classifier such as SVM23 or locally linear embedding (LLE)24, and then the classifier is used for classification of MCI-C and MCI-NC. In our experiment, CN and AD are used as auxiliary dataset to pre-train the classifier, and then MCI-C and MCI-NC are also used for training.
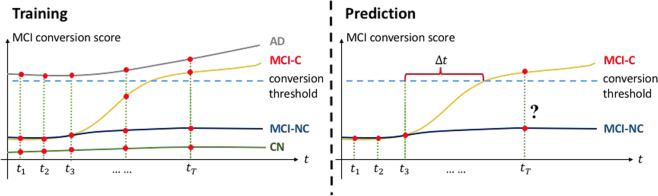
We tested the classifier on MCI patients to predict the conversion after Δt from baseline (6, 12, 18, and 24 months) as shown in Fig. 1. Due to the nature of our data the sample size available for training varies over Δt (Fig. 2). For example, if AD occurs early from the baseline visit, then we have relatively fewer training samples because we have a smaller data window to predict on. At each prediction time (Δt), we ran 5-fold cross-validation 10 times in which every fold has the same ratio of MCI-C and MCI-NC subjects. MCI samples were partitioned into 5 subsets, and one subset was selected for testing, while the remaining subsets were used for training.
Figure 1.
An example using longitudinal data for MCI conversion prediction. Contrary to the experiment with baseline visit data, longitudinal data of individuals in all stages (CN, MCI, and AD) was used for training a classifier. Then, portion of longitudinal data was taken to the classifier to predict AD progression after Δt.
Figure 2.
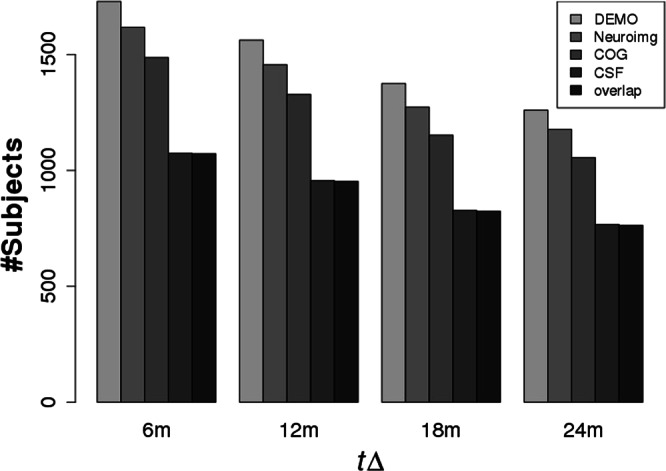
The number of subjects available in demographic data, neuroimaging data, cognitive performance, and CSF biomarkers over ∆t.
Comparison of prediction of MCI to AD conversion using cross-sectional data at baseline and longitudinal data
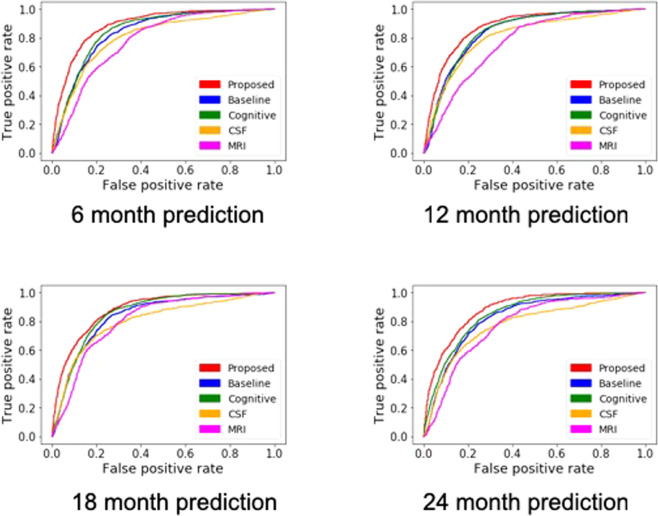
To evaluate the advantage of using longitudinal data, we first compared the performances of two schemes: “baseline” and “proposed” (Figs 3 and 4). Intuitively, data from multiple time points has more information than data at a single time point. Thus, the GRU analyzes the temporal changes in cognitive performance and CSF to extract features (which are not contained in baseline visit data) for the correct MCI conversion prediction. As shown in Table 4(a,b), the prediction model based on longitudinal data shows better performance than the model using only cross-sectional data at baseline. In particular, sensitivity is an important measure for the prediction task in which identifying true positive rate is crucial25. In prediction of MCI conversion, a classifier with higher true positive rate is more applicable for timely treatment.
Figure 3.
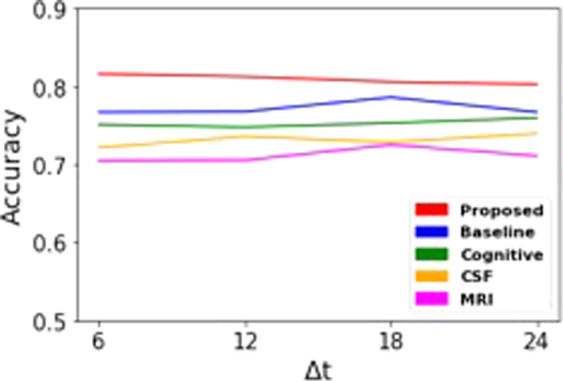
Predictive performances with “proposed”, “baseline”, and “single modal”. Abbreviations: COG: cognition performance biomarkers; CSF: cerebrospinal fluid biomarkers; NeuroImg: MRI biomarkers.
Figure 4.
ROC curves from the “proposed”, “baseline”, and each “single modal” method. (a) 6 month prediction. (b) 12 month prediction. (c) 18 month prediction. (d) 24 month prediction.
Table 4.
Prediction performance based on Δt over different schemes.
| Δt | 6 m | 12 m | 18 m | 24 m |
|---|---|---|---|---|
| (a) Multimodal longitudinal data (proposed) | ||||
| ACC (mean ±sd) | 0.81 ± 0.03 | 0.81 ± 0.03 | 0.79 ± 0.03 | 0.80 ± 0.03 |
| SEN (mean ± sd) | 0.84 ± 0.07 | 0.84 ± 0.05 | 0.82 ± 0.07 | 0.81 ± 0.10 |
| SPE (mean ± sd) | 0.80 ± 0.04 | 0.80 ± 0.04 | 0.79 ± 0.04 | 0.80 ± 0.04 |
| (b) Multimodal baseline visit data (baseline) | ||||
| ACC (mean ±sd) | 0.76 ± 0.03 | 0.76 ± 0.03 | 0.78 ± 0.03 | 0.76 ± 0.03 |
| SEN (mean ± sd) | 0.81 ± 0.07 | 0.82 ± 0.07 | 0.81 ± 0.07 | 0.80 ± 0.08 |
| SPE (mean ± sd) | 0.80 ± 0.04 | 0.75 ± 0.03 | 0.77 ± 0.05 | 0.76 ± 0.03 |
| (c) Cognitive performance data (single modal) | ||||
| ACC (mean ±sd) | 0.74 ± 0.06 | 0.75 ± 0.05 | 0.74 ± 0.04 | 0.74 ± 0.04 |
| SEN (mean ± sd) | 0.81 ± 0.07 | 0.78 ± 0.14 | 0.76 ± 0.20 | 0.76 ± 0.20 |
| SPE (mean ± sd) | 0.75 ± 0.08 | 0.71 ± 0.05 | 0.71 ± 0.06 | 0.71 ± 0.06 |
Comparison of prediction of MCI to AD conversion using single modal and multimodal data
For evaluating the effectiveness of multimodal data integration, we compared the performances of “proposed” and “single modal” experiments. Figure 3 shows the accuracies of “proposed” and models with single modality of data. We removed the accuracy from the model with demographic data because the prediction performance was too low. The model using cognitive performance was observed to be the most accurate among models that use each single modality of data. Even though the sample size for neuroimaging data was larger than those of cognitive performance and CSF biomarkers (Fig. 3), the model with neuroimaging data showed less accuracy. This is because cognitive performance is a longitudinal data which takes advantage of giving relatively closer data record to MCI conversion point. However, model with cognitive performance shows extremely high variance of sensitivity for predicting 18 and 24 months. It is observed that model only with cognitive performance not a stable predictor for long period of prediction while integrating other biomarkers can alleviate the high variance in proposed.
Discussion
We proposed an integrative approach for the prediction of MCI to AD conversion using a deep learning approach, more specifically, a multi-modal recurrent neural network. Our method takes advantages of longitudinal and multi-modal nature of available data to discover nonlinear patterns associated with MCI progression. To evaluate the advantages of our proposed method, we compared performance outputs from three schemes: “baseline”, “single modal”, and “proposed”. As observed in Fig. 4, “baseline” and “single modal” with cognitive test biomarkers show similar performances over prediction periods. Using longitudinal data or combining multimodal data are effective ways for increasing predictive power thus, it seems natural for combining longitudinal multimodal data (“proposed”) to show the best performance. In Table 5, as predicted further periods, the reliability of performance improvement is lower due to the lack of positive samples. However, specificities of proposed model showed enhanced performance over competing methods consistently. In addition, the prediction results of our model were compared to those of previous studies with machine learning approaches (Table 6). Our method showed comparable prediction ability even though we had a highly unbalanced ratio of positive and negative samples. Specifically, the sensitivity of our model shows higher performance while specificity is lower. Moreover, The balanced accuracy26, which is a measure of accuracy considering sensitivity and specificity shows 0.82 for our model and 0.81 for27.
Table 5.
Performance comparison between different models. P-values are calculated using a paired t-test between the proposed and each competing method.
| Model | accuracy (p-value) | sensitivity (p-value) | specificity (p-value) |
|---|---|---|---|
| (a) 6 month prediction. | |||
| Proposed | 0.81 | 0.84 | 0.80 |
| Baseline | 0.76 (1.16e-08) | 0.81 (0.07) | 0.75 (1.87e-10) |
| Single modal (cognitive performance) | 0.74 (3.17e-09) | 0.81 (8.12e-05) | 0.70 (2.68e-14) |
| Single modal (CSF) | 0.72 (4.65e-17) | 0.78 (8.12e-05) | 0.70 (2.68e-14) |
| Single modal (MRI) | 0.70 (4.68e-24) | 0.75 (6.71e-10) | 0.68 (2.05e-21) |
| (b) 12 month prediction | |||
| Proposed | 0.81 | 0.84 | 0.80 |
| Baseline | 0.76 (1.77e-07) | 0.82 (0.057) | 0.75 (3.65e-07) |
| Single modal (cognitive performance) | 0.75 (2.70e-08) | 0.78 (0.007) | 0.71 (1.73e-09) |
| Single modal (CSF) | 0.73 (3.47e-20) | 0.78 (1.40e-05) | 0.72 (3.90e-16) |
| Single modal (MRI) | 0.70 (7.29e-22) | 0.73 (1.03e-12) | 0.69 (2.07e-16) |
| (c) 18 month prediction | |||
| Proposed | 0.79 | 0.82 | 0.79 |
| Baseline | 0.78 (0.08) | 0.81 (0.64) | 0.77 (0.15) |
| Single modal (cognitive performance) | 0.74 (2.43e-06) | 0.76 (0.03) | 0.71 (7.72e-08) |
| Single modal (CSF) | 0.72 (9.84e-12) | 0.78 (0.07) | 0.71 (6.55e-11) |
| Single modal (MRI) | 0.72 (5.26e-14) | 0.76 (0.16e-04) | 0.71 (2.20e-10) |
| (d) 24 month prediction | |||
| Proposed | 0.80 | 0.81 | 0.80 |
| Baseline | 0.76 (2.12e-05) | 0.80 (0.57) | 0.76 (8.95e-05) |
| Single modal (cognitive performance) | 0.74 (4.69e-07) | 0.76 (0.15) | 0.72 (1.20e-09) |
| Single modal (CSF) | 0.73 (1.06e-09) | 0.73 (0.001) | 0.74 (3.16e-07) |
| Single modal (MRI) | 0.71 (1.18e-12) | 0.76 (0.03) | 0.70 (2.53e-10) |
Table 6.
A list of previous models that train a classifier mainly using MCI samples.
| Method | Subjects (MCI-C/MCI-NC) | Data source | ACC | SEN | SPE |
|---|---|---|---|---|---|
| SVM7 | 43/48 | MRI, PET, CSF | 0.73 | 0.68 | 0.73 |
| SVM8 | 43/56 | MRI, FDG-PET, CSF | 0.79 | 0.84 | 0.72 |
| SVM9 | 35/50 | MRI, PET, cognitive score | 0.78 | 0.79 | 0.78 |
| Gaussian process39 | 47/96 | MRI, PET, CSF, APOE genotype | 0.68 | 0.90 | 0.52 |
| Hierarchical ensemble25 | 70/61 | MRI | 0.79 | 0.86 | 0.78 |
| Deep neural network27 | 235/409 | MRI, PET | 0.82 | 0.79 | 0.83 |
| Proposed | 134/561 | Cognitive score, MRI, CSF biomarker, demographic data | 0.81 | 0.84 | 0.80 |
The biggest advantage of our approach is that irregular longitudinal data can be used. One of the major problems when dealing with longitudinal data is that a preprocessing step is required for handling variable-length of sequential data and missing values. In previous studies, the fixed length of time points was collected by taking data that fell within a certain time window. Additionally, an additional feature extraction phase is required to produce a fixed-size feature representation. In the first training step, separate GRU components make an encoding process, where longitudinal data are transformed into a vector containing AD-sensitive features. Thanks to the structure of GRU, our approach is capable of accepting any irregular length of data as an input without preprocessing.
In addition, our method can make full use of available subjects from each modality for training our classifier. This is a huge advantage in the face of data scarcity. As seen in Fig. 2, the number of subjects with CSF data is smallest in the overlapping sample. Traditional approaches can use only the overlapping samples while non-overlapping samples were abandoned. In our case, non-overlapping samples contribute to training the individual GRU component it belongs to for the better representation learning. Furthermore, additional modality data are easily integrated into the model. Contrary to the kernel-based integration, concatenation-based integration method can incorporate other domains of data such as multi-modal neuroimaging and genomic data without any prior knowledge. Thus, next we will integrate multi-modal neuroimaging and genomics data for learning features that might be useful in predicting early MCI to AD conversion.
Although there are some strengths as described above, our approach has some limitations. In the first training step, the input of each modality was transformed into a feature vector that is optimized for MCI conversion prediction only by single modality. Thus, features that are irrelevant to AD progression with respect to the single modality will be filtered out. However, if there are features that cannot be extracted by single modality but only can be explained by a combination of multi-modality of data then those are also likely to be filtered out. This is because parameters in GRUs are not updated against the final prediction result. In other words, parameter optimization for the second training step does not affect the parameters in each GRU for feature extraction, thus each GRU cannot learn from the final prediction based on the combined features. To solve this problem, we will link GRUs to logistic regressions at the second step so that GRU learns feature representation from multi-modality as well as single modality. In addition, we plan to modify the structure of our model making it possible for individual GRU components to extract integrative features. We are currently investigating this possibility as a sequel to this work.
Methods
Recurrent Neural Network
Recurrent Neural Network (RNN) is a class of deep learning architecture used when sequential data can be considered. In natural language processing (NLP), speech recognition, and anomaly detection in time series, RNN is popularly used for analyzing the sequence of words and time series data28. The advantage of applying RNN is that variable-length sequence can be processed to exploit temporal patterns hidden in the given sequence. In the sentiment analysis task, for example, the goal is to classify the sentiment (good or bad) of a given sentence. The classifier needs to take a sentence (a sequence of words) as an input, understand the context in it, and return a correct sentiment as an output29. For the prediction task that detects initial diagnosis of heart failure in30, RNN takes time series of electronic health records (EHRs) using 12 to 18 month observation window. In these cases where variable-length input should be dealt with RNN is an appropriate candidate to use.
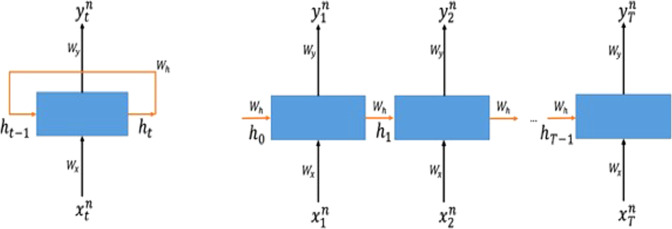
An RNN processes one element of an input sequence at a time and updates its memory state that implicitly contains information about the history of all the past elements of the sequence31. The hidden state is represented as a Euclidean vector (i.e., a sequence of real numbers) and is updated recursively from the input at the given step and the previous value of the hidden state (Fig. 5).
Figure 5.
Illustration of recurrent neural network. RNN is composed of input, memory state, and output, each of which has a weight parameter to be learned for a given task. The memory state (blue box) takes the input and computes the output based on the memory state from the previous step and the current input (left). Since the RNN has a feedback loop, variable-length input and output sequence can be represented as an “unfolded” sequence (right).
Suppose we have N number of subjects, each of which has a sequence where is a data record of n-th sample and the t-th element in a sequence and T is the length of the sequence. The corresponding sequence of output is recursively computed as:
| 1 |
| 2 |
Wh, Wx, and Wy are the weight matrices to extract task-specific features from the previous memory state ht−1, the t-th input xt and the current memory state ht, respectively. As can be seen from the equations, the memory states ht and the input xt are all represented as Euclidean vectors as well. Therefore, the dynamics of the entire RNN are captured by a sequence of matrix-vector multiplications, followed by elementwise non-linearity applications. The tanh function is a non-linear activation function taking the form of . The role of the non-linear activation function is to endow the RNN with higher representational power. is the predicted output resulting from the computation of the network. This final result is computed by the function σ, which is known as the softmax function. The role of the softmax function is to turn an arbitrary vector into a probability vector via the following operation:
where ui is the i-th element of the vector u. In equation (2), we abuse notation to express elementwise application of the above expression.
| 3 |
| 4 |
In our model, the last output sequence provided by the RNN is treated as the probability vector for classification, and the cross-entropy loss function (equation (3)) is used to quantify how “far away” our n-th prediction is from the n-th ground truth label yn. That is, we choose the optimal parameters that minimize the cross-entropy loss of the given data (equation (4)). The algorithm we use to optimize the parameters is Backpropagation Through Time (BPTT)32, which updates the weights in the RNN to minimize the given loss function.
However, when the task requires long sequences of input to be processed, training an RNN is difficult33. This is called the long-term dependency problem. Variants of RNN such as Long Short-Term Memory (LSTM) and Gated Recurrent Unit (GRU) have been developed and practically used to solve this problem34,35. In the proposed model, we use GRU for each modality of data to process multiple time points of the input. The detailed structure of GRU is described in the supplementary.
Multi-modal GRU for MCI conversion prediction
Our problem can be considered as a sequential data classification. The classification objective is to predict whether an individual with MCI at baseline is converted to AD or not using sequence data, which consist of four modalities including cognitive performance, CSF, and MRI biomarkers as well as demographic information. Even though demographic data and MRI biomarkers are not longitudinal data we will consider them as length-one sequential data.
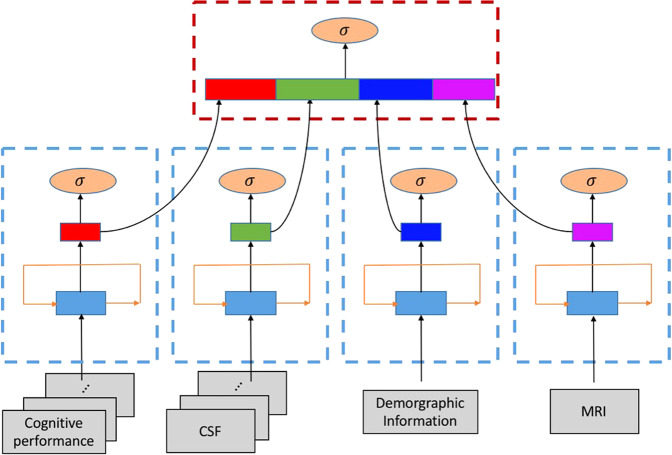
To apply a GRU-based classification algorithm to our problem, we need to design a model that can incorporate the four modalities of data. The main idea of our model is to separately build GRU feature extractors for each modality and integrate the extracted four feature vectors at the end. Our model is comprised of two training steps: (1) learning a single GRU for each modality of data, and (2) learning the integrative feature representation to make the final prediction. At the first training step, a single GRU is trained separately for each modality in which the classification objective is to predict conversion to AD from MCI. Using GRUs is essential to take longitudinal data and transform them into a fixed-size vector. This is quite similar to the approach proposed in36 that maps the input sequence into fixed-length representation. In the second step, MCI conversion is predicted based on the four vectors produced from each GRU components. For merging four vectors, we select concatenation-based data integration, which is conceptually the simplest method to integrate multiple sources of data into a single vector37. For the final prediction, l1-regularized logistic regression38 is used for the classification between MCI-C and MCI-NC. The overview of our proposed method is illustrated in Fig. 6.
Figure 6.
Overview of the proposed method. Our proposed method contains multiple GRU components that accept each modality of the dataset. At the first training step (blue dashed rectangle), each GRU component takes both time series or non-time series data to produce fixed-size feature vectors. And then the vectors are concatenated to form an input for the final prediction in the second training step (red dashed rectangle).
Conclusion
Here, we proposed a multi-modal deep learning approach to study the prediction of MCI to AD conversion using longitudinal cognitive performance and CSF biomarkers as well as cross-sectional neuroimaging and demographic data at baseline. We applied multiple GRUs to use longitudinal multi-domain data and all subjects with each modality data. Our results showed that we achieved the better prediction accuracy of MCI to AD conversion by incorporating longitudinal multi-domain data. A multi-modal deep learning approach has potential to identify persons at risk of developing AD who might benefit most from a clinical trial or as a stratification approach within clinical trials.
Acknowledgements
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found http://adni.loni.usc.edu/wp-content/themes/freshnews-dev-v2/documents/policy/ADNI_Acknowledgement_List%205-29-18.pdf. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www. fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Samples from the National Cell Repository for AD (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (AIG), were used in this study. Additional support for data analysis was provided by NLM R01 LM012535, NIA R03 AG054936, and the Pennsylvania Department of Health (#SAP 4100070267). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. This research was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education [NRF- 2016R1D1A1B03933875].
Author Contributions
The study was conceived by Kim, Nho, Kang, and Sohn. Experiments were designed and performed by all authors. The manuscript was written by Lee and Nho. All authors revised the manuscript and approved the final version prior to submission.
Data Availability
Demogra phic information, neuroimaging data, APOE genotype, CSF measurements, neuropsychological test scores, and diagnostic information are publicly available from the ADNI data repository (http://adni.loni.usc.edu).
Competing Interests
The authors declare no competing interests.
Footnotes
*A comprehensive list of consortium members appears at the end of the paper
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Garam Lee and Kwangsik Nho contributed equally.
Contributor Information
Kyung-Ah Sohn, Email: kasohn@ajou.ac.kr.
Dokyoon Kim, Email: dkim@geisinger.edu.
for Alzheimer’s Disease Neuroimaging Initiative:
Michael W. Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, Jr., William Jagust, John Q. Trojanowki, Arthur W. Toga, Laurel Beckett, Robert C. Green, Andrew J. Saykin, John Morris, Leslie M. Shaw, Zaven Khachaturian, Greg Sorensen, Maria Carrillo, Lew Kuller, Marc Raichle, Steven Paul, Peter Davies, Howard Fillit, Franz Hefti, Davie Holtzman, M. Marcel Mesulam, William Potter, Peter Snyder, Tom Montine, Ronald G. Thomas, Michael Donohue, Sarah Walter, Tamie Sather, Gus Jiminez, Archana B. Balasubramanian, Jennifer Mason, Iris Sim, Danielle Harvey, Matthew Bernstein, Nick Fox, Paul Thompson, Norbert Schuff, Charles DeCArli, Bret Borowski, Jeff Gunter, Matt Senjem, Prashanthi Vemuri, David Jones, Kejal Kantarci, Chad Ward, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Chet Mathis, Susan Landau, Nigel J. Cairns, Erin Householder, Lisa Taylor-Reinwald, Virginia Lee, Magdalena Korecka, Michal Figurski, Karen Crawford, Scott Neu, Tatiana M. Foroud, Steven Potkin, Li Shen, Kelley Faber, Sungeun Kim, Lean Tha, Richard Frank, John Hsiao, Jeffrey Kaye, Joseph Quinn, Lisa Silbert, Betty Lind, Raina Carter, Sara Dolen, Beau Ances, Maria Carroll, Mary L. Creech, Erin Franklin, Mark A. Mintun, Stacy Schneider, Angela Oliver, Lon S. Schneider, Sonia Pawluczyk, Mauricio Beccera, Liberty Teodoro, Bryan M. Spann, James Brewer, Helen Vanderswag, Adam Fleisher, Daniel Marson, Randall Griffith, David Clark, David Geldmacher, John Brockington, Erik Roberson, Marissa Natelson Love, Judith L. Heidebrink, Joanne L. Lord, Sara S. Mason, Colleen S. Albers, David Knopman, Kris Johnson, Hillel Grossman, Effie Mitsis, Raj C. Shah, Leyla deToledo-Morrell, Rachelle S. Doody, Javier Villanueva-Meyer, Munir Chowdhury, Susan Rountree, Mimi Dang, Ranjan Duara, Daniel Varon, Maria T. Greig, Peggy Roberts, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Marilyn Albert, Chiadi Onyike, Daniel D’Agostino, II, Stephanie Kielb, James E. Galvin, Brittany Cerbone, Christina A. Michel, Dana M. Pogorelec, Henry Rusinek, Mony J. de Leon, Lidia Glodzik, Susan De Santi, Kyle Womack, Dana Mathews, Mary Quiceno, P. Murali Doraiswamy, Jeffrey R. Petrella, Salvador Borges-Neto, Terence Z. Wong, Edward Coleman, Allan I. Levey, James J. Lah, Janet S. Cella, Jeffrey M. Burns, Russell H. Swerdlow, William M. Brooks, Steven E. Arnold, Jason H. Karlawish, David Wolk, Christopher M. Clark, Liana Apostolova, Kathleen Tingus, Ellen Woo, Daniel H. S. Silverman, Po H. Lu, George Bartzokis, Charles D. Smith, Greg Jicha, Peter Hardy, Partha Sinha, Elizabeth Oates, Gary Conrad, Neill R. Graff-Radford, Francine Parfitt, Tracy Kendall, Heather Johnson, Oscar L. Lopez, MaryAnn Oakley, Donna M. Simpson, Martin R. Farlow, Ann Marie Hake, Brandy R. Matthews, Jared R. Brosch, Scott Herring, Cynthia Hunt, Anton P. Porsteinsson, Bonnie S. Goldstein, Kim Martin, Kelly M. Makino, M. Saleem Ismail, Connie Brand, Ruth A. Mulnard, Gaby Thai, Catherine Mc-Adams-Ortiz, Christopher H. van Dyck, Richard E. Carson, Martha G. MacAvoy, Pradeep Varma, Howard Chertkow, Howard Bergman, Chris Hosein, Sandra Black, Bojana Stefanovic, Curtis Caldwell, Ging-Yuek Robin Hsiung, Howard Feldman, Benita Mudge, Michele Assaly, Elizabeth Finger, Stephen Pasternack, Irina Rachisky, Dick Trost, Andrew Kertesz, Charles Bernick, Donna Munic, Kristine Lipowski, Masandra Weintraub, Borna Bonakdarpour, Diana Kerwin, Chuang-Kuo Wu, Nancy Johnson, Carl Sadowsky, Teresa Villena, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Reisa A. Sperling, Keith A. Johnson, Gad Marshall, Jerome Yesavage, Joy L. Taylor, Barton Lane, Allyson Rosen, Jared Tinklenberg, Marwan N. Sabbagh, Christine M. Belden, Sandra A. Jacobson, Sherye A. Sirrel, Neil Kowall, Ronald Killiany, Andrew E. Budson, Alexander Norbash, Patricia Lynn Johnson, Thomas O. Obisesan, Saba Wolday, Joanne Allard, Alan Lerner, Paula Ogrocki, Curtis Tatsuoka, Parianne Fatica, Evan Fletcher, Pauline Maillard, John Olichney, Owen Carmichael, Smita Kittur, Michael Borrie, T.-Y. Lee, Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Adrian Preda, Dana Nguyen, Pierre Tariot, Anna Burke, Nadira Trncic, Adam Fleisher, Stephanie Reeder, Vernice Bates, Horacio Capote, Michelle Rainka, Douglas W. Scharre, Maria Kataki, Anahita Adeli, Earl A. Zimmerman, Dzintra Celmins, Alice D. Brown, Godfrey D. Pearlson, Karen Blank, Karen Anderson, Laura A. Flashman, Marc Seltzer, Mary L. Hynes, Robert B. Santulli, Kaycee M. Sink, Leslie Gordineer, Jeff D. Williamson, Pradeep Garg, Franklin Watkins, Brian R. Ott, Henry Querfurth, Geoffrey Tremont, Stephen Salloway, Paul Malloy, Stephen Correia, Howard J. Rosen, Bruce L. Miller, David Perry, Jacobo Mintzer, Kenneth Spicer, David Bachman, Elizabether Finger, Stephen Pasternak, Irina Rachinsky, John Rogers, Dick Drost, Nunzio Pomara, Raymundo Hernando, Antero Sarrael, Susan K. Schultz, Laura L. Boles Ponto, Hyungsub Shim, Karen Ekstam Smith, Norman Relkin, Gloria Chaing, Michael Lin, Lisa Ravdin, Amanda Smith, Balebail Ashok Raj, and Kristin Fargher
References
- 1.Alzheimer’s A. 2015 Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11:332–384. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Albert MS, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jack CR, Jr., et al. Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:257–262. doi: 10.1016/j.jalz.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperling RA, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen RC, et al. Mild cognitive impairment: clinical characterization and outcome. Archives of neurology. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 6.Tábuas-Pereira M, et al. Prognosis of Early-Onset vs. Late-Onset Mild Cognitive Impairment: Comparison of Conversion Rates and Its Predictors. Geriatrics. 2016;1:11. doi: 10.3390/geriatrics1020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang D, Shen D, Alzheimer’s Disease Neuroimaging I. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage. 2012;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng B, Liu M, Zhang D, Munsell BC, Shen D. Domain Transfer Learning for MCI Conversion Prediction. IEEE Trans Biomed Eng. 2015;62:1805–1817. doi: 10.1109/TBME.2015.2404809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D, Shen D, Initiative ASDN. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PloS one. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nho K, et al. Automatic Prediction of Conversion from Mild Cognitive Impairment to Probable Alzheimer’s Disease using Structural Magnetic Resonance Imaging. AMIA Annu Symp Proc. 2010;2010:542–546. [PMC free article] [PubMed] [Google Scholar]
- 11.Wee CY, Yap PT, Shen D, Alzheimer’s Disease Neuroimaging I. Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Hum Brain Mapp. 2013;34:3411–3425. doi: 10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolz R, et al. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS One. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho Y, Seong JK, Jeong Y, Shin SY, Alzheimer’s Disease Neuroimaging I. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, et al. A Graph-Based Integration of Multimodal Brain Imaging Data for the Detection of Early Mild Cognitive Impairment (E-MCI) Multimodal Brain Image Anal (2013) 2013;8159:159–169. doi: 10.1007/978-3-319-02126-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewers M, et al. Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging. 2012;33:1203–1214. doi: 10.1016/j.neurobiolaging.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heister D, et al. Predicting MCI outcome with clinically available MRI and CSF biomarkers. Neurology. 2011;77:1619–1628. doi: 10.1212/WNL.0b013e3182343314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saykin AJ, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2015;11:792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saykin AJ, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nho K, et al. Voxel and surface-based topography of memory and executive deficits in mild cognitive impairment and Alzheimer’s disease. Brain imaging and behavior. 2012;6:551–567. doi: 10.1007/s11682-012-9203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falahati F, Westman E, Simmons A. Multivariate data analysis and machine learning in Alzheimer’s disease with a focus on structural magnetic resonance imaging. J Alzheimers Dis. 2014;41:685–708. doi: 10.3233/JAD-131928. [DOI] [PubMed] [Google Scholar]
- 21.Westman E, Aguilar C, Muehlboeck JS, Simmons A. Regional magnetic resonance imaging measures for multivariate analysis in Alzheimer’s disease and mild cognitive impairment. Brain Topogr. 2013;26:9–23. doi: 10.1007/s10548-012-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Tosun D, Weiner MW, Schuff N, Alzheimer’s Disease Neuroimaging I. Locally linear embedding (LLE) for MRI based Alzheimer’s disease classification. Neuroimage. 2013;83:148–157. doi: 10.1016/j.neuroimage.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20:273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 24.Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. science. 2000;290:2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 25.Huang M, et al. Longitudinal measurement and hierarchical classification framework for the prediction of Alzheimer’s disease. Scientific reports. 2017;7:39880. doi: 10.1038/srep39880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodersen, K. H., Ong, C. S., Stephan, K. E. & Buhmann, J. M. In Pattern recognition (ICPR), 2010 20th international conference on. 3121–3124 (IEEE).
- 27.Lu D, Popuri K, Ding GW, Balachandar R, Beg MF. Multimodal and Multiscale Deep Neural Networks for the Early Diagnosis of Alzheimer’s Disease using structural MR and FDG-PET images. Scientific reports. 2018;8:5697. doi: 10.1038/s41598-018-22871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng, L., Hinton, G. & Kingsbury, B. In Acoustics, Speech and Signal Processing (ICASSP), 2013 IEEE International Conference on. 8599–8603 (IEEE).
- 29.Tang, D., Qin, B. & Liu, T. In Proceedings of the 2015 conference on empirical methods in natural language processing. 1422–1432.
- 30.Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. Journal of the American Medical Informatics Association. 2017;24:361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 32.Guo, J. Backpropagation through time. Unpubl. ms., Harbin Institute of Technology (2013).
- 33.Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994;5:157–166. doi: 10.1109/72.279181. [DOI] [PubMed] [Google Scholar]
- 34.Cho, K. et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv preprint arXiv:1406.1078 (2014).
- 35.Hochreiter S, Schmidhuber J. Long short-term memory. Neural computation. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 36.Srivastava, N., Mansimov, E., Salakhudinov, R. Unsupervised learning of video representations using LSTMs. In: 2015 International Conference on Machine Learning. 843–852 (2015).
- 37.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nature reviews. Genetics. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 38.Tibshirani, R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological), 267–288 (1996).
- 39.Young J, et al. Accurate multimodal probabilistic prediction of conversion to Alzheimer’s disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Demogra phic information, neuroimaging data, APOE genotype, CSF measurements, neuropsychological test scores, and diagnostic information are publicly available from the ADNI data repository (http://adni.loni.usc.edu).